Computational Analysis of the Effect of Dietary Interventions on the Gut Microbiome Composition in Parkinson’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Studies for Simulation
Selection of Metabolic Models for Microbiome Reconstruction
2.2. Competition and Complementarity Assembly
2.3. Generation of a Computational Model of the Bacterial Composition of the Colon
2.4. Availability of Bacterial Metabolites to the Host
2.5. Topological Analysis of Networks
2.6. Statistical Analysis
3. Results
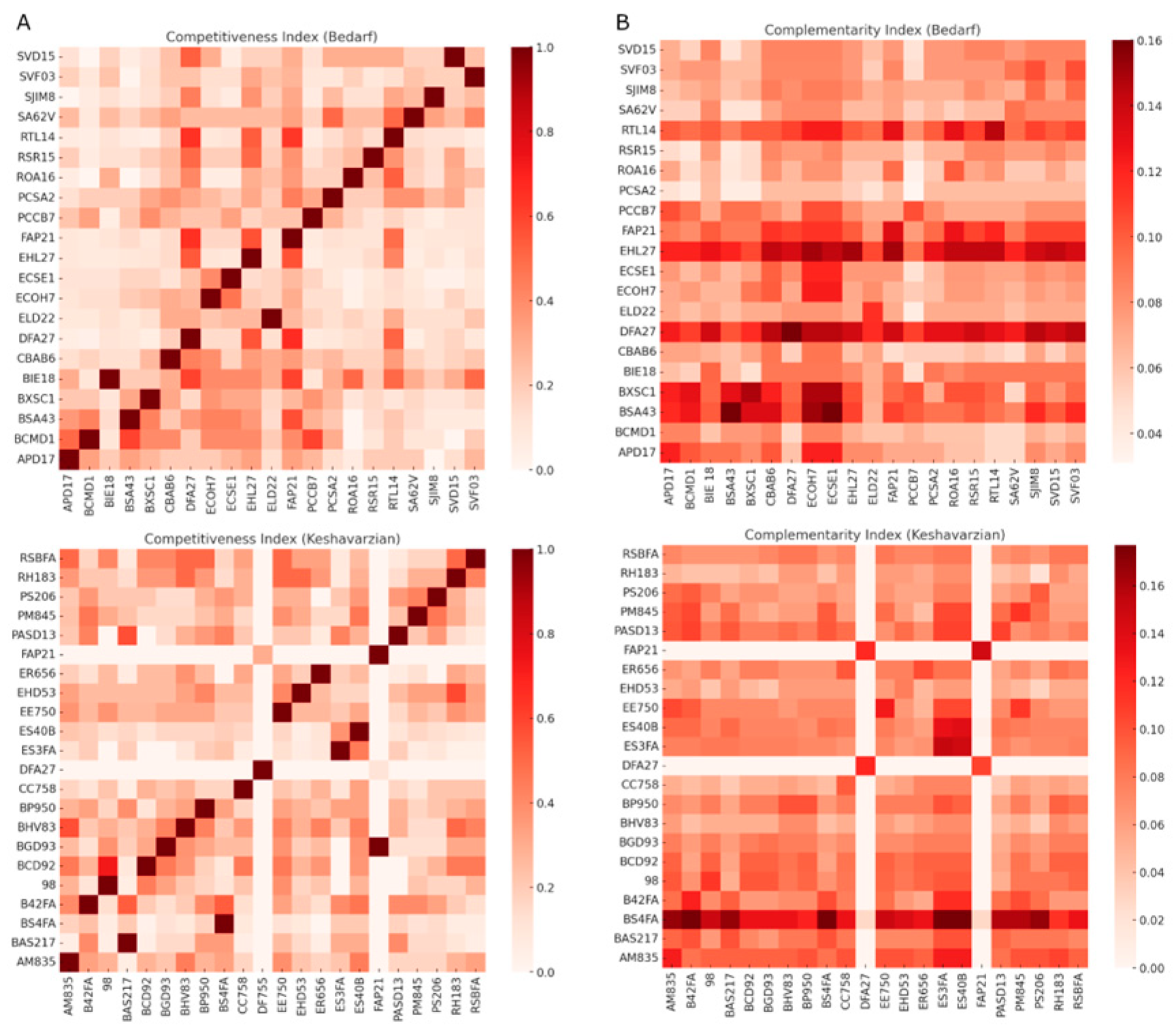
3.1. Competition Indices Outweighed Complementarity in Bacterial Communities
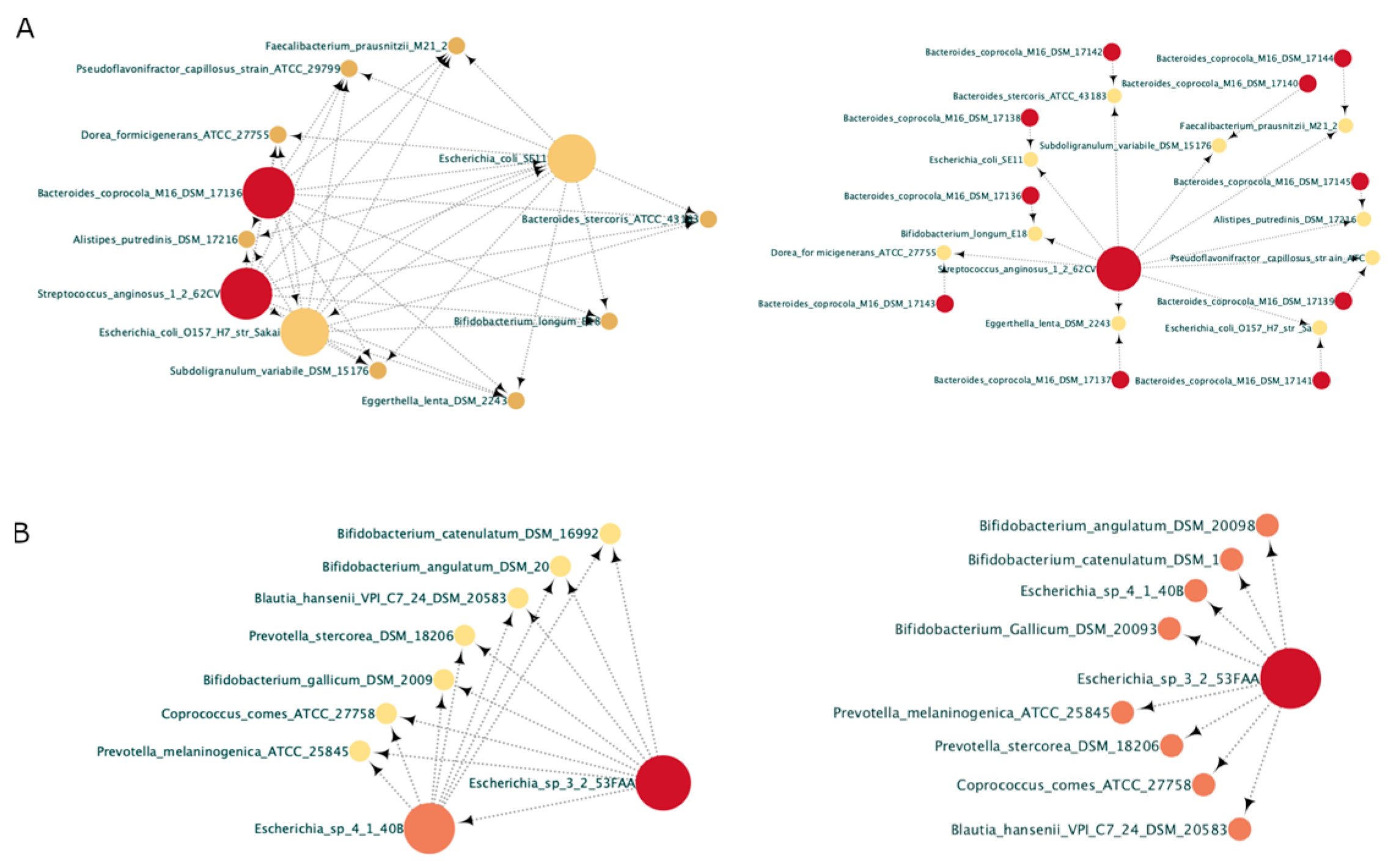
3.2. In the Parkinsonian Context, Pro-Inflammatory Bacteria Are Opportunistic
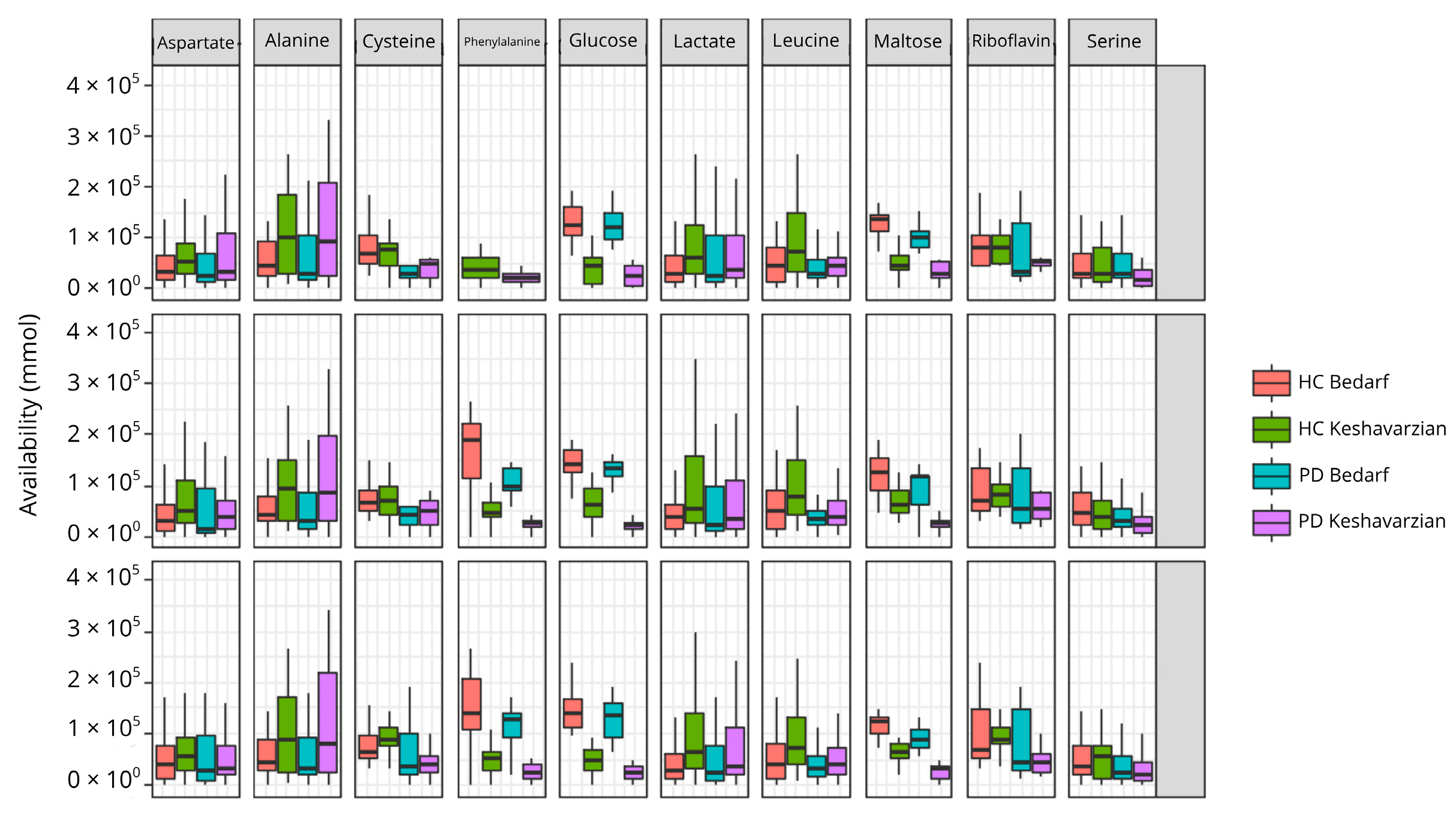
3.3. Metabolic Exchange Networks Leave Certain Metabolites Less Available in PD
3.4. Changes in PD in the Individual Amino Acid Production Network
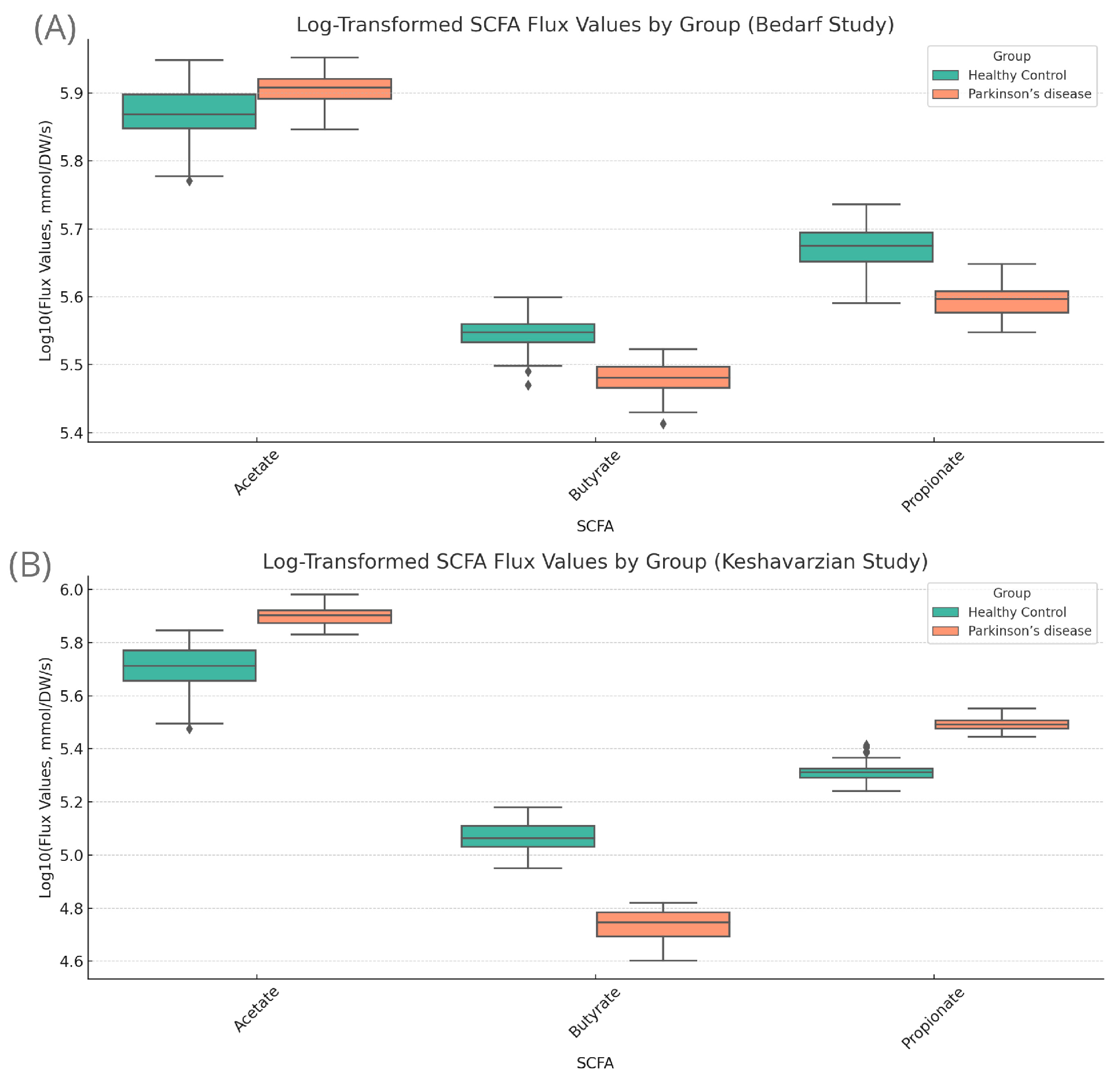
3.5. Butyrate Dynamics Mediated by Diet
3.6. In Search of a Healthy Diet for PD, Specifically for AGCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sveinbjornsdottir, S. The clinical symptoms of Parkinson’s disease. J. Neurochem. 2016, 139, 318–324. [Google Scholar] [CrossRef]
- Ascherio, A.; Schwarzschild, M.A. The epidemiology of Parkinson’s disease: Risk factors and prevention. Lancet Neurol. 2016, 15, 1257–1272. [Google Scholar] [CrossRef]
- Forero-Rodríguez, J.; Zimmermann, J.; Taubenheim, J.; Arias-Rodríguez, N.; Caicedo-Narvaez, J.D.; Best, L.; Mendieta, C.V.; López-Castiblanco, J.; Gómez-Muñoz, L.A.; Gonzalez-Santos, J.; et al. Changes in Bacterial Gut Composition in Parkinson’s Disease and Their Metabolic Contribution to Disease Development: A Gut Community Reconstruction Approach. Microorganisms 2024, 12, 325. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e12. [Google Scholar] [CrossRef] [PubMed]
- Scheperjans, F.; Aho, V.; Pereira, P.A.B.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Salim, S.; Ahmad, F.; Banu, A.; Mohammad, F. Gut microbiome and Parkinson’s disease: Perspective on pathogenesis and treatment. J. Adv. Res. 2023, 50, 83–105. [Google Scholar] [CrossRef]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut Microbiota and Metabolome Alterations Associated with Parkinson’s Disease. mSystems 2020, 5, 1–15. [Google Scholar] [CrossRef]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef]
- Unger, M.M.; Spiegel, J.; Dillmann, K.-U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Baert, F.; Matthys, C.; Maselyne, J.; Van Poucke, C.; Van Coillie, E.; Bergmans, B.; Vlaemynck, G. Parkinson’s disease patients’ short chain fatty acids production capacity after in vitro fecal fiber fermentation. Npj Park. Dis. 2021, 7, 72. [Google Scholar] [CrossRef]
- Shin, C.; Lim, Y.; Lim, H.; Ahn, T. Plasma Short-Chain Fatty Acids in Patients With Parkinson’s Disease. Mov. Disord. 2020, 35, 1021–1027. [Google Scholar] [CrossRef]
- Jansma, J.; El Aidy, S. Understanding the host-microbe interactions using metabolic modeling. Microbiome 2021, 9, 16. [Google Scholar] [CrossRef]
- Sen, F.; Ganim, M.A.; Baloglu, M.C.; Aygun, A.; Sayiner, H.S.; Altunoglu, Y.C.; Kandemirli, F.; Demirkan, B.; Kuyuldar, E.; Bulut, E. Synergistic and Antagonistic Effects of Phenylalanine and Various Antibiotics on the Growth of Pathogenic Bacteria. BioNanoScience 2019, 9, 446–452. [Google Scholar] [CrossRef]
- Babaei, P.; Shoaie, S.; Ji, B.; Nielsen, J. Challenges in modeling the human gut microbiome. Nat. Biotechnol. 2018, 36, 682–686. [Google Scholar] [CrossRef]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017, 9, 39. [Google Scholar] [CrossRef]
- Hopfner, F.; Künstner, A.; Müller, S.H.; Künzel, S.; Zeuner, K.E.; Margraf, N.G.; Deuschl, G.; Baines, J.F.; Kuhlenbäumer, G. Gut microbiota in Parkinson disease in a northern German cohort. Brain Res. 2017, 1667, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Zheng, W.; He, Y.; Tang, W.; Wei, X.; He, R.; Huang, W.; Su, Y.; Huang, Y.; Zhou, H.; et al. Gut microbiota in patients with Parkinson’s disease in southern China. Parkinsonism Relat. Disord. 2018, 53, 82–88. [Google Scholar] [CrossRef]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Noronha, A.; Modamio, J.; Jarosz, Y.; Guerard, E.; Sompairac, N.; Preciat, G.; Daníelsdóttir, A.D.; Krecke, M.; Merten, D.; Haraldsdóttir, H.S.; et al. The Virtual Metabolic Human database: Integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019, 47, D614–D624. [Google Scholar] [CrossRef] [PubMed]
- Osorio, D.; Botero, K.; Andrés, P.V.; Mendoza-Mejía, N.; Felipe, R.R.; Barreto, G.; González, J. g2f as a Novel Tool to Find and Fill Gaps in Metabolic Networks. R J. 2021, 13, 28–37. [Google Scholar] [CrossRef]
- Gelius-Dietrich, G.; Desouki, A.A.; Fritzemeier, C.J.; Lercher, M.J. Sybil —Efficient Constraint-Based Modelling in R. BMC Syst. Biol. 2013, 7, 125. [Google Scholar] [CrossRef]
- Levy, R.; Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 2013, 110, 12804–12809. [Google Scholar] [CrossRef]
- Bauer, E.; Zimmermann, J.; Baldini, F.; Thiele, I.; Kaleta, C. BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput. Biol. 2017, 13, e1005544. [Google Scholar] [CrossRef]
- Wahlgren, M.; Axenstrand, M.; Håkansson, Å.; Marefati, A.; Pedersen, B.L. In Vitro Methods to Study Colon Release: State of the Art and An Outlook on New Strategies for Better In-Vitro Biorelevant Release Media. Pharmaceutics 2019, 11, 95. [Google Scholar] [CrossRef]
- Read, N.W.; Al-Janabi, M.N.; Holgate, A.M.; Barber, D.C.; A Edwards, C. Simultaneous measurement of gastric emptying, small bowel residence and colonic filling of a solid meal by the use of the gamma camera. Gut 1986, 27, 300–308. [Google Scholar] [CrossRef]
- Szarka, L.A.; Camilleri, M. Methods for the Assessment of Small-Bowel and Colonic Transit. Semin. Nucl. Med. 2012, 42, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Guyton, A.C.; Hall, J.E. Compendio de Fisiología Médica, 12th ed.; Elsevier España: Barcelona, España, 2012. [Google Scholar]
- Bauer, E.; Thiele, I. From metagenomic data to personalized in silico microbiotas: Predicting dietary supplements for Crohn’s disease. Npj Syst. Biol. Appl. 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Bowness, R.; Chaplain, M.A.; Powathil, G.G.; Gillespie, S.H. Modelling the effects of bacterial cell state and spatial location on tuberculosis treatment: Insights from a hybrid multiscale cellular automaton model. J. Theor. Biol. 2018, 446, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal vol. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Machado, D.; Maistrenko, O.M.; Andrejev, S.; Kim, Y.; Bork, P.; Patil, K.R.; Patil, K.R. Polarization of microbial communities between competitive and cooperative metabolism. Nat. Ecol. Evol. 2021, 5, 195–203. [Google Scholar] [CrossRef]
- Moya, A.; Ferrer, M. Functional Redundancy-Induced Stability of Gut Microbiota Subjected to Disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [CrossRef]
- Liu, M.; Bing, G. Lipopolysaccharide Animal Models for Parkinson’s Disease. Park. Dis. 2011, 2011, 1–7. [Google Scholar] [CrossRef]
- Ohno, M.; Hasegawa, M.; Hayashi, A.; Caballero-Flores, G.; Alteri, C.J.; Lawley, T.D.; Kamada, N.; Núñez, G.; Inohara, N. Lipopolysaccharide O structure of adherent and invasive Escherichia coli regulates intestinal inflammation via complement C3. PLoS Pathog. 2020, 16, e1008928. [Google Scholar] [CrossRef]
- Rusconi, B.; Sanjar, F.; Koenig, S.S.K.; Mammel, M.K.; Tarr, P.I.; Eppinger, M. Whole Genome Sequencing for Genomics-Guided Investigations of Escherichia coli O157:H7 Outbreaks. Front. Microbiol. 2016, 7, 327089. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, C.; Xiong, Y.; Xu, X.; Lan, R.; Wang, H.; Yao, X.; Bai, X.; Liu, X.; Meng, Q.; et al. Global Transcriptional and Phenotypic Analyses of Escherichia coli O157:H7 Strain Xuzhou21 and Its pO157_Sal Cured Mutant. PLoS ONE 2013, 8, e65466. [Google Scholar] [CrossRef]
- Monk, J.M.; Charusanti, P.; Aziz, R.K.; Lerman, J.A.; Premyodhin, N.; Orth, J.D.; Feist, A.M.; Palsson, B.Ø. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl. Acad. Sci. USA 2013, 110, 20338–20343. [Google Scholar] [CrossRef] [PubMed]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, F.; Cardenas, M.A.N.; Cardenas, F.; León, L.A. La Enfermedad de Parkinson: Etiología, Tratamientos y Factores Preventivos. Univ. Psychol. 2017, 15, 1–26. [Google Scholar] [CrossRef]
- Figura, M.; Kuśmierska, K.; Bucior, E.; Szlufik, S.; Koziorowski, D.; Jamrozik, Z.; Janik, P. Serum amino acid profile in patients with Parkinson’s disease. PLoS ONE 2018, 13, e0191670. [Google Scholar] [CrossRef]
- Jameson, G.N.L. Iron, cysteine and Parkinson’s disease. Monatshefte Chem.-Chem. Mon. 2011, 142, 325–329. [Google Scholar] [CrossRef]
- Martínez-Banaclocha, M. Cysteine Network (CYSTEINET) Dysregulation in Parkinson’s Disease: Role of N-acetylcysteine. Curr. Drug Metab. 2016, 17, 368–385. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, C.; Junqueira, V. High doses of riboflavin and the elimination of dietary red meat promote the recovery of some motor functions in Parkinson’s disease patients. Braz. J. Med. Biol. Res. 2003, 36, 1409–1417. [Google Scholar] [CrossRef]
- Marashly, E.T.; Bohlega, S.A. Riboflavin Has Neuroprotective Potential: Focus on Parkinson’s Disease and Migraine. Front. Neurol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Bäumler, A.J. Colonocyte metabolism shapes the gut microbiota. Science 2018, 362, eaat9076. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, J.; Wang, F.; Ding, G.; Chen, W.; Fang, R.; Yao, Y.; Pang, M.; Lu, Z.-Q.; Liu, J. Sodium butyrate exerts neuroprotective effects by restoring the blood-brain barrier in traumatic brain injury mice. Brain Res. 2016, 1642, 70–78. [Google Scholar] [CrossRef]
- Kakoty, V.; C, S.K.; Dubey, S.K.; Yang, C.-H.; Taliyan, R. Neuroprotective Effects of Trehalose and Sodium Butyrate on Preformed Fibrillar Form of α-Synuclein-Induced Rat Model of Parkinson’s Disease. ACS Chem. Neurosci. 2021, 12, 2643–2660. [Google Scholar] [CrossRef]
- Paiva, I.; Pinho, R.; Pavlou, M.A.; Hennion, M.; Wales, P.; Schütz, A.-L.; Rajput, A.; Szegő, É.M.; Kerimoglu, C.; Gerhardt, E.; et al. Sodium butyrate rescues dopaminergic cells from alpha-synuclein-induced transcriptional deregulation and DNA damage. Hum. Mol. Genet. 2017, 26, 2231–2246. [Google Scholar] [CrossRef]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. eBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Jobin, C. GPR109a: The Missing Link between Microbiome and Good Health? Immunity 2014, 40, 8–10. [Google Scholar] [CrossRef][Green Version]
- Prasad, K.N.; Bondy, S.C. Dietary fibers and their fermented short-chain fatty acids in prevention of human diseases. Bioact. Carbohydr. Diet. Fibre 2019, 17, 100170. [Google Scholar] [CrossRef]
- Cuervo, A.; Salazar, N.; Ruas-Madiedo, P.; Gueimonde, M.; González, S. Fiber from a regular diet is directly associated with fecal short-chain fatty acid concentrations in the elderly. Nutr. Res. 2013, 33, 811–816. [Google Scholar] [CrossRef]
- Fu, Y.; Moscoso, D.I.; Porter, J.; Krishnareddy, S.; Abrams, J.A.; Seres, D.; Chong, D.H.; Freedberg, D.E. Relationship Between Dietary Fiber Intake and Short-Chain Fatty Acid–Producing Bacteria During Critical Illness: A Prospective Cohort Study. J. Parenter. Enter. Nutr. 2020, 44, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.; Wang, B.; Jahani, P.S.; Hurrell, B.P.; Banie, H.; Muench, G.R.A.; Maazi, H.; Helou, D.G.; Howard, E.; Galle-Treger, L.; et al. Dietary Fiber-Induced Microbial Short Chain Fatty Acids Suppress ILC2-Dependent Airway Inflammation. Front. Immunol. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Maraki, M.I.; Yannakoulia, M.; Stamelou, M.; Stefanis, L.; Xiromerisiou, G.; Kosmidis, M.H.; Dardiotis, E.; Hadjigeorgiou, G.M.; Sakka, P.; Anastasiou, C.A.; et al. Mediterranean diet adherence is related to reduced probability of prodromal Parkinson’s disease. Mov. Disord. 2019, 34, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Molsberry, S.; Bjornevik, K.; Hughes, K.C.; Healy, B.; Schwarzschild, M.; Ascherio, A. Diet pattern and prodromal features of Parkinson disease. Neurology 2020, 95, e2095–e2108. [Google Scholar] [CrossRef] [PubMed]
- Alcalay, R.N.; Gu, Y.; Mejia-Santana, H.; Cote, L.; Marder, K.S.; Scarmeas, N. The association between Mediterranean diet adherence and Parkinson’s disease. Mov. Disord. 2012, 27, 771–774. [Google Scholar] [CrossRef]
- Kalampokini, S.; Becker, A.; Fassbender, K.; Lyros, E.; Unger, M.M. Nonpharmacological Modulation of Chronic Inflammation in Parkinson’s Disease: Role of Diet Interventions. Park. Dis. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Metcalfe-Roach, A.; Yu, A.C.; Golz, E.; Cirstea, M.; Sundvick, K.; Kliger, D.; Foulger, L.H.; Mackenzie, M.; Finlay, B.B.; Appel-Cresswell, S. MIND and Mediterranean Diets Associated with Later Onset of Parkinson’s Disease. Mov. Disord. 2021, 36, 977–984. [Google Scholar] [CrossRef]



| Study | Diet | Healthy Controls (mmol) | Parkinson’s (mmol) | p-Value |
|---|---|---|---|---|
| Bedarf | Fiber | 4.12003 ± 1.5456 | 3.97040 ± 1.2456 | 0.03 |
| Mediterranean | 4.111063 ± 1.4450 | 4.037472 ± 1.3235 | 0.423 | |
| Vegan | 4.107646 ± 1.5344 | 3.888306 ± 1.6568 | 0.006 | |
| Kashavarzian | Fiber | 8.81435 ± 2.2341 | 8.319964 ± 2.1324 | 2.02 × 10−42 |
| Mediterranean | 8.628139 ± 2.1568 | 3.760015 ± 2.2756 | 1.68 × 10−45 | |
| Vegan | 8.713214 ± 2.2341 | 3.767233 ± 2.3876 | 6.23 × 10−46 |
| Study | Diet | Healthy Controls (mmol) | Parkinson’s (mmol) | p-Value |
|---|---|---|---|---|
| Bedarf | Fiber | 5.571× 10−9 ± 1.2× 10−5 | 2.449273× 10−9 ± 1.2× 10−4 | 0.007 |
| Mediterranean | 1.0356× 10−9 ± 1.2× 10−4 | 1.821× 10−9 ± 1.2× 10−4 | 0.488 | |
| Vegan | 4.225× 10−10 ± 1.3× 10−5 | 1.391× 10−9 ± 1.2× 10−4 | 3.605 × 10−5 | |
| Kashavarzian | Fiber | 3.440652 ± 5.412 | 7.250732 ± 5.018 | 2.036 × 10−28 |
| Mediterranean | 3.363205 ± 6.324 | 6.495293 ± 6.578 | 2.506 × 10−31 | |
| Vegan | 3.481832 ± 8.951 | 6.495293 ± 5.610 | 3.304 × 10−28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franyer, L.; Adrian, G.M.; Oscar, B.; Janneth, G.; Andrés, P. Computational Analysis of the Effect of Dietary Interventions on the Gut Microbiome Composition in Parkinson’s Disease. Bacteria 2025, 4, 59. https://doi.org/10.3390/bacteria4040059
Franyer L, Adrian GM, Oscar B, Janneth G, Andrés P. Computational Analysis of the Effect of Dietary Interventions on the Gut Microbiome Composition in Parkinson’s Disease. Bacteria. 2025; 4(4):59. https://doi.org/10.3390/bacteria4040059
Chicago/Turabian StyleFranyer, López, García Macias Adrian, Beltran Oscar, González Janneth, and Pinzón Andrés. 2025. "Computational Analysis of the Effect of Dietary Interventions on the Gut Microbiome Composition in Parkinson’s Disease" Bacteria 4, no. 4: 59. https://doi.org/10.3390/bacteria4040059
APA StyleFranyer, L., Adrian, G. M., Oscar, B., Janneth, G., & Andrés, P. (2025). Computational Analysis of the Effect of Dietary Interventions on the Gut Microbiome Composition in Parkinson’s Disease. Bacteria, 4(4), 59. https://doi.org/10.3390/bacteria4040059