Abstract
This study employed machine learning algorithms to identify lead compounds that inhibit the antibiotic targets DNA gyrase and Dihydrofolate reductase in Escherichia coli, and identified new, multifaceted antimicrobial compounds. This study used three separate datasets: (1) 326 Escherichia coli DNA gyrase inhibitors and 132 non-inhibitors, (2) 346 Escherichia coli Dihydrofolate reductase inhibitors and 176 non-inhibitors, and (3) 18387 non-specific drug-like chemicals. All datasets were then processed using ECFP-4 fingerprints and split into train, test, and validation datasets according to a 70–15–15 train-test-validation split. We explored the potential of six different classification algorithms, all optimized with Bayesian optimization. Our results indicate that the Gradient Boosting Classifier (GBC) performed the best at identifying a compound’s efficacy towards DNA gyrase with an accuracy, precision, recall, F1-score, and AUC of 0.91, 0.92, 0.86, 0.88, and 0.933, respectively. The Random Forest Classifier (RFC) performed optimally for identifying a compound’s effectiveness towards Dihydrofolate reductase with an accuracy, precision, recall, F1-score, and AUC of 0.86, 0.83, 0.85, 0.84, and 0.944, respectively. As a result, the GBC and RFC were used to search for compounds that inhibited both DNA gyrase and Dihydrofolate reductase. Out of 18387 compounds, we identified five novel compounds that have a predicted probability greater than 95% to inhibit both DNA gyrase and Dihydrofolate reductase, suggesting a high antimicrobial potential. The models evaluated in this study, particularly the GBC and RFC models, hold tremendous promise in computationally screening large libraries of compounds for antimicrobial potential.
1. Introduction
1.1. Background on Antibiotics and Antibiotic Resistance
Amidst the recent explosion of antibiotic use in both humans and agriculture, antibiotic resistance in bacterial strains has begun to spike. This has led to the advent of “superbugs”, bacteria that have developed resistance to multiple antibiotics [1]. As a result, there have been numerous research efforts in recent years aiming to identify new antibiotics.
1.2. Recent Advances in Computational Drug Discovery: Applications to Antimicrobial Compounds
Recent advances in machine learning and computational biology have demonstrated the potential to accelerate computational drug discovery by filtering the chemical space for target molecules [2,3]. Previous researchers have demonstrated the efficacy of random forest classification models when predicting for life-extending chemicals [4]. Furthermore, most recently, researchers have demonstrated the potential of deep learning models, particularly in identifying novel antibiotics that are successful in vivo against a wide range of bacterial infections, indicating the potential for computational methods to revolutionize antibiotic bacteria [5].
1.3. DNA Gyrase and Dihydrofolate Reductase as Antimicrobial Targets
Many current antimicrobial compounds operate by inhibiting the function of key proteins that are vital to bacterial function [6]. This study focused on two such proteins proposed by prior literature as antimicrobial targets: DNA gyrase and Dihydrofolate reductase. DNA gyrase functions as topoisomerase in bacteria, aiding in the process of ATP-dependent negative supercoiling of DNA in bacteria [7]. Previous successful antibiotic classes such as Coumarins and Quinolones have modulated the function of DNA gyrase, leading to antimicrobial function via the breakdown of bacterial function [8]. Similarly, Dihydrofolate reductase has also been a popular target for antimicrobial agents due to its crucial role in nucleotide synthesis [9].
1.4. Purpose
In an effort to speed up antibiotic discovery, this study demonstrated the promise of machine learning classification models in multifaceted antimicrobial compound screening and identification.
2. Materials and Methods
2.1. Datasets and Dataset Preprocessing
The breakdown of the three datasets used in this research are displayed in Table 1. The datasets consisting of Escherichia coli DNA gyrase and Dihydrofolate reductase inhibitors were sourced from ChEMBL [10]. The dataset consisting of 18,387 non-specific drug-like chemicals was sourced from Zinc 15 [11].

Table 1.
Dataset Breakdown.
All compounds in datasets were characterized using ECFP-4 fingerprints. All datasets were then split into train, test, and validation datasets according to a 70–15–15 train–test–validation split.
2.2. Machine Learning Models
This study employed six classification models in total: logistic regressions (LR), support vector machines (SVM), random forests (RFC), k-nearest neighbors (K-NN), AdaBoost (ADA), and Gradient Boosting (GBC). All models were evaluated using accuracy, prevision, recall, F1-score, and area under curve (AUC).
2.3. Bayesian Optimization
This study also implemented Bayesian optimization in order to optimize all six classification models. For each of the classification algorithms, Bayesian optimization was run to optimize the parameters. The models were optimized using the validation dataset in order to minimize overfitting when evaluating model metrics with the test dataset.
3. Results
3.1. Machine Learning Model Evaluation
3.1.1. DNA gyrase Machine Learning Model Evaluation
All six optimized machine learning models were trained on DNA gyrase inhibitors and evaluated using accuracy, prevision, recall, F1-score, and AUC. The gradient-boosting algorithm performed the best with an accuracy, precision, recall, F1-score, and AUC of 0.91, 0.92, 0.86, 0.88, and 0.933, respectively (Table 2, Figure 1 and Figure 2).

Table 2.
DNA gyrase Machine Learning Model Accuracy Metrics.
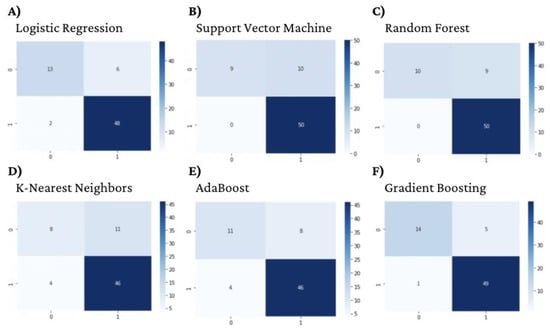
Figure 1.
DNA gyrase Machine Learning Model Confusion Matrices.
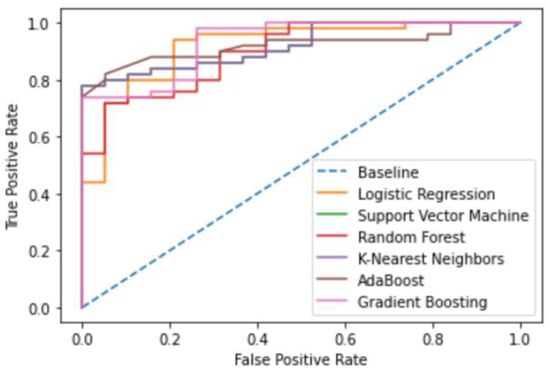
Figure 2.
DNA gyrase Machine Learning Model receiver operating characteristic (ROC) curves.
3.1.2. Dihydrofolate reductase Machine Learning Model Evaluation
All six optimized machine learning models were trained on Dihydrofolate reductase inhibitors and evaluated using accuracy, prevision, recall, F1-score, and AUC. The random forest algorithm performed the best, with an accuracy, precision, recall, F1-score, and AUC of 0.91, 0.92, 0.86, 0.88, and 0.933, respectively (Table 3, Figure 3 and Figure 4).

Table 3.
Dihydrofolate reductase Machine Learning Model Accuracy Metrics.
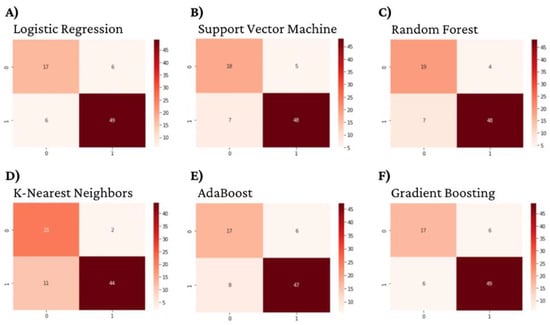
Figure 3.
Dihydrofolate reductase Machine Learning Model Confusion Matrices.
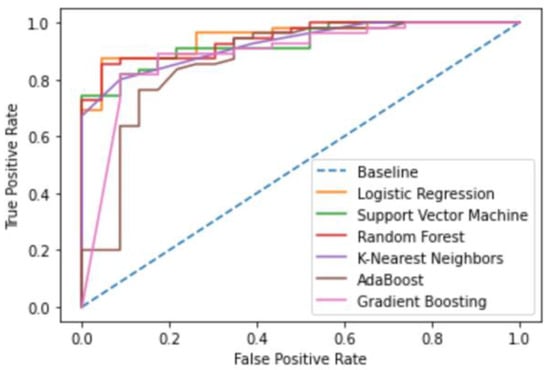
Figure 4.
Dihydrofolate reductase Machine Learning Model ROC curves.
3.2. Identification and Analysis of Novel Antimicrobial Ligands
By implementing the best performing model for identifying DNA gyrase inhibitors (gradient boosting) and the best performing model for identifying Dihydrofolate reductase (random forest), this study used each model to identify novel compounds that are predicted to inhibit both DNA gyrase and Dihydrofolate reductase. Using both models, five compounds were identifed that had an average predicted probability greater than 0.97. The best performing compound is CN(Cc1cnc2nc(N)nc(N)c2n1)c1ccc(C(=O)N[C@@H](CCC(=O)NO)C(=O)O)cc1, with a predicted probability of 0.9988515310206159 to inhibit DNA gyrase, a predicted probability of 0.9897304236200257 to inhibit Dihydrofolate reductase, and an averaged predicted probability of 0.9942909773203208 (Table 4).

Table 4.
Novel Antimicrobial Compounds and Probabilistic Analyses.
4. Discussion
Of the six models trained on DNA gyrase, gradient boosting was the most accurate and had the highest F1-score. Unlike the some of the other algorithms, the Bayesian optimization of gradient-boosting models drastically changes the model performance and metrics (i.e., number of trees, learning rate, and maximum depth), greatly enhancing its performance. The use of gradient boosting is further enhanced as the DNA gyrase dataset consists of few outliers, and overall computation time was not a major constraint [12].
For the Dihydrofolate reductase models, the random forest model nominally outperformed other models. With the optimal models from both DNA gyrase and Dihydrofolate reductase, the compounds identified had a probability of 0.97 to inhibit both DNA gyrase and Dihydrofolate reductase.
Since the precision in both models are either similar or higher than respective recall metrics, the false positivity rate will be comparable to the accuracy metrics of the models, ensuring that the chosen compounds maintain a high probability of being effective.
5. Conclusions
This study evaluated the efficacy of machine learning models at identifying novel antimicrobial compounds. The machine learning models evaluated in this study, particularly the gradient-boosting and random forest models, performed very well and hold tremendous potential in computationally screening large libraries of compounds for antimicrobial potential. Furthermore, the compounds identified in this study hold promise as potential, novel antimicrobial compounds. Future investigations should explore alternative classification approaches to antimicrobial compound screening. The compounds identified in this study should also be researched further in vivo to identify additional antimicrobial potential.
Author Contributions
Conceptualization, J.S. and D.V.; methodology, J.S. and D.V.; software, J.S. and D.V.; validation, J.S. and D.V.; formal analysis, J.S.; investigation, J.S.; resources, J.S.; data curation, J.S.; writing—original draft preparation, J.S. and D.V.; writing—review and editing, J.S. and D.V.; visualization, J.S.; supervision, J.S.; project administration, J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
The authors would like to acknowledge their friends, families, and mentors for their endless support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. P T A Peer-Rev. J. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Reviews. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [CrossRef] [PubMed]
- Kapsiani, S.; Howlin, B.J. Random forest classification for predicting lifespan-extending chemical compounds. Sci. Rep. 2021, 11, 13812. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Browne, J.J.; Van Crugten, J.; Hasan, M.F.; Liu, L.; Barkla, B.J. In Silico, Molecular Docking and In Vitro Antimicrobial Activity of the Major Rapeseed Seed Storage Proteins. Front. Pharmacol. 2020, 11, 1340. [Google Scholar] [CrossRef] [PubMed]
- Reece, R.J.; Maxwell, A. DNA gyrase: Structure and function. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 335–375. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, A. DNA gyrase as a drug target. Trends Microbiol. 1997, 5, 102–109. [Google Scholar] [CrossRef]
- Zhang, Y.; Chowdhury, S.; Rodrigues, J.V.; Shakhnovich, E. Development of antibacterial compounds that constrain evolutionary pathways to resistance. eLife 2021, 10, e64518. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Modeling 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobot. 2013, 7, 21. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).