Abstract
Plant and fruit diseases significantly impact agricultural economies by diminishing crop quality and yield. Developing precise, automated detection techniques is crucial to minimize losses and drive economic growth. We introduce YOLO-AppleScab, integrating Content-Aware ReAssembly of FEature () architecture into YOLOv7 for enhanced apple fruit detection and disease classification. The model achieves impressive metrics: F1, recall, and precision of 89.75%, 85.20%, and 94.80%, and a mean average precision of 89.30% at 0.5. With 64% efficiency, this model’s integration with YOLOv7 improves detection, promising economic benefits by accurately detecting apple scab disease and reducing agricultural damage.
1. Introduction
Plant and fruit diseases greatly impact agriculture, causing lower crop yields and increased costs [1]. Climate change, globalization, and agricultural practice changes have led to more disease incidents [2]. Researchers are developing better ways to detect, diagnose, and treat these diseases [3,4], like by using remote sensing [5], genomics [6], monitoring systems [7], and artificial intelligence (AI) [8]. AI has also enabled disease detection and treatment using robots [9], involving two steps: computer vision-based fruit detection and robot-guided treatment. Fruit detection is especially challenging [10].
The methods for detecting high-quality fruits include bio-molecular sensing, hyperspectral/multispectral imaging, and traditional vision technology. Traditional image processing, like binarization, struggles with complex backgrounds [11]. Researchers have used methods like multithreshold segmentation [11], artificial neural networks (ANN) [12], support vector machines (SVM), and convolutional neural networks (CNNs) [13,14] for disease identification. CNNs excel in image recognition tasks due to their deep learning capabilities.
Developing an automatic disease diagnosis system using image processing and neural networks can reduce fruit damage [15]. Deep learning, especially CNNs, has led to effective image recognition models [15]. Various architectures like AlexNet [16], GoogLeNet [17], VGGNet [18], and ResNet [19] have been employed [20]. AI’s rise prompted research on applying machine learning to agriculture [21]. This study proposes a detection model that integrates architecture into YOLOv7 to better identify healthy and scab apples in challenging conditions.
This work explores integrating these technologies to enhance detection accuracy [22]. The experimental results validate the model’s effectiveness in maintaining real-time processing speeds [22]. Such advances are crucial for food security and sustainable agriculture [23].
2. Material and Methodology of the Proposed Research
2.1. You Only Look Once Series (YOLO Series)
The YOLO (You Only Look Once) framework, shown in Figure 1, divides the input image into an S S grid, with each grid cell responsible for object detection. It generates B-bounding boxes and confidence scores for each grid cell to indicate the probability of an object’s presence. The framework also uses class probability maps from these scores to detect and classify objects accurately, streamlining object detection into a single process for real-time and precise results.
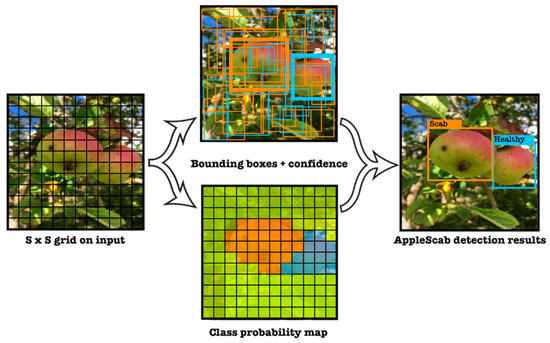
Figure 1.
YOLO model detection. The classification task is addressed while dealing with regression. Therefore, each interested class (Healthy, Scab) is represented under different colors to illustrate the YOLO detection model.
2.2. Rectangular Bounding Box and Function
Each grid cell predicts , and class probabilities, totaling 5 values. The score gauges object presence and calculates Intersection over Union () with the ground truth () box. If the cell offset is (), and box prior is , the prediction is calculated using Equations (1)–(5). This grid-based approach efficiently detects objects and reduces computation. For specific tasks, custom bounding boxes like R-Bboxes can enhance detection; in our case, apple fruits are targeted. Figure 2 illustrates the R-Bboxes prediction:
where is sigmoid function.
where and is the Intersection over Union between the predicted box and the ground truth.
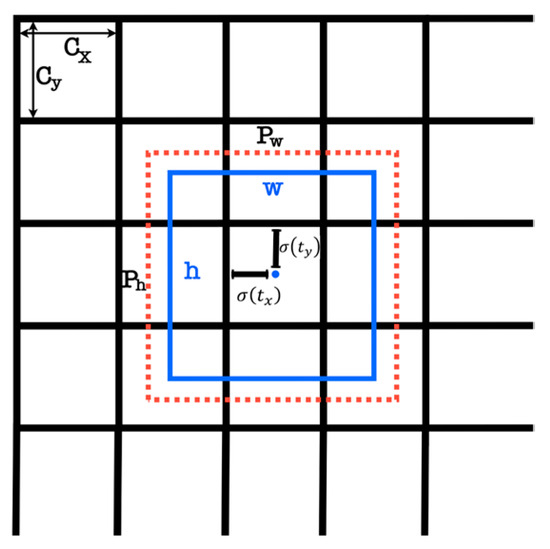
Figure 2.
Prediction of bounding box. YOLOv7 will predict the width and height of the box as offsets from cluster centroids and center coordinates of the box relative to the location of the filter application using a sigmoid function. The red dotted indicates the prior anchor, and the blue square is the prediction.
The loss function remains the same as in the YOLOv4 model; the Complete IoU () loss function is given by Equation (6):
where represents the Euclidean distance between the center points of the prediction box and the , and c represents the diagonal distance of the smallest closed area that can simultaneously contains the prediction box and the ground truth.
Equations (7) and (8) presented the formulas of and as follows:
and
2.3. Content-Aware ReAssembly of Feature:
In deep neural networks, spatial feature upsampling is crucial for tasks like resolution enhancement and segmentation. YOLOv7 introduces , a novel feature upsampling method that efficiently combines information over a wide receptive field, adapts to specific instances, and improves the performance in various tasks.
2.4. Image Acquisition
The image dataset was collected in orchards using smartphones (12 MP, 13 MP, 48 MP) and a digital compact camera (10 MP), capturing apples at various stages of development and damage. The images were taken from different viewpoints, times of day, and lighting conditions. The dataset, named AppleScabLDs, was curated by manually reviewing and sorting images of healthy and diseased (apple scab disease) apples. Subsets were created with and without scab symptoms, excluding images with visual noise. This meticulous selection process ensured noise would not affect disease detection. The model’s performance was evaluated using proper metrics and reported results. The dataset consisted of 297 images: 237 in the training set (200 healthy apples, 206 with scabs) and 60 in the test set (55 healthy apples, 49 with scabs). Samples from the dataset in various environments are shown in Figure 3.
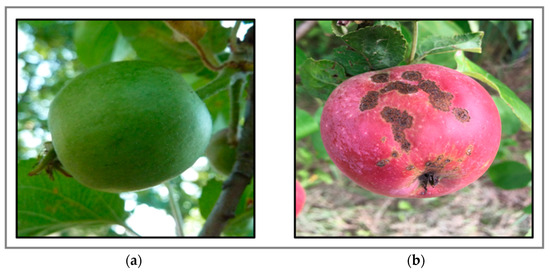
Figure 3.
Apple fruit samples from dataset AppleScabLDs: (a) healthy apple fruit, and (b) infected apple by scab disease.
2.5. The Proposed YOLO-AppleScab Model
An overview of the proposed apple fruit with a scab disease detection model is shown in Figure 4. On the SOTA of the YOLOv7 architecture model, a architecture was incorporated for better feature reuse and representation. Furthermore, the R-Bbox can derive a more accurate IoU between the predictions, which is called YOLO-AppleScab.
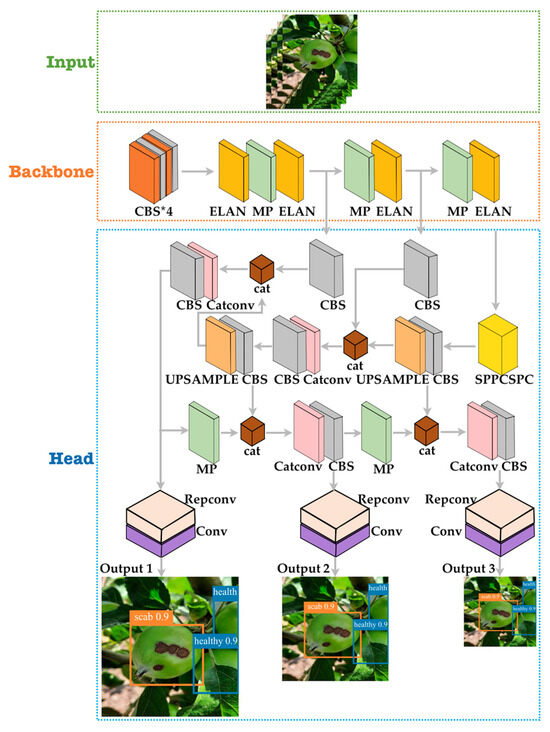
Figure 4.
An overview of the proposed model of architecture incorporated in YOLOv7 network architecture.
2.6. Experiment Setup
The experiments were conducted on a computer with the following specifications: 11th Gen Intel® Core™ i5-11400H, 64-bit 2.70 GHz dodeca-core CPUs and a NVIDIA GeForce RTX 3050 GPU, Santa Clara, CA, USA. Table 1 presents the basic configuration of the local computer.

Table 1.
The basic configuration of the local computer.
In the binary classification problem, according to the combination of the sample’s true class and the model’s prediction class, the sample can be divided into 4 types: , , , and . A series of experiments were conducted to evaluate the performance of the proposed method. The indexes for evaluation of the trained model are defined by Equations (9) and (10) as follows:
where , , and are abbreviations for true positives (correct detection), false negative (miss), and false positive (false detection).
3. Results and Discussion
3.1. The Network Visualization
Understanding deep neural networks can be complex; yet, they grasp vital visual cues. Figure 5 illustrates the 32 feature maps from YOLOv7’s upsample and the 32 feature maps from in the proposed model. Stage 53 generates 32 maps from YOLOv7’s upsample, while stage 65 presents ’s 32 maps. These maps unveil captured visual insights, with ’s maps being richer, depicting diverse edges. This underscore ’s role in enhancing the model’s feature representation, showcasing its capability in extracting detailed visual elements.
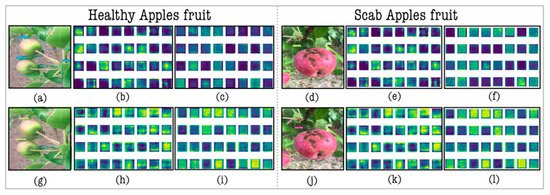
Figure 5.
Feature maps activation in Upsample and layers. (a,d) YOLOv7 prediction; (b,e) stage 53 Upsample feature maps; (c,f) stage 65 Upsample feature maps; (g,j) YOLO-AppleScab prediction; (h,k) stage 53 feature maps; (i,l) stage 65 feature maps. Each color represents a distinct outcome of the convoluted feature map at the present layer.
3.2. Performance of the Proposed Model under Different Lighting Conditions
The efficiency of the proposed model in different illumination conditions was examined in this study. Images were recorded in the morning (09:00–10:00), noon (12:00–14:00), and afternoon (16:00–17:00) to provide a variety of natural light conditions. Because it was hard to clearly separate images according to the time, the image was divided into two groups, as shown in Table 2: strong light and soft light.

Table 2.
The detection performance of the proposed model.
Among all the apples evaluated, 48 were present under strong light and the remaining 56 were in soft light. This study examines two subclasses under strong and soft light. Strong light included 20 healthy and 28 scab-infected apples; soft light had 29 healthy and 27 scab-infected apples. A healthy identification rate was 90% under strong light and 75.86% under soft light. Scab disease detection was 96.43% under strong light and 85.18% under soft light. False detections were 5% healthy and 0% scab under strong light, and 6% false and 7.41% scab under soft light, mainly from the background.
3.3. Comparison of Different State-of-the-Art Algorithms
Table 3 displays key results, including precision, recall, F1 score, , and , for apple fruit detection: healthy and scab-infected classes. This information assesses the model’s effectiveness by class, revealing strengths and weaknesses in detection. and provide overall accuracy. This table is vital for evaluating the model’s performance and identifying improvement areas for future iterations. Table 4 offers a comprehensive analysis of classification metrics, evaluating the accuracy of YOLOv3 [24], YOLOv4 [24], YOLOv7, and Faster RCNN [25] against our proposed method. The efficacy of our YOLO-AppleScab model was validated against other SOTA.

Table 3.
Classification metrics. Comparison with different SOTA of YOLOv3, YOLOv4, YOLOv7 and the proposed method for the two studied classes (healthy and scab) that apple represents. The input image size is 416 416.

Table 4.
Classification metrics. Comparison with different SOTA of YOLOv3, YOLOv4, YOLOv7 and Faster R-CNN is used for benchmarking. The input image size is 416 416. The are expressed in percentages. Two classes (healthy and scab) represent the apple condition.
Table 5 details precision, recall, F1 score, , , and average CPU time per image. YOLO-AppleScab excels, achieving 89.30% precision, 64% , and a detection time of 0.1752 s per image. Its superior performance underscores its prowess in detecting apples with scab disease, offering real-time detection capabilities suitable for robot-assisted disease detection in fruits.

Table 5.
A comparison of different state-of-the-art detection methods.
4. Conclusions
This study introduces YOLO-Apple Scab detection, leveraging YOLOv7 to classify healthy and scab-infected apples. This method minimizes challenges like overlap and illumination changes using architecture for feature extraction, enhancing model learning. The experiments validate its efficiency. Incorporating boosts the F1 score by around 4.2%, while maintaining the performance under diverse lighting. Notably, strong light yields a 90% accurate identification of healthy apples (over 14% more than soft light), and 96.43% for scab-infected apples (over 11% higher). This method surpasses other SOTA techniques, showcasing the potential for broader applications in apple disease detection. Future work aims to enhance detection in occlusion and variable light and explore treatment applications based on disease type, possibly integrating this algorithm into a robot for versatile detection and treatment at various growth stages.
Supplementary Materials
The presentation materials can be downloaded at: https://www.mdpi.com/article/10.3390/IOCAG2023-16688/s1.
Author Contributions
J.C.N. and J.S. participated in validating the data, curating it, creating visualizations, and labeling the database, and played significant roles in the development of the methodology, conception, software, and provision of resources. J.C.N. supervised this work, wrote the first draft, and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset of AppleScabFDs used in this project was provided by the Institute of Horticulture (LatHort). The dataset is available here [29].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lu, Y.; Lu, R. Non-Destructive Defect Detection of Apples by Spectroscopic and Imaging Technologies: A Review. Trans. ASABE 2017, 60, 1765–1790. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Pandey, M.K.; Bohra, A.; Singh, V.K.; Thudi, M.; Saxena, R.K. Toward the Sequence-Based Breeding in Legumes in the Post-Genome Sequencing Era. Theor. Appl. Genet. 2019, 132, 797–816. [Google Scholar] [CrossRef] [PubMed]
- Sikati, J.; Nouaze, J.C. YOLO-NPK: A Lightweight Deep Network for Lettuce Nutrient Deficiency Classification Based on Improved YOLOv8 Nano. Eng. Proc. 2023, 58, 31. [Google Scholar] [CrossRef]
- Ali, A.; Imran, M. Remotely Sensed Real-Time Quantification of Biophysical and Biochemical Traits of Citrus (Citrus sinensis L.) Fruit Orchards—A Review. Sci. Hortic. 2021, 282, 110024. [Google Scholar] [CrossRef]
- He, W.; Liu, M.; Qin, X.; Liang, A.; Chen, Y.; Yin, Y.; Qin, K.; Mu, Z. Genome-Wide Identification and Expression Analysis of the Aquaporin Gene Family in Lycium Barbarum during Fruit Ripening and Seedling Response to Heat Stress. Curr. Issues Mol. Biol. 2022, 44, 5933–5948. [Google Scholar] [CrossRef]
- Nouaze, J.C.; Kim, J.H.; Jeon, G.R.; Kim, J.H. Monitoring of Indoor Farming of Lettuce Leaves for 16 Hours Using Electrical Impedance Spectroscopy (EIS) and Double-Shell Model (DSM). Sensors 2022, 22, 9671. [Google Scholar] [CrossRef]
- Liu, G.; Nouaze, J.C.; Touko Mbouembe, P.L.; Kim, J.H. YOLO-Tomato: A Robust Algorithm for Tomato Detection Based on YOLOv3. Sensors 2020, 20, 2145. [Google Scholar] [CrossRef]
- Cubero, S.; Marco-Noales, E.; Aleixos, N.; Barbé, S.; Blasco, J. RobHortic: A Field Robot to Detect Pests and Diseases in Horticultural Crops by Proximal Sensing. Agriculture 2020, 10, 276. [Google Scholar] [CrossRef]
- Tian, Y.; Li, E.; Liang, Z.; Tan, M.; He, X. Diagnosis of Typical Apple Diseases: A Deep Learning Method Based on Multi-Scale Dense Classification Network. Front. Plant Sci. 2021, 12, 698474. [Google Scholar] [CrossRef]
- Zou, X.-b.; Zhao, J.-w.; Li, Y.; Holmes, M. In-Line Detection of Apple Defects Using Three Color Cameras System. Comput. Electron. Agric. 2010, 70, 129–134. [Google Scholar] [CrossRef]
- Zarifneshat, S.; Rohani, A.; Ghassemzadeh, H.R.; Sadeghi, M.; Ahmadi, E.; Zarifneshat, M. Predictions of Apple Bruise Volume Using Artificial Neural Network. Comput. Electron. Agric. 2012, 82, 75–86. [Google Scholar] [CrossRef]
- Omrani, E.; Khoshnevisan, B.; Shamshirband, S.; Saboohi, H.; Anuar, N.B.; Nasir, M.H.N.M. Potential of Radial Basis Function-Based Support Vector Regression for Apple Disease Detection. Measurement 2014, 55, 512–519. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early Detection and Classification of Plant Diseases with Support Vector Machines Based on Hyperspectral Reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Gongal, A.; Amatya, S.; Karkee, M.; Zhang, Q.; Lewis, K. Sensors and Systems for Fruit Detection and Localization: A Review. Comput. Electron. Agric. 2015, 116, 8–19. [Google Scholar] [CrossRef]
- Kzizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks|Communications of the ACM. Assoc. Comput. Mach. 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. In Proceedings of the IEEE Computer Society, Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar]
- Zhang, K.; Wu, Q.; Liu, A.; Meng, X. Can Deep Learning Identify Tomato Leaf Disease? Adv. Multimed. 2018, 2018, 10. [Google Scholar] [CrossRef]
- Bhange, M.A.; Hingoliwala, H.A. A Review of Image Processing for Pomegranate Disease Detection. Int. J. Comput. Sci. Inf. Technol. 2014, 6, 92–94. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6517–6525. [Google Scholar]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Wan, S.; Goudos, S. Faster R-CNN for Multi-Class Fruit Detection Using a Robotic Vision System. Comput. Netw. 2019, 168, 107036. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2015; Volume 28. [Google Scholar]
- Kodors, S.; Lacis, G.; Sokolova, O.; Zhukovs, V.; Apeinans, I.; Bartulsons, T. AppleScabFDs Dataset. Available online: https://www.kaggle.com/datasets/projectlzp201910094/applescabfds (accessed on 17 February 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).