Abstract
Late blight is a destructive disease of solanaceous crops such as tomato (Solanum lycopersicum L.), caused by the Oomycete Phytophthora infestans (Mont.) de Bary. Late blight is generally controlled by fungicide applications, which quickly become ineffective due to the appearance of new P. infestans genotypes that can overcome the resistance of improved tomato cultivars and cause total production losses. The aim of this study is to assess the resistance level of tomato cultivars under controlled conditions and inoculations were carried out on detached leaflets (cvs. Trakia, Saint Pierre and Marmande) using inoculums of the major P. infestans clonal lineages found in Algeria such as EU_13_A2 (n = 1), EU_23_A1 (n = 2) and EU_2_A1 (n = 1) (three replicates of each isolate). This investigation showed that the choice of resistant cultivars can help control late blight and provide economic and environmental advantages by reducing the use of inputs.
1. Introduction
Tomato (Solanum lycopersicum L.) is an important crop cultivated globally in tropical to temperate regions. It is the second-most-consumed vegetable after the potato [1]. In Algeria, tomato is the fourth most important crop after potato, melon and onion. It is grown throughout the year, with winter cultivation taking place in greenhouses and summer cultivation in open fields [2]. Often, diseases limit tomato production. But late blight caused by the oomycete Phytophthora infestans (Mont.) de Bary is the most damaging disease in this crop [3]. This oomycete attacks all aerial parts of the plant. It causes leaf and stem necrosis, fruit loss and ultimately plant death [4].
In Algeria, three major clonal lineages of P. infestans (EU_13_A2, EU_2_A1 and EU_23_A1) have been identified in commercial potato and tomato production regions [5]. However, most methods of controlling this pathogen are based on the application of expensive fungicides, which can be less effective when weather conditions are favorable for pathogen spread [6] or the emergence of new P. infestans genotypes resistant to fongicides.
When it is difficult to control P. infestans with fungicides, resistant cultivars provide an alternative means of disease control [7].
This study was conducted to evaluate the behaviour of three tomato cultivars against the major late blight clonal lineages using the detached leaflets test conducted under in vitro conditions and the aggressiveness components were measured including the incubation and latency period, lesion area and sporangia production. It is important to identify and characterise new sources of resistance and to develop new resistant cultivars to make it easier to control late blight and offer economic and environmental benefits through reduced inputs.
2. Materials and Methods
2.1. Plant Material
Three cultivars (Marmande, Saint Pierre and Trakia) were evaluated for their resistance to P. infestans under controlled conditions. The cultivars were grown in a glasshouse with a 20–25 °C day/night cycle. Leaflets were harvested after nine weeks.
2.2. Phytophthora Infestans Isolates
Four isolates were selected for this investigation (Table 1). The isolates were obtained from samples of infected potato and tomato using isolation techniques that consist of placing small pieces of fresh samples of leaves, stems and fruits infected with P. infestans on potato slices and then putting them into closed Petri dishes. These were subsequently incubated at 15 °C in the dark. After 4 to 5 days, the mycelium obtained was purified by repeated transfers into pea agar, which had been amended with antibiotics (30 mg of Rifamycin and 200 mg of Ampicillin). Pure cultures were kept at a temperature of 15 °C. After isolation, genotyping was performed using 17 SSR loci [5]. Subsequently, the lineages were named according to the classic European nomenclature as defined by [8].

Table 1.
Characteristics of isolates used in the detached-leaflet experiment.
2.3. Inoculation
Under controlled conditions, leaflets aged nine weeks were harvested and detached leaflets were inoculated on the abaxial side with a 20 μL drop of sporangial suspension (5 × 104 sporangia mL−1). The inoculum was produced from P. infestans mycelium that had been grown on pea agar for 3 weeks. Two leaflets were placed on sterilised moist paper in Petri dishes and incubated at 18 °C in a growth chamber for 16 h in the light (three replicates for each isolate).
2.4. Aggressiveness Components
The incubation period (IP) was evaluated by daily observations of the first symptoms, while the latency period (LP) was expressed through daily observations of the initial sporangia production. Subsequently, lesion area (LA) was measured five days after inoculation, according to the formula provided by [9]; the lesion area was calculated as LA = 1/4 × π × length × width of necrosis. Sporangia production (SP) was assessed seven days after inoculation. The infected leaflets were washed in 10 mL of sterilised distilled water and the number of sporangia were quantified with a Malassez cell. The sporangia were expressed as the number of sporangia per mL.
2.5. Data Analysis
All statistical analyses, including analysis of variance and Tukey HSD test, were performed using the software R v.3.3.2. (The R Foundation for Statistical Computing, Vienna, Austria, 2016).
3. Results
Isolates produced a shorter incubation period on cv. Marmande (Table 2), but longer on cv. Saint Pierre, ranging from 3 days (EU_13_A2, cv. Marmande) to 5 days (EU_23_A1, cv. Saint Pierre). The same results were noticed with a latency period that ranged from 3.5 days for (EU_13_A2, cv. Marmande) to 6 days for (EU_2_A1, cv. Saint Pierre). The EU_13_A2 and EU_2_A1 clonal lineages showed significant results with all cultivars (p ≤ 0.001). However, there was no significant difference in the incubation and latency periods between EU_23_A1 clonal lineages and cultivars (p ≥ 0.001). A short incubation and latency period means that the pathogen is able to attack a cultivar more quickly, indicating its susceptibility, as in the case of cv. Marmande.

Table 2.
Mean values of aggressiveness components.
The lesion area was larger on the cv. Marmande with all isolates compared to the Saint Pierre and Trakia cultivars, where sizes ranged from 68.56 mm2 (EU_23_A1, cv. Trakia) to 377.07 mm2 (EU_23_A1, cv. Marmande). Furthermore, sporangia production was more significant in the cv. Marmande with isolates of the EU_13_A2 and EU_23_A1 lineages, compared to the Saint Pierre and Trakia cultivars, while the sporulation rate varied from 3.1 × 104 sporangia mL−1 (isolate EU_23_A1, cv. Trakia) to 45.73 × 104 sporangia mL−1 (isolate 23_A2, cv. Marmande) (Figure 1).
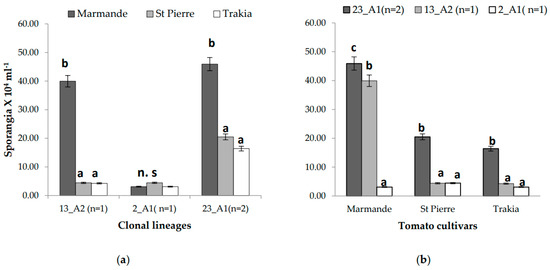
Figure 1.
The rate of sporulation production expressed as the number of sporangia × 104 mL−1: (a) Comparison between tomato cultivars; (b) comparison of P. infestans clonal lineages. Letters a–c: classification of means into homogeneous groups.
The EU_13_A2 and EU_23_A1 clonal lineages showed significant results for the lesion area and sporulation with all the cultivars, whereas the EU_2_A1 lineage had no significant effect on all the cultivars tested (p = 0.49 for lesion area and p = 0.83 for sporulation). This result suggests that the isolates have different levels of virulence (Figure 1).
4. Discussion
The data of the aggressiveness components obtained from the detached leaflet assay can be a reliable predictor of cultivars’ and isolates’ behaviour under field conditions [10]. In our case, the cv. Marmande was very susceptible, compared with cvs. Saint Pierre and Trakia, which showed a good level of resistance to all P. infestans clonal lineages.
The sensitivity of Marmande can be explained by the fact that this cultivar is extensively cultivated, and its partial resistance has been overcome by local P. infestans populations in Algeria. In contrast, the resistance of the other two cultivars (cvs. Saint Pierre and Trakia) was higher due to their limited cultivation. Our results are in agreement with those of [11], who observed the adaptation of the P. infestans population to the locally dominant cultivars, which had overcome their partial resistance.
We also noticed that the isolates behaved differently according to their genotype, with some of them proving to be very aggressive, such as EU_23_A1. This can be explained by the adaptation of this genotype to tomato cultivars [2]. Compared with the EU_13_A2 genotype, which was less aggressive on tomato cultivars except for the Marmande. The reason is that this genotype has only been found on potato under field conditions in Algeria, so it is well adapted to potato [5]. Similarly, certain lineages such as EU_13_A2 and EU_6_A1 in Europe are identified as potato specialists [12]. Therefore, they do not adapt to substrates other than potato, and their virulence decreases on tomato [13].
5. Conclusions
Our experiments were conducted under controlled conditions. However, in the field, many factors such as cultivar–pathogen interactions and climate can affect a cultivar’s resistance to a pathogen, causing cultivars to move from resistant to susceptible. It is essential to assess cultivars’ behaviour, particularly those that have exhibited a notable level of pathogen resistance in the field. Searching for cultivars with resistance to late blight would be a positive step towards enhancing tomato production. This approach would also reduce the need to use fungicides, which can adversely affect both the environment and human health.
Author Contributions
This research was carried out by S.B.; the protocol was established by S.B. and the isolates were isolated by S.B. and L.B.; the methodology: S.B.; the supervision: Z.B., the writing of this article was carried out by S.B., the reading of this article was carried out by all the authors. All authors have read and agreed to the published version of the manuscript.
Funding
This investigation was supported by the PoH-MED project, Potato Health–Managed for Efficiency and Durability, funded under the ARIMNet (Agricultural Research in the Mediterranean Area) ERA-Net (KBBE 219262).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Dataset available on request from the authors.
Acknowledgments
The experiments were carried out in the laboratory of Phytopathology and molecular biology, National Higher School of Agronomy, ENSA, Algiers. The authors would like to thank Mabon R. (INRAE, IGEPP) for his help with the isolate characterisation, Corbière R. (INRAE, IGEPP) and Andrivon D. (INRAE, IGEPP) for their advice and constructive discussions during the experiments.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Kalia, P.; Palanisamy, M. Tomato. In Alien Gene Transfer in Crop Plants, Achievements and Impacts; Pratap, A., Kumar, J., Eds.; Studium Press LLC: New Delhi, India, 2014; Volume 2, pp. 347–380. [Google Scholar]
- Belkhiter, S.; Beninal, L.; Khedidji, H.; Krimi, A.; Bouznad, Z. Aggressiveness and host adaptation of some Algerian Phytophthora infestans clonal lineages on potato and tomato. Pak. J. Phytopathol. 2019, 31, 147–154. [Google Scholar]
- Fry, W.E. Phytophthora infestans, the plant (and R gene) destroyer. Mol. Plant Pathol 2008, 9, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, M.; Kozik, E.U.; Foolad, M.R. Late blight of tomato. In Translational Genomics for Crop Breeding; Varshney, R.K., Tuberosa, R., Eds.; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 2013; pp. 241–265. [Google Scholar]
- Beninal, L.; Bouznad, Z.; Corbière, R.; Belkhiter, S.; Mabon, R.; Taoutaou, A.; Keddad, A.; Runno-Paurson, E.; Andrivon, D. Distribution of major clonal lineages EU_13_A2, EU_2_A1, and EU_23_A1 of Phytophthora infestans associated with potato late blight across crop seasons and regions in Algeria. Plant Pathol. 2021, 71, 458–469. [Google Scholar] [CrossRef]
- Foolad, M.R.; Merk, H.L.; Ashrafi, H. Genetics, Genomics and Breeding of Late Blight and Early Blight Resistance in Tomato. Crit. Rev. Plant Sci. 2008, 27, 75–107. [Google Scholar] [CrossRef]
- McGrath, M.T. Late Blight Management in Tomato with Resistant Varieties. eOrganic10822. 2015. Available online: https://eorganic.org/node/10822 (accessed on 9 September 2023).
- Cooke, D.E.L.; Cano, L.M.; Raffaele, S.; Bain, R.A.; Cooke, L.R.; Etherington, G.J.; Deahl, K.L.; Farrer, R.A.; Gilroy, E.M.; Goss, E.M.; et al. Genome analyses of an aggressive and invasive lineage of the rish potato famine pathogen. PLoS Pathog. 2012, 8, e1002940. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.G.A.A.; Van Dooijeweert, W.; Govers, F.; Kamoun, S.; Colon, L.T. The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans. Planta 2000, 210, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Foolad, M.R.; Sullenberger, M.T.; Ohlson, E.W.; Gugino, B.C. Response of 17 accessions within tomato wild species, Solanum pimpinellifolium to late blight. Plant Breed 2014, 133, 401–411. [Google Scholar] [CrossRef]
- Montarry, J.; Andrivon, D.; Glais, I.; Corbière, R.; Mialdea, G.; Delmotte, F. Microsatellite markers reveal two admixed genetic groups and an ongoing displacement within the French population of the invasive plant pathogen Phytophthora infestans. Mol. Ecol. 2010, 19, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Kröner, A.; Mabon, R.; Corbière, R.; Montarry, J.; Andrivon, D. The coexistence of generalist and specialist clonal lineages in natural populations of the Irish Famine pathogen Phytophthora infestans explains local adaptation to potato and tomato. Mol. Ecol. 2017, 26, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Michalska, A.M.; Sobkowiak, S.; Flis, B.; Zimnoch-Guzowska, E. Virulence and aggressiveness of Phytophthora infestans isolates collected in Poland from potato and tomato plants identified no strong specificity. Eur. J. Plant Pathol. 2016, 144, 325–336. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).