Main Mechanical Forces to Analyse the Chemical Interactions Shaping Backbone Torsion Angles in DNA Tertiary Structures
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Travers, A.; Muskhelishvili, G. DNA structure and function. FEBS J. 2025, 282, 2279–2295. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Schneider, B.; Neidle, S.; Berman, H.M. Conformations of the sugar-phosphate backbone in helical DNA crystal structures. Biopolymers 1997, 42, 113–124. [Google Scholar] [CrossRef]
- Larocca, M.; Foglia, F.; Cilibrizzi, A. Dihedral Angle Calculations to Elucidate the Folding of Peptides through its Main Mechanical Forces. Biochemistry 2019, 58, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Larocca, M.; Floresta, G.; Cilibrizzi, A. Principles that Rule the Calculation of Dihedral Angles in Secondary Structures: The cases of an α-helix and a β-sheet. J. Mol. Struct. 2021, 1229, 129802. [Google Scholar] [CrossRef]
- Larocca, M. Polypeptides folding: Rules for the calculation of the backbone dihedral angles ϕ starting from the amino acid sequence. ChemRxiv 2020, chemrxiv.12000132.v2. [Google Scholar] [CrossRef]
- Larocca, M.; Floresta, G.; Verderese, D.; Cilibrizzi, A. ‘Main Mechanical Forces-Chemical Interactions′ Interplay as a Tool to Elucidate Folding Mechanisms. Pept. Sci. 2024, 116, e24365. [Google Scholar] [CrossRef]
- Larocca, M.; Floresta, G.; Verderese, D.; Cilibrizzi, A. Dominant Chemical Interactions Governing the Folding Mechanism of Oligopeptides. Int. J. Mol. Sci. 2024, 25, 9586. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-G.; Robinson, H.; Wang, A.H.-J. High-resolution A-DNA crystal structures of d(AGGGGCCCCT). Eur. J. Biochem. 1999, 261, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Stone, M.P. Solution structure of an oligodeoxynucleotide containing the human n-ras codon 61 sequence refined from 1H NMR using molecular dynamics restrained by nuclear Overhauser effects. Chem. Res. Toxicol. 1995, 8, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Gessner, R.V.; Frederick, C.A.; Quigley, G.J.; Rich, A.; Wang, A.H. The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG). J. Biol. Chem. 1989, 264, 7921–7935. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiong, Y.; Xiao, Y. 3dDNA: A Computational Method of Building DNA 3D Structures. Molecules 2022, 27, 5936. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chiu, T.P.; Rohs, R. Predicting DNA structure using a deep learning method. Nat. Commun. 2024, 15, 1243. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M., Jr.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Hunter, C.A.; Sanders, J.K.M. The nature of pi.-.pi. interactions. J. Am. Chem. Soc. 1990, 112, 5525–5534. [Google Scholar] [CrossRef]
- Liao, T.-C.; Ma, T.-Z.; Chen, S.-B.; Cilibrizzi, A.; Zhang, M.-J.; Li, J.-H.; Zhou, C.-Q. Human telomere double G-quadruplex recognition by berberine-bisquinolinium imaging conjugates in vitro and in cells. Int. J. Biol. Macromol. 2020, 158, 1299–1309. [Google Scholar] [CrossRef]
- Obara, P.; Wolski, P.; Pańczyk, T. Insights into the Molecular Structure, Stability, and Biological Significance of Non-Canonical DNA Forms, with a Focus on G-Quadruplexes and i-Motifs. Molecules 2024, 29, 4683. [Google Scholar] [PubMed]
- Tateishi-Karimata, H.; Sugimoto, N. Chemical biology of non-canonical structures of nucleic acids for therapeutic applications. Chem. Commun. 2020, 56, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
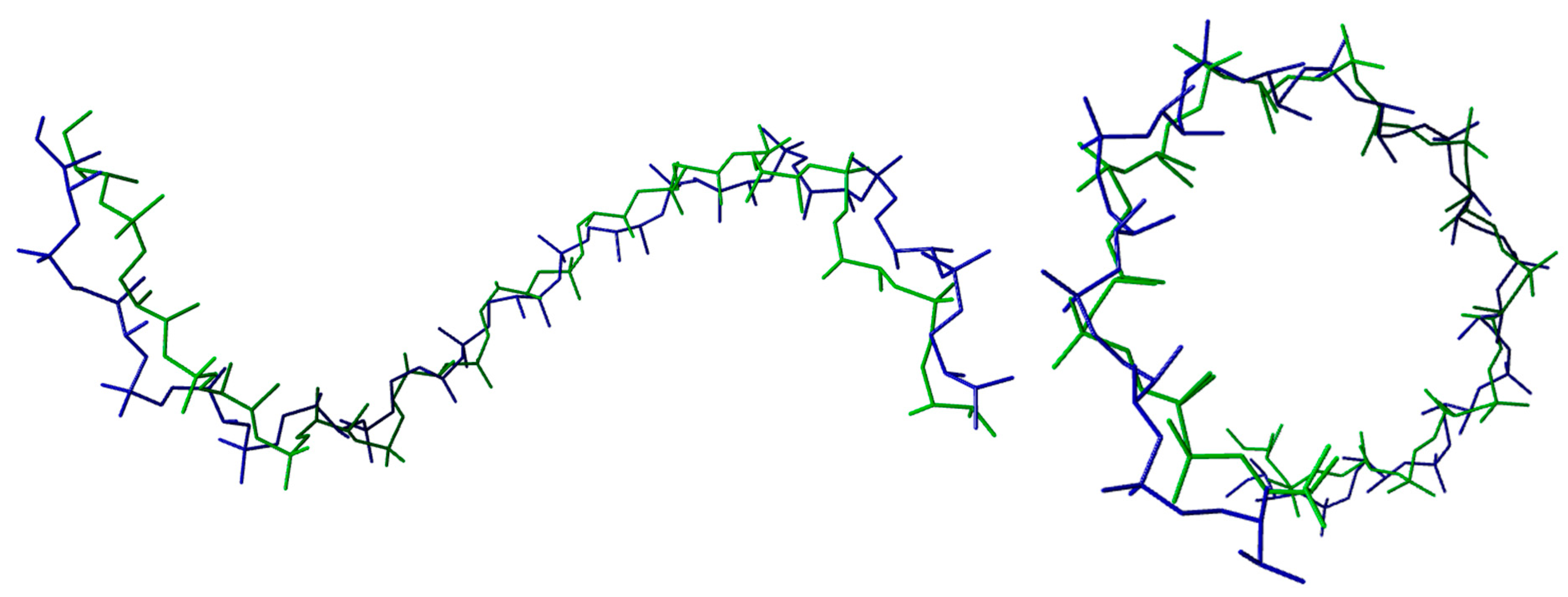
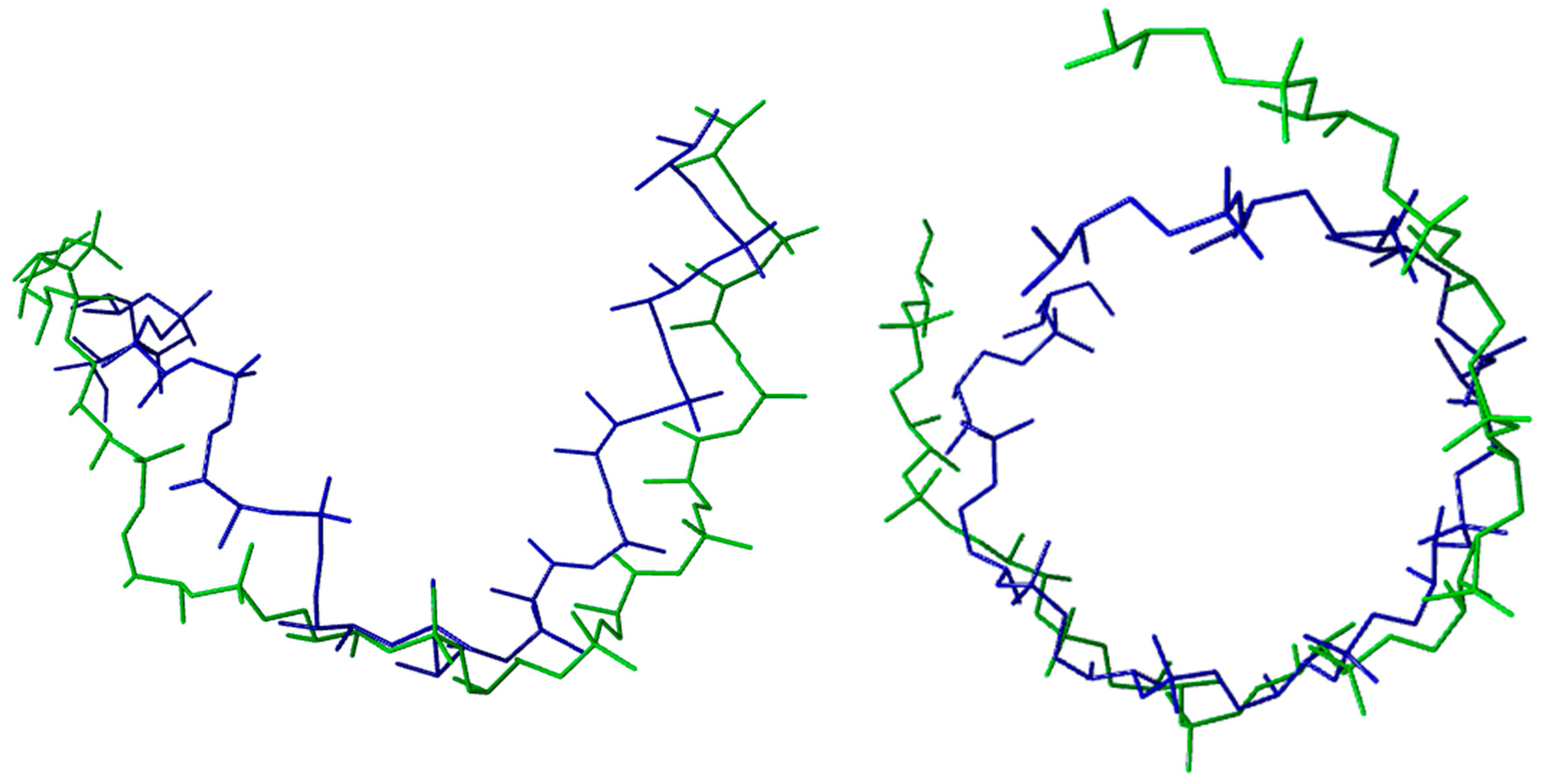
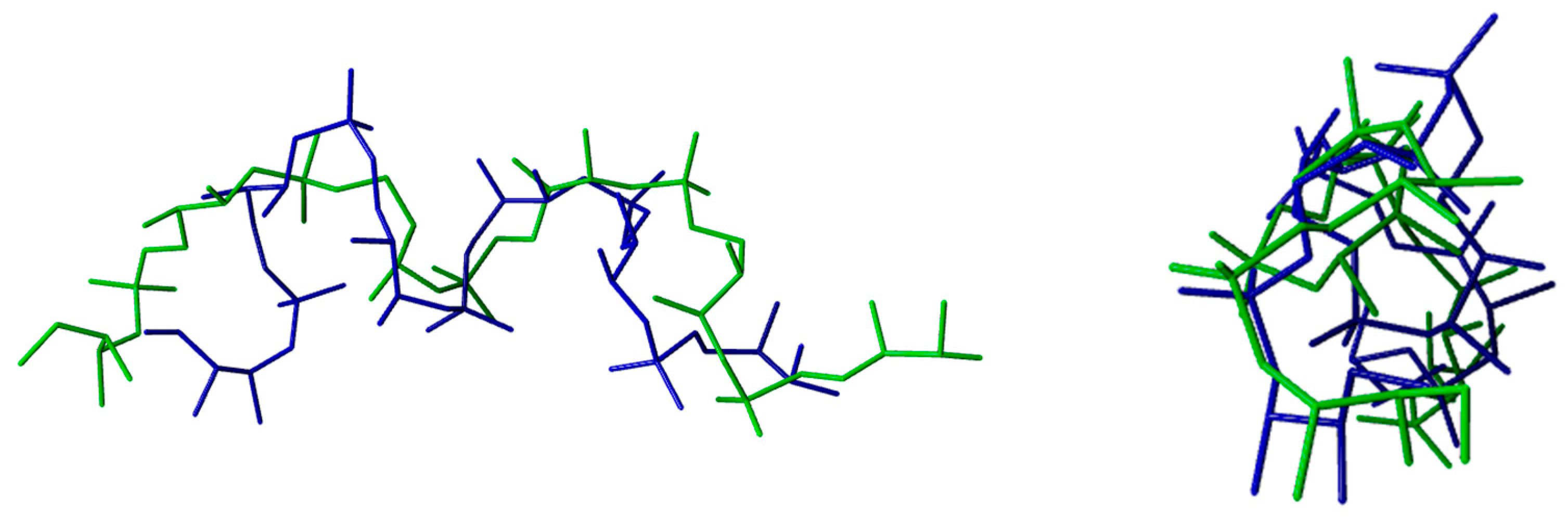
| DNA TERTIARY STRUCTURE | ||||||
|---|---|---|---|---|---|---|
| A-DNA | B-DNA | Z-DNA | ||||
| AT or TA | GC or CG | AT or TA | GC or CG | PURINES | PYRIMIDINES | |
| Equations to Calculate the Sugar-Phosphate Backbone Torsion Angles | α° = 360° − P (8) | α° = 360° − P (14) | α° = 360° − P (20) | α° = 360° − P (26) | α° = P (32) | α° = 360° − (P + H1 + H2 + H3) (38) |
| β° = P + π + H1 + H2 + H3 (9) | β° = P + π + H1 + H2 + H3 (15) | β° = π + H1 + H2 + 30° (21) | β° = π + H1 + H2 + H3 (27) | β° = (P/2) + π + H1 + H2 + H3 (33) | β° = P + π + H1 + H2 + H3 (39) | |
| γ° = π (10) | γ° = π (16) | γ° = π (22) | γ° = π (28) | γ° = (P/3) + π + H1 + H2 + H3 (34) | γ° = π (40) | |
| δ° = (P/2) + π (11) | δ° = (P/2) + π (17) | δ° = π + H1 + H2 + 30° (23) | δ° = π + H1 + H2 + H3 (29) | δ° = H1 + H2 + H3 (35) | δ° = π + H1 + H2 + H3 (41) | |
| ε° = P + π + H1 + H2 + 30° (12) | ε° = P + π + H1 + H2 + H3 (18) | ε° = 2P + π + H1 + H2 (24) | ε° = 2P + π + H1 + H2 + H3 (30) | ε° = P + π + H1 + H2 + H3 (36) | ε° = P + (2∙π) + H1 + H2 + H3 (42) | |
| ζ° = 360° − (H1 + H2) (13) | ζ° = 360° − (H1 + H2) (19) | ζ° = P + π + H1 + H2 (25) | ζ° = 360° − [(P/2) +π + H1 + H2 + H3] (31) | ζ° = 360° − π (37) | ζ° = (π + H1 + H2 + H3)/2 (43) | |
| A-DNA (PDB CODE: 440D) | ||||||
| Average Torsion Angles (°) | α | β | γ | δ | ε | ζ |
| Calculated | 295 ± 3 | 181 ± 4 | 50 ± 3 | 82 ± 3 | 213 ± 5 | 294 ± 1 |
| Experimental | 293 ± 17 | 174 ± 4 | 56 ± 14 | 81 ± 7 | 203 ± 12 | 289 ± 12 |
| BII-DNA (PDB CODE: 1AGH) | ||||||
| Average Torsion Angles (°) | α | β | γ | δ | ε | ζ |
| Calculated | 293 ± 3 | 146 ± 4 | 49 ± 4 | 146 ± 4 | 249 ± 5 | 180 ± 4 |
| Experimental | 298 ± 15 | 146 ± 8 | 48 ± 11 | 144 ± 7 | 246 ± 15 | 174 ± 14 |
| Z-DNA (PDB CODE: 1DCG)—PURINES | ||||||
| Average Torsion Angles (°) | α | β | γ | δ | ε | ζ |
| Calculated | 69 ± 3 | 191 ± 3 | 180 ± 2 | 99 ± 1 | 226 ± 4 | 302 ± 1 |
| Experimental | 71 ± 13 | 183 ± 9 | 179 ± 9 | 95 ± 8 | 240 ± 9 | 301 ±16 |
| Z-DNA (PDB CODE: 1DCG)—PYRIMIDINES | ||||||
| Average Torsion Angles (°) | α | β | γ | δ | ε | ζ |
| Calculated | 191 ± 2 | 219 ± 7 | 50 ± 7 | 150 ± 8 | 269 ± 14 | 75 ± 4 |
| Experimental | 201 ± 20 | 225 ± 16 | 54 ± 13 | 141 ± 8 | 267 ± 9 | 75 ± 9 |
| Chemical Interactions | Phosphate Groups Electrostatic Repulsion | Intra-Strand π-π Stacking | H-Bond 1 | H-Bond 2 | H-Bond 3 (Only for GC or CG Base Pairs) |
|---|---|---|---|---|---|
| Average Interatomic Distances (Å) for A-DNA Structure (PDB CODE: 440D) | 6.31 ± 0.81 | 4.76 ± 0.44 | 1.89 ± 0.09 | 1.89 ± 0.07 | 1.80 ± 0.12 |
| Average Interatomic Distances (Å) for BII-DNA Structure (PDB CODE: 1AGH) | 6.98 ± 0.79 | 4.65 ± 0.58 | 1.86 ± 0.10 | 1.85 ± 0.04 | 1.88 ± 0.06 |
| Average Interatomic Distances (Å) for Z-DNA Structure (PDB CODE: 1DCG): PURINES | 7.63 ± 1.03 | 6.37 ± 0.14 | 1.86 ± 0.04 | 1.92 ± 0.01 | 1.86 ± 0.05 |
| Average Interatomic Distances (Å) for Z-DNA Structure (PDB CODE: 1DCG): PYRIMIDINES | 7.52 ± 0.95 | 4.92 ± 1.35 | 1.85 ± 0.03 | 1.95 ± 0.04 | 1.93 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larocca, M.; Floresta, G.; Verderese, D.; Cilibrizzi, A. Main Mechanical Forces to Analyse the Chemical Interactions Shaping Backbone Torsion Angles in DNA Tertiary Structures. AppliedChem 2025, 5, 26. https://doi.org/10.3390/appliedchem5040026
Larocca M, Floresta G, Verderese D, Cilibrizzi A. Main Mechanical Forces to Analyse the Chemical Interactions Shaping Backbone Torsion Angles in DNA Tertiary Structures. AppliedChem. 2025; 5(4):26. https://doi.org/10.3390/appliedchem5040026
Chicago/Turabian StyleLarocca, Michele, Giuseppe Floresta, Daniele Verderese, and Agostino Cilibrizzi. 2025. "Main Mechanical Forces to Analyse the Chemical Interactions Shaping Backbone Torsion Angles in DNA Tertiary Structures" AppliedChem 5, no. 4: 26. https://doi.org/10.3390/appliedchem5040026
APA StyleLarocca, M., Floresta, G., Verderese, D., & Cilibrizzi, A. (2025). Main Mechanical Forces to Analyse the Chemical Interactions Shaping Backbone Torsion Angles in DNA Tertiary Structures. AppliedChem, 5(4), 26. https://doi.org/10.3390/appliedchem5040026








