Abstract
The aim of this review was to investigate the effectiveness of the Human Oral Microbe Identification Microarray (HOMIM) in identifying and quantifying bacterial species of the oral microbiome in periodontal disease. The search for articles was conducted in CENTRAL, CINAHL, MEDLINE and EMBASE by two reviewers, and included articles published in English between January 1990 and December 2021. The selected articles were human observational studies in adults between 18 and 65 years, presenting specific predefined keywords. Articles were initially selected by title and abstract; articles that met the inclusion criteria were analyzed for methodological quality using a detailed checklist for quality assessment. Data were extracted and reported using the PRISMA tool. The study design, sample, follow-up period, collection and microbial analysis methods, statistical treatment, results and discussion were quality assessed and risk of bias was evaluated using the Cochrane Risk-of-Bias tool. A narrative synthesis approach was used to synthesize and interpret the extracted data. From the initial search, 2931 articles were retrieved; 51 of these were then selected after screening by title and abstract. Subsequently, 8 articles met the inclusion after full-text reading and were classed according to methodological quality as high (2), moderate (3) or low (3). Studies included in this review were of high and medium quality. Data from the Human Oral Microbe Identification Microarray (HOMIM) provide much more robust results, showing major shifts between periodontal health and periodontal disease. Compared to earlier techniques such as Denaturing Gradient Gel Electrophoresis (DGGE), HOMIM represents a more effective approach for quantification due to its high sensitivity; thus, it is able to identify a high prevalence of periodontal pathogens and novel species in low abundance. The literature provides moderate evidence that the Human Oral Microbe Identification Microarray (HOMIM) is more effective in identifying and quantifying bacterial species of the oral microbiome in periodontal disease, compared to earlier molecular and non-molecular methods such as Denaturing Gradient Gel Electrophoresis (DGGE) and a culture-based approach with phenotypic tests.
1. Introduction
Periodontal disease is a multifactorial, chronic inflammatory disease of the mouth involving the gingiva, teeth and alveolar bone, initiated and sustained by an aberrant host immune response against resident bacterial biofilm on the teeth [1]. It is usually characterized by loss of connective tissue attached to the teeth and alveolar bone loss thus if left untreated can cause exfoliation of the tooth [2]. Gingivitis and periodontitis are the most frequent types of periodontal diseases; the former is characterized by inflammation confined to the gingiva and is reversible with good oral hygiene, while the latter is mostly irreversible and is usually characterized by extended inflammation, resulting in tissue destruction and alveolar bone resorption [3]. Furthermore, in periodontitis, periodontal pockets are formed between the gingiva and the tooth due to the breakdown of collagen fibers of the periodontal ligament as a result of the tissue destruction mentioned above [2]. Research shows that periodontitis is a highly prevalent disease globally, with its mildest form having a prevalence of 45–50% in adults and its most severe form estimated to affect 10–15% of adults in most populations [4]. Current research unequivocally confirms diabetes as a major risk factor for periodontitis, with the prevalence of periodontitis estimated to be two to three times higher in diabetics than in an otherwise healthy population [5].
The terms “oral microbiome”, “oral microflora” or “oral microbiota” are commonly used to describe the microbial community within the human oral cavity [6]. Research has identified over 700 bacteria species in the human oral cavity, with 400 identified from the periodontal pockets and 300 from other oral sites such as the tongue, oral mucous membranes and carious lesions [7]. These bacteria can be classified into different categories based on their Gram stain results (Gram-positive or Gram-negative bacteria), their shape (coccus, bacillus or spirochetes) and their tolerance to oxygen (aerobic, facultative anaerobes, microaerobic or obligate anaerobes) [8]. Some known oral pathogens are believed to contribute to the development of oral diseases such as dental caries and periodontal diseases [9], as well as systemic diseases such as Diabetes Mellitus, cardiovascular diseases and the development of tumors [6]. In healthy populations, organisms such as Streptococcus salivarius and Rothia mucilaginosa are usually predominant in the oral microbiome [10]. However, with the development of periodontitis, organisms such as Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola and Aggregatibacter actinomycetemcomitans become more dominant within the oral community [11].
The isolation and identification of oral organisms can be difficult because oral microorganisms are numerous and composed of diverse species and new genera and species are constantly being discovered, while the classification of some previously discovered species changes with time. Earlier methods such as Denaturing Gradient Gel Electrophoresis (DGGE) have been used to identify oral bacteria, and DGGE was first introduced to microbial ecology by a study in 1993 [12]. This technique separates short-to-medium-length Polymerase Chain Reaction (PCR)-amplified DNA fragments according to their melting point [13]. The technique works on the basis that DNA fragments of the same size but with differing base pair sequences can be separated, and this separation by DGGE relies on the electrophoretic mobility of partially denatured DNA molecules in a polyacrylamide gel [14]. Studies have, however, shown that current 16S rRNA sequence hybridization methods such as Human Oral Microbe Identification Microarray (HOMIM) are more effective in providing a comprehensive representation of the oral bacterial community [7,15,16,17]. HOMIM, first introduced in 2008, is a custom array-based approach that utilizes specially designed probes to detect over 300 of the most prevalent bacterial species [17]. Briefly, 16S rRNA-based oligonucleotide probes are covalently attached to aldehyde-coated slides [7]. The 16S rRNA gene are PCR amplified from bacterial DNA extracts and are labelled with fluorescent dye, producing a fluorescent signal when the bacterial DNA hybridizes to a specific spot on the slide [15]. The output data from the HOMIM assay are usually merged onto the Human Oral Microbiome Database (HOMD), which is based on a curated 16S rRNA gene-based provisional naming scheme that provides comprehensive information on the prokaryote species present in the human oral cavity [9].
This review will focus on investigating the effectiveness of the Human Oral Microbe Identification Microarray (HOMIM), a current molecular profiling technology used for identifying and quantifying bacterial species of the oral microbiome in periodontal disease. The objective of this study is to compare this current profiling technology to earlier identification methods such as Denaturing Gradient Gel Electrophoresis (DGGE).
2. Materials and Methods
The following databases were searched by two independent researchers (S.J., E.B.) from their January 1990 records through March 2021: MEDLINE via PubMed, EMBASE, The Cochrane Central Register of Controlled Trials (CENTRAL) and CINAHL. The strategy was developed for MEDLINE using controlled vocabulary, with words derived from “Medical Subject Headings” (MeSH) associated with free terms relevant to the topic in question (Table 1). The search strategy and the flow of information through the different phases of the systematic review were established according to the PRISMA statement for systematic reviews and metanalysis. In total, 18 articles were selected, evaluated and classified by two independent readers (S.J., E.A.); the full-text article was obtained whenever a study seemed to meet the inclusion criteria, but complete information was lacking. By using a pre-defined data-extraction form, the reviewers independently extracted the data on characteristics of the study population, length of follow-up, interventions and outcomes. The results of both readers were compared, and eventual differences were resolved by discussion.

Table 1.
Summary of keywords used for database search.
Initially, all articles were selected by title and abstract; articles with duplicate records were considered only once. The publications selected were essentially observational clinical studies conducted in humans. They were required to quantify and characterize the bacterial species and periodontal pathogens in the oral microbiome using HOMIM or DGGE. Afterwards, inclusion and exclusion criteria were applied. The following inclusion criteria were applied: observational clinical studies in humans aged between 18 and 65 years; presence of periodontal disease; DNA extraction of samples collected from the subgingival plaque in the mouth. Conversely, the exclusion criteria were inclusion of humans under 18 years or over 70 years; inclusion of patients with systemic diseases or under any condition that could influence oral microbiota or periodontal support tissues; antibiotic therapy three months before and during the study; absence of periodontal disease; studies written in any other language than English. At this stage, if articles did not meet any exclusion criteria but met part of the inclusion criteria, they were included.
Subsequently, full text articles were read and those that met the inclusion and exclusion criteria were carefully analyzed and qualified according to their methodological aspects, as described in Table 2. A detailed checklist for quality assessment was adapted for this review, based on the study design, sample, follow-up period, collection and DNA extraction methods, statistical treatment, results and discussion [18]. The selected articles were finally classified according to the total score after qualification. Their methodological quality was classified as high (score 7 to 8), moderate (score from 5 to 6.9) or low (score from 0 to 4.9) (Table 3). Those classified as low were excluded. A hand search was performed to complement the previous searches, by which the references of the selected articles were analyzed. The Cochrane Collaboration tool for assessing risk of bias was used in the included studies [19]. The following domains were evaluated as having low, high or unclear risk of bias: random sequence generation, allocation concealment, blinding, incomplete outcome data, selective outcome reporting and other potential bias.

Table 2.
Methodological quality score [18].

Table 3.
Quality assessment of studies.
A narrative synthesis was used to systematically explore the similarities and differences between results of different studies, identifying data patterns in associations between study characteristics and outcomes. Furthermore, different interventions, outcomes, study designs and the rationale of their effects were explored and analyzed to integrate and synthesize data.
3. Results
The electronic search retrieved a total of 2931 records from the following databases: CENTRAL (224), CINAHL (0), MEDLINE (2704) and EMBASE (3). After excluding duplicates and records marked as ineligible by automation tools, there were 261 records, 230 of which did not meet the inclusion criteria; 31 were selected for full reading. A manual search of the references from the 31 articles was performed, retrieving an additional 20 new titles. Titles and abstracts not related to the topic were initially excluded. Having selected articles based on the inclusion and exclusion criteria, a total of 51 full text articles have been assessed for eligibility according to all selection criteria; 43 studies were excluded at this stage. After careful reading and quality assessment, 8 articles were categorized according to methodological quality as follows: high (2), moderate (3), low (3). Low-quality articles were excluded from this systematic review. Consequently, 5 studies were included (Figure 1). The general characteristics of the included studies are presented in Table 4. Furthermore, risk of bias for the included articles appeared to be low.
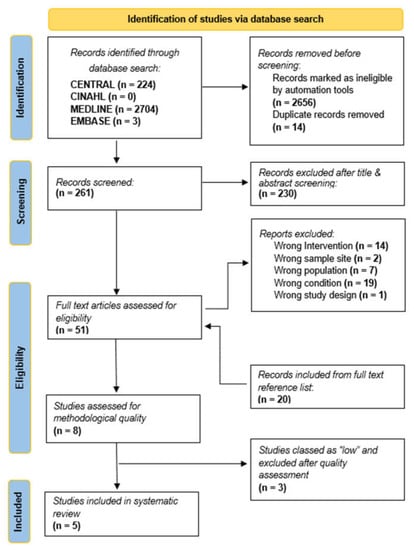
Figure 1.
PRISMA flow chart.

Table 4.
Study characteristics and results of individual studies.
4. Discussion
The oral cavity is colonized by a large and diverse group of bacteria, which form biofilm communities in several habitats within the mouth, including the tooth, subgingival sulcus, tongue, buccal mucosa and tonsils [22,26]. More than half of these species have been detected in the subgingival habitat, many of which have not yet been formally named, or are novel species that cannot yet be grown or are difficult to grow in culture [25,26,28]. Although most of these organisms are commensal, numerous oral bacterial species have been associated with oral disease and oral health, including those that cannot be cultivated in vitro [25,28].
Various microbiological studies presented in the retrieved articles revealed different aspects of the supragingival plague and important changes in the subgingival plaque of patients with periodontal disease [26]. Although the articles included in this review reported that HOMIM was an effective method for quantifying the oral community in periodontal disease, it is relevant to indicate that insufficient methodological information was provided. For instance, only two publications outlined the design and ethical information of the study [25,26]. However, in all included studies, collection of the biological material was carefully monitored to avoid interference from other sites in the mouth [20,22,25,26]. Methodologically poor articles [21,24,27] were characterized by similarities that contributed to their low score and then exclusion from the review. These studies did not describe the control of factors that might influence the collection of biological material, which is an essential aspect of the study. As diverse organisms associated with periodontal disease and periodontal health are characteristic of different oral habitats, isolation of the area and previous removal of other biological material surrounding the collection site is essential [13,20,25,26].
Periodontal health is usually associated with supragingival Gram-positive microbiota that consists mainly of diverse species of Streptococci and Actinomyces [25,26]. Moreover, they are also predominant in gingivitis; however, the number of Gram-negative bacteria, such as Fusobacterium and Bacteroides, increases [6]. On the other hand, in periodontitis, the microflora is dominated by Gram-negative anaerobes, with increased spirochetes [25,26].The Human Oral Microbe Identification Microarray (HOMIM) provides a semi-quantitative identification of oral microbiome bacterial species [24,25,26] Earlier methods such as Denaturing Gradient Gel Electrophoresis (DGGE) have been used to study microbial population dynamics in periodontal disease [20,22,28,29,30]. This molecular technique, while useful for analyzing bacterial communities and studying shifts in microbial composition at a population level, is often limited by numerous factors, particularly low detection limit and difficulties associated with species identification based on gel positioning [20,22,28,29,30].
For this review, included articles were classified as having high to moderate methodological quality [20,22,23,25,26]. The studies classed as having moderate methodological quality reported on the use of Denaturing Gradient Gel Electrophoresis to provide a visual representation of bacterial diversity in periodontal disease. The first study [20] aimed to examine the bacterial community of the subgingival plaque using DGGE. Samples were collected from the subgingival plaque of four patients using sterile paper points, and DNA was extracted using the InstaGene Matrix kit. Furthermore, Polymerase Chain Reaction (PCR) was performed using specifically designed primers and genomic DNAs of typical periodontal bacteria. The generated 16S rDNA fragments were separated by denaturing gel. Results indicated that DGGE was able to show distinct banding patterns observed among several samples from identical subjects, but the bands were not always observed at the species-specific positions of periodontal bacteria. The authors in this study highlight the difficulty of the DGGE method to detect bacteria with a low abundance in various samples. This method of bacterial identification assumes that DNA is extracted equally from all bacterial species. Moreover, its reliability depends on the quality and reproducibility of bacteria sample processing and DNA extraction. Thus, any organisms forming < 1% of the microbiota may not be represented. This limitation is widely reported in other studies that have used this method to analyze subgingival microbiota in health and disease [28,29,30].
Similarly, in the second study [22], the authors highlight the limitations of DGGE due to its low sensitivity and difficulties in identifying species based on their position in the gel. Their research aimed to study the microbial population dynamics in the subgingival pocket of 15 subjects with untreated advanced periodontitis (n = 9) or periodontal health (n = 6). After the supragingival plaque was removed, subgingival plaque was collected by inserting sterile endodontic absorbing points. DNA extraction was carried out using the phenol/chloroform method and part of the bacterial 16S rRNA was PCR amplified and separated by DGGE. Samples were evaluated at baseline, 1 day after and 3 months after treatment; DGGE banding profile showed that treatment resulted in a decrease in the diversity of the population. DGGE results also indicated that after 3 months, a microbial population 33–47% different from the population before treatment had re-established, highlighting shifts in composition and diversity in the microbial population. Despite DGGE’s effectiveness in providing a fingerprint representative of the microbial flora, the culture-independent, PCR-based method can only detect up to 30–40 bands, thus presenting with low resolution and sensitivity issues as reported in another study evaluating changes in oral bacterial composition [28]. Due to the limitations of this method, most studies simply report the relative position of amplicons, while others provide an estimation of denaturing percentage for comparisons due to the limited number of computer programs capable of acquiring DGGE gel images, transferring them to specially designed analytical software and recording the banding patterns [28].
The third study, [23] similar to the previous two studies, confirms the limitations of DGGE, particularly with its inability to detect bacteria below a certain threshold due to its low sensitivity. The authors of this study aimed to investigate the microbial population in the subgingival community, using DGGE. Specifically, the study examined whether primer choice affected DGGE results and assessed the most appropriate primer pairs for DGGE analysis. Firstly, the authors analyzed the DGGE profiles of different 16S rDNA regions of three periodontal strains (P. gingivalis ATCC 33277, F. nucleatum ATCC 25586 and P. nigrescens ATCC 33563) using the target regions (V3, V3–V5 and V6–V8). These regions were cloned into plasmid vectors and the constructed plasmids was used as templates for PCR-DGGE analysis templates in the study. Moreover, the study included non-smoking adults with chronic periodontitis (n = 6), between the ages of 29 and 52 years. Subgingival samples were collected from periodontal pockets using sterile curettes with a probing depth and clinical attachment loss of more than 5 mm at baseline after removal of supragingival plaque. Following mechanical debridement, patients were examined six weeks later and their periodontium was found to have improved significantly. Again, subgingival plaque was sampled from the same pockets (the probing depth was decreased by 2 or 3 mm). Using a bacterial genomic DNA extraction kit (TIANGEN), microbial DNA was extracted and used for PCR amplification of the target fragments. The results suggested that V3–V5 and V6–V8 fragments may be suitable for community analysis of subgingival bacteria; however, it was concluded that 16S rDNA of the V3 region may cause over-estimation of subgingival bacterial populations in DGGE analysis due to multiple banding patterns. Further analysis with the V3–V5 and V6–V8 fragments suggested that, in chronic periodontitis, periodontal bacteria may recolonize within 6 weeks after mechanical debridement with a population very similar to the baseline as there were no significant differences in banding patterns between the two groups. However, these changes would not be identified using DGGE, possibly due to its low sensitivity; thus, there is a need for further analysis with quantitative methods [23]. The authors were successful in identifying the potential of some targeted regions of 16S rDNA for DGGE analysis, but they highlight the necessity for careful consideration of the regions used in the analysis as it is currently impossible to predict which regions would yield different results for species identification in DGGE analysis of the same sample. This, in itself, poses another limitation for DGGE as there is potential for different primers to affect the results generated, as well as certain regions making it difficult to estimate bacterial population due to multiple-band appearance for a single pathogenic bacterium.
Only two studies which described the study design and ethics were classed as having high methodological quality. These studies [25,26] were conducted by the same research group; however, they presented different objectives and distinct samples. Both studies showed evidence of controlled microbial collection by prior removal of the supragingival plaque with a sterile gauze to preserve sample quality. Meanwhile, subgingival biofilm samples were collected using sterile periodontal curettes. For DNA extraction, samples were placed in separate 1.5 mL tubes containing 50 μL of TE (50 mM Tris-HCl, 1 mM EDTA, pH 7.6), 44 μL of each sample was then taken and mixed with 0.5% Tween 20 and 1 μL of Proteinase K (10 mg/mL).
Colombo et al. [25] carried out a study in 2009, which aimed to analyze and compare the baseline subgingival microbiota of subjects with refractory periodontitis (RP) to those in subjects with treatable periodontitis (GR) or periodontal health (PH) using the Human Oral Microbe Identification Microarray (HOMIM). A total of 67 subjects were measured at baseline (17 RP individuals, 30 GR individuals and 20 PH individuals), and analyzed for the presence of 300 bacterial species using HOMIM. Results indicated a distinct microbial profile in RP patients compared to patients in GR and PH groups. In addition, more species were detected in diseased patients (GR or RP) than those without disease (PH). The authors report that the HOMIM technique allowed for the detection of about 300 species, including cultivable and not-yet cultivable species. Moreover, in periodontal sites losing attachment, HOMIM effectively identified a high prevalence of periodontal pathogens and novel species in low abundance, particularly S. intermedius/constellatus, S. anginosus, P. micra, Selenomonas spp., S. parasanguinis, Streptococcus sp. OT 070/071, F. alocis, D. invisus, D. pneumosintes, C. rectus/concisus, TM7 spp. OT 346/356/437, Treponema socranskii, Treponema maltophilum, Bacteroidetes sp. OT 274/272, Prevotella tannerae, T. forsythia, Eubacterium spp., G. sanguinis, Porphyromonas endodontalis, Peptostreptococcus sp. OT 113, Desulfobulbus sp. OT 041, P. stomatis, S. moorei, Sphaerocytophaga sp. OT 337, P. gingivalis, Megashaera micronuciformis, S. satelles, Prevotella oralis, Mogibacterium timidum, Anaerococcus geminatus, Atopobium rimae, Atopobium parvulum and P. alactolyticus.
In the last study included in this review [22], the authors compared the changes in the subgingival microbiota of 47 subjects with refractory periodontitis (RP) (n = 17) or treatable periodontitis (GR) (n = 30) before and after periodontal therapy using HOMIM. Subgingival plaque samples were taken at baseline and 15 months, the HOMIM technique was used to analyze the samples for the presence of over 300 species. HOMIM results indicated that the majority of the evaluated species decreased in prevalence in both groups after treatment. However, only a small subset of organisms was affected significantly. Furthermore, HOMIM data identified several species that increased or persisted in high frequency in RP but reduced significantly in GR, including Bacteroidetes sp., Porphyromonas endodontalis, Porphyromonas gingivalis, Prevotella spp., Tannerella forsythia, Dialister spp., Selenomonas spp., Catonella morbi, Eubacterium spp., Filifactor alocis, Parvimonas micra, Peptostreptococcus sp. OT113, Fusobacterium sp. OT203, Pseudoramibacter alactolyticus, Streptococcus intermedius or Streptococcus constellatus, and Shuttlesworthia satelles. Furthermore, HOMIM analysis was effective in identifying novel species in subjects with RP, suggesting that different microbial profiles, including not only combinations of known species but also novel species, and consequently, significant differences in treatment response.
The results presented in this systematic review indicate that there is moderate scientific evidence that the HOMIM is significantly effective in identifying periodontal pathogens of the oral microbiome. For the HOMIM analysis in both studies [25,26], a total of 400 16S rRNA-based, reverse capture oligonucleotide probes printed on aldehyde-coated slides were used to target over 300 bacterial taxa, suggesting a potential limitation of the technology, as it is possible that HOMIM is only able to recognize bacterial taxa/clusters that have a target probe present on the microarray slides. However, refinement of the HOMIM technology will provide a better identification tool for the oral microbiome and will be beneficial to the understanding of periodontal pathogens in the oral cavity. Moreover, the evidence presented in this review has identified the main limitation for DGGE as its inability to detect species when abundance is below a certain level. Due to the importance of these low-abundance species, a more comprehensive technology such as HOMIM is best to understand the complexity of the periodontal disease process and probable multifactorial etiology.
5. Conclusions
To our knowledge, this is the first systematic review regarding the significance and effectiveness of HOMIM in quantifying the human oral microbiome. A search of the literature found moderate evidence that the Human Oral Microbe Identification Microarray (HOMIM) is significantly effective in identifying and quantifying bacterial species of the oral microbiome in periodontal disease. Despite the limitations associated with this current molecular profiling technology, HOMIM expanded oral bacterial species identification compared to earlier methods such as DGGE. Furthermore, the species probes utilized in HOMIM provided a more comprehensive representation of the oral bacterial community, critical for future characterization of oral microbes in periodontal disease. It is important for future work to explore the effectiveness of Human Oral Microbe Identification using Next-Generation Sequencing (HOMINGS), a successor technology of HOMIM, for identifying and quantifying the human oral microbiome.
Author Contributions
S.U.: Database search, paper screening, quality assessment, data synthesis, contribution to abstract, main body, results and discussion. E.A.: Article screening, quality assessment, data synthesis. S.S.: Contribution to main body and discussion. A.V.: Review, edit and contribution to abstract, main body and discussion. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Uzoma Igwe (Department of Applied Science, University of the West of England) for her assistance as an independent reviewer for this paper.
Conflicts of Interest
The authors report no commercial, proprietary or financial interest in the products or companies described in this article.
Abbreviations
The following abbreviations are used in this manuscript:
| DGGE | Denaturing Gradient Gel Electrophoresis |
| HOMD | Human Oral Microbe Database |
| HOMIM | Human Oral Microbe Identification Microarray |
| HOMINGS | Human Oral Microbe Identification using Next-Generation Sequencing |
| MESH | Medical Subject Headings |
| PH | Periodontal Health |
| PCR | Polymerase Chain Reaction |
| RP | Refractory Periodontitis |
References
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial population by DGGE analysis of polymerase chain reaction amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 1993, 62, 2676–2680. [Google Scholar]
- Nazir, M.A. Prevalence of periodontal disease, its association with systemic diseases and prevention. Int. J. Health Sci. 2017, 11, 72–80. [Google Scholar]
- Casanova, L.; Hughes, F.J.; Preshaw, P.M. Diabetes and periodontal disease: A two-way relationship. Br. Dent. J. 2014, 217, 433–437. [Google Scholar] [CrossRef]
- Persson, G.R. Diabetes and periodontal disease: An update for health care providers. Diabetes Spectr. 2011, 24, 195–198. [Google Scholar] [CrossRef]
- Sanz, M.; Ceriello, A.; Buysschaert, M.; Chapple, I.; Demmer, R.T.; Graziani, F.; Herrera, D.; Jepsen, S.; Lione, L.; Madianos, P.; et al. Scientific evidence on the links between periodontal diseases and diabetes: Consensus report and guidelines of the joint workshop on periodontal diseases and diabetes by the International Diabetes Federation and the European Federation of Periodontology. J. Clin. Periodontol. 2018, 45, 138–149. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol. 2015, 60, 69–80. [Google Scholar] [CrossRef]
- Lourenço, T.G.B.; Heller, D.; Silva-Boghossian, C.M.; Cotton, S.L.; Paster, B.J.; Colombo, A.P.V. Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 2015, 41, 1027–1036. [Google Scholar] [CrossRef]
- Teeuw, W.J.; Kosho, M.X.; Poland, D.C.; Gerdes, V.E.; Loos, B.G. Periodontitis as a possible early sign of diabetes mellitus. BMJ Open Diabetes Res. Care 2017, 5, e000326. [Google Scholar] [CrossRef]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell. 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Yu, G.; Dye, B.A.; Gail, M.H.; Shi, J.; Klepac-Ceraj, V.; Paster, B.J.; Wang, G.Q.; Wei, W.Q.; Fan, J.H.; Qiao, Y.L.; et al. The association between the upper digestive tract microbiota by HOMIM and oral health in a population-based study in Linxian, China. BMC Public Health 2014, 14, 1–7. [Google Scholar] [CrossRef]
- Preshaw, P.M.; Alba, A.L.; Herrera, D.; Jepsen, S.; Konstantinidis, A.; Makrilakis, K.; Taylor, R. Periodontitis and diabetes: A two-way relationship. Diabetologia 2012, 55, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS. Med. 2019, 6, e1000097. [Google Scholar]
- Gafan, G.P.; Spratt, D.A. Denaturing gradient gel electrophoresis gel expansion (DGGEGE)–an attempt to resolve the limitations of co-migration in the DGGE of complex polymicrobial communities. FEMS Microbiol. Lett. 2005, 253, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Gafan, G.P.; Lucas, V.S.; Roberts, G.J.; Petrie, A.; Wilson, M.; Spratt, D.A. Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J. Clin. Microbiol. 2005, 43, 3971–3978. [Google Scholar] [CrossRef] [PubMed]
- Seerangaiyan, K.; Jüch, F.; Winkel, E.G. Tongue coating: Its characteristics and role in intra-oral halitosis and general health—A review. J. Breath. Res. 2018, 12, 034001. [Google Scholar] [CrossRef]
- Heller, D.; Helmerhorst, E.J.; Gower, A.C.; Siqueira, W.L.; Paster, B.J.; Oppenheim, F.G. Microbial diversity in the early in vivo-formed dental biofilm. Appl. Environ. Microbiol. 2016, 82, 1881–1888. [Google Scholar] [CrossRef]
- Ahn, J.; Yang, L.; Paster, B.J.; Ganly, I.; Morris, L.; Pei, Z.; Hayes, R.B. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS. ONE 2011, 6, e22788. [Google Scholar] [CrossRef]
- Freitas, A.O.A.D.; Marquezan, M.; Nojima, M.D.C.G.; Alviano, D.S.; Maia, L.C. The influence of orthodontic fixed appliances on the oral microbiota: A systematic review. Dental. Press. J. Orthod. 2014, 19, 46–55. [Google Scholar] [CrossRef]
- Higgins, J.; Green, S. Cochrane Handbook for Systematic Reviews of Interventions, The Cochrane Collaboration; Available online: https://training.cochrane.org/handbook (accessed on 3 July 2022).
- Fujimoto, C.; Maeda, H.; Kokeguchi, S.; Takashiba, S.; Nishimura, F.; Arai, H.; Fukui, K.; Murayama, Y. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J. Periodontal Res. 2003, 38, 440–445. [Google Scholar] [CrossRef]
- Zijnge, V.; Welling, G.W.; Degener, J.E.; van Winkelhoff, A.J.; Abbas, F.; Harmsen, H.J. Denaturing gradient gel electrophoresis as a diagnostic tool in periodontal microbiology. J. Clin. Microbiol. 2006, 44, 3628–3633. [Google Scholar] [CrossRef]
- Zijnge, V.; Harmsen, H.J.M.; Kleinfelder, J.W.; Van Der Rest, M.E.; Degener, J.E.; Welling, G.W. Denaturing gradient gel electrophoresis analysis to study bacterial community structure in pockets of periodontitis patients. Oral Microbiol. Immunol. 2003, 18, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhou, Y.; Li, C.; Li, Y.; Jiang, Y.; Huang, Z.; Liang, J.; Shu, R. Denaturing gradient gel electrophoresis analysis with different primers of subgingival bacterial communities under mechanical debridement. Microbiol. Immunol. 2010, 54, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Mougeot, J.L.C.; Stevens, C.B.; Cotton, S.L.; Morton, D.S.; Krishnan, K.; Brennan, M.T.; Lockhart, P.B.; Paster, B.J.; Bahrani Mougeot, F.K. Concordance of HOMIM and HOMINGS technologies in the microbiome analysis of clinical samples. J. Oral Microbiol. 2016, 8, 30379. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.P.V.; Boches, S.K.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.; et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodonl. 2009, 80, 1421–1432. [Google Scholar] [CrossRef]
- Colombo, A.P.V.; Bennet, S.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.E.; et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: Comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J. Periodontol. 2012, 83, 1279–1287. [Google Scholar] [CrossRef]
- Aspiras, M.B.; Barros, S.P.; Moss, K.L.; Barrow, D.A.; Phillips, S.T.; Mendoza, L.; de Jager, M.; Ward, M.; Offenbacher, S. Clinical and subclinical effects of power brushing following experimental induction of biofilm overgrowth in subjects representing a spectrum of periodontal disease. J. Clin. Periodontol. 2013, 40, 1118–1125. [Google Scholar] [CrossRef]
- Zhou, C.; Trivedi, H.M.; Chhun, N.; Barnes, V.M.; Saxena, D.; Tao, X.; Yihong, L. Using DGGE and 16S rRNA gene sequence analysis to evaluate changes in oral bacterial composition. Chin. J. Dent. Res. 2011, 14, 95–103. [Google Scholar]
- Ireland, A.J.; Soro, V.; Sprague, S.V.; Harradine, N.W.T.; Day, C.; Al-Anezi, S.; Jenkinson, H.F.; Sherriff, M.; Dymock, D.; Sandy, J.R. The effects of different orthodontic appliances upon microbial communities. Orthod. Craniofac. Res. 2014, 17, 115–123. [Google Scholar] [CrossRef]
- Ledder, R.G.; Gilbert, P.; Huws, S.A.; Aarons, L.; Ashley, M.P.; Hull, P.S.; McBain, A.J. Molecular analysis of the subgingival microbiota in health and disease. App. Environ. Microbiol. 2007, 73, 516–523. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).