Abstract
Drought stress negatively affects plant growth and development, thus reducing plant productivity. Therefore, understanding the molecular mechanisms underlying drought stress responses is essential for crop improvement. The plant-specific NAM/ATAF1,2/CUC2 (NAC) transcription factors play important roles in the drought stress response. Here, we show that rice (Oryza sativa) ONAC054, a membrane-bound NAC transcription factor, is involved in the drought stress response. We found that onac054 mutants were sensitive, whereas ONAC054-overexpressing (ONAC054-OX) plants were tolerant to drought stress. Under drought stress conditions, several genes associated with abscisic acid (ABA) synthesis and signaling were downregulated in onac054 mutants but upregulated in ONAC054-OX plants. Among these genes, the TRANSCRIPTION FACTOR RESPONSIBLE FOR ABA REGULATION 1 (TRAB1), which encodes an ABA-inducible bZIP transcription factor, was directly activated by ONAC054. On the other hand, the expression of ONAC054 was directly activated by several ABA-responsive elements (ABRE)-binding factors (ABFs) in an ABA-dependent manner, indicating that ONAC054 acts as an enhancer of ABA-induced drought stress tolerance. Additionally, the overexpression of ONAC054 in rice greatly improved grain yield under drought stress conditions, indicating that the overexpression of ONAC054 could facilitate the improvement of drought stress tolerance in rice and other crops.
1. Introduction
In an agricultural field and a natural ecosystem, environmental stresses such as salinity, extreme temperatures, drought, and nutrient deficiency seriously threaten plant health by negatively affecting plant survival, biomass production, and crop yield, thus reducing the average yield of most major crop plants by more than 50% worldwide [1,2]. Since the area of water deficiency-affected land is dramatically increasing across the world, plant breeding has emerged as one of the most effective methods for the development of drought-tolerant crop plants. Extensive research conducted to date shows that plants employ highly complex adaptation mechanisms, intricately regulated by multiple genes, proteins, and metabolites, to survive under water deficit conditions [3]. For instance, drought stress triggers the accumulation of phytohormones, such as abscisic acid (ABA), jasmonic acid (JA), and salicylic acid (SA), and other metabolites, including sugar derivatives and amino acids (such as proline and glutamine) in plants, which facilitates their acclimation to drought stress [4,5,6]. Furthermore, drought stress modulates the expression of genes associated with drought stress responses [7], and a number of studies show that altering the expression or activity of such drought stress-responsive genes can lead to the development of drought-tolerant plants [3,8]. For instance, some ABA-inducible bZIP transcription factors (TFs), called ABA-responsive element (ABRE)-binding factors (ABFs), interact with the ABRE motif in the promoters of downstream genes to induce their expression [9]. In Arabidopsis thaliana, dehydration and/or ABA treatment strongly upregulated three ABF genes: ABF2/ABRE-BINDING PROTEIN 1 (AREB1), ABF3, and ABF4/AREB2 [10]. In addition, transgenic Arabidopsis plants overexpressing these ABF genes exhibited greater tolerance to drought stress than the wild type [11,12].
The NAM/ATAF1,2/CUC2 (NAC) proteins comprise one of the largest families of plant-specific TFs, encoded by 106 and 149 genes in the model plant species Arabidopsis and rice (Oryza sativa), respectively [13,14]. NAC proteins harbor a highly conserved DNA-binding domain, termed the NAC domain, in their N-terminal region [15]. However, the C-terminal region of NAC family proteins is diverse, suggesting that the C-terminal region plays a critical role in determining the cis-element-specificity of NACs in their respective target genes. Furthermore, some NAC TFs harbor a transmembrane domain (TMD) in their C-terminal region, which determines their subcellular localization. Under moderate growth conditions, TMD-containing NAC TFs localize to the endoplasmic reticulum (ER); however, under specific stress conditions, the TMD is cleaved off, which allows the NAC TFs to translocate to the nucleus. Thus, the NAC proteins exhibit conditional TF activity [16,17].
NAC TFs are involved in multiple developmental processes, such as shoot meristem development [18], lateral root formation [19], phytohormone signaling [19,20], and leaf senescence [21,22]. In addition, several NAC TFs in Arabidopsis and crop plants are involved in drought stress responses. Three Arabidopsis NAC genes, ANAC019, ANAC055, and ANAC072, were induced by the drought stress treatment, and transgenic plants overexpressing these NAC genes exhibited increased tolerance to drought stress, indicating that these three NAC TFs act as enhancers of drought stress responses [23]. The Arabidopsis NAC TF ANAC096 also positively regulates the drought stress response by specifically binding to the ABRE motif [12] in the promoters of several drought stress-responsive genes [24]. Similarly, in rice, overexpression of several NAC genes, including STRESS-RESPONSIVE NAC1, ONAC045, and ONAC066, which have been identified as positive regulators of drought stress responses, improved drought stress tolerance [25,26,27]. Several Arabidopsis TMD-containing NAC TFs, such as ANAC016, are also involved in drought stress responses; anac016 knockout mutants exhibited tolerance to drought stress, whereas ANAC016-overexpressing transgenic plants were sensitive to drought stress [28]. Under drought stress conditions, TMD cleavage allows ANAC016 to translocate to the nucleus, where it directly represses the transcription of AREB1 [28], which encodes a key TF involved in the stress-responsive ABA signaling pathway [12]. In Arabidopsis, other TMD-containing NAC TFs, including NAC WITH TRANSMEMBRANE MOTIF 1-LIKE4 (NTL4) and NTL6, also act as positive and negative regulators of drought stress responses [29,30]. Together, these studies indicate that TMD-containing NAC TFs play crucial roles in drought stress responses. Nevertheless, the functions of TMD-containing NAC TFs in crop plants are not yet fully understood.
We previously showed that a rice TMD-containing NAC TF, ONAC054, plays an important role in the regulation of ABA-induced leaf senescence. We reported that the nuclear import of ONAC054 requires the ABA-dependent cleavage of its C-terminal TMD [31]. Once in the nucleus, ONAC054 directly activates the transcription of ABA INSENSITIVE5 (OsABI5), encoding a key TF involved in ABA-mediated leaf senescence [32], and NON-YELLOWING COLORING1, encoding a chlorophyll catabolic enzyme [33]. Furthermore, the ONAC054 transcript (termed ONAC054α) has an alternatively spliced variant, ONAC054β, which carries a 7-nt insertion in-between the sequences corresponding to intron 5 and exon 6 of the ONAC054 gene. The insertion of these 7-nt creates a premature stop codon in the deduced amino acid sequence, resulting in the production of the TMD-lacking ONAC054β protein [31]. While both ONAC054α and ONAC054β are strongly induced by ABA, the transcript level of ONAC054β increases at a much faster pace than that of ONAC054α, indicating that the two ONAC054 isoforms form an intricate regulatory cascade to control the induction of ABA-induced leaf senescence.
In this study, we show that ONAC054 also participates in drought stress responses by modulating the expression of a number of genes associated with ABA metabolism and signaling. Additionally, a subset of rice ABF TFs, including OsbZIP23, OsABF2, OsABF4, and TRAB1, could bind to the ABRE sequence in the ONAC054 gene promoter and directly activate its transcription. Furthermore, overexpression of ONAC054α and ONAC054β greatly improved rice grain yield under drought stress conditions. Finally, we discuss the significance of the OsABF–ONAC054 regulatory module in the drought stress response in rice.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Plants of onac054 T-DNA insertion knockout mutant lines (onac054-1, PFG_3A-07241; onac054-2, PFG_3A-07240), ONAC054-overexpressing transgenic lines (ONAC054α-OX and ONAC054β-OX), and parental wild-type (WT) japonica rice (Oryza sativa) cultivar Dongjin were grown in a growth chamber at 28 °C/25 °C day/night temperature under long-day (LD; 16 h light/8 h dark) photoperiod and cool-white fluorescent light (400 μmol m−2 s−1). Onac054 knockout (onac054-KO) mutant lines, which were obtained from the Crop Biotech Institute at Kyung Hee University (South Korea), and ONAC054-OX lines have been described previously [31].
2.2. Abiotic Stress Treatments
Rice seedlings were initially grown in soil (Honenagri Co., Ltd., Niigata, Japan) in a growth chamber for 14 days under LD conditions. Then, seedlings were subjected to dehydration or treated with 200 mM PEG6000 (Sigma-Aldrich, St. Louis, MO, USA) for 5 days. During the 5 days of dehydration stress, leaves were collected on the indicated days and used for further experiments. Subsequently, the seedlings were rehydrated, and their survival rate was determined 3 days after rehydration.
2.3. Measurement of Physiological Traits
The ion leakage rate of seedlings was measured as described previously [34]. In brief, three 1 cm2 leaf discs from each treatment were immersed in 6 mL of 0.4 M mannitol at room temperature for 3 h with gentle shaking. The initial conductivity of the solution was measured with a conductivity meter (CON6METER). Then, the samples were incubated at 85 °C for 20 min. The ion leakage rate was calculated by dividing the initial conductivity by the total conductivity and expressed as a percentage.
The malondialdehyde (MDA) content of seedlings was determined as described previously [35].
2.4. RNA Extraction and Gene Expression Analysis
Total RNA was isolated from rice seedlings using the ISOSPIN Plant RNA Kit (Nippon Gene, Tokyo, Japan) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 1 μg of total RNA using SuperScriptTM II reverse transcriptase and oligo(dT)15 primers (Invitrogen, Waltham, MA, USA) in a 20 μL reaction mixture. Then, quantitative PCR with reverse transcription (RT-qPCR) was performed on the StepOnePlus System (Applied Biosystems, Waltham, MA, USA), with each reaction containing 1 μL of reverse transcriptase, 5 μL of 2× KAPA SYBR Fast qPCR mixture (KAPA biosystems, Waltham, MA, USA), and 0.25 mM of forward and reverse gene-specific primers (Table S1). Transcript levels of each gene were normalized relative to those of UBIQUITIN 5 (OsUBQ5).
2.5. Chromatin Immunoprecipitation (ChIP) Assays
The 35S:GFP-ONAC054α and 35S:GFP-ONAC054β plasmids, previously constructed in the pMDC43 binary vector [36], were transfected into rice protoplasts, as described previously [37]. The transfected protoplasts were then subjected to crosslinking for 20 min in 1% (v/v) formaldehyde under a vacuum. The chromatin complexes were isolated and sonicated, as previously described [38], with slight modifications. After crosslinking and nuclei isolation/lysis, DNA was subjected to ten 30 s sonication pulses using BIORUPTOR II (Cosmo Bio, Tokyo, Japan) at high power mode, with a 30 s interval between two pulses. Protein G agarose conjugated with anti-Myc polyclonal antibody (Millipore, Waltham, MA, USA) was used for immunoprecipitation. DNA recovered from agarose beads was purified using the DNeasy Plant Mini Kit (Qiagen, Hilden, The Netherlands). Then, quantitative PCR (qPCR) was performed using sequence-specific primers (Table S1), as described above.
2.6. Protoplast Transient Expression Assay
To construct reporter plasmids, the promoter fragment of TRAB1 (−2000 to −1 bp, relative to the translation start site) was cloned into the pJD301 vector upstream of the luciferase (LUC) reporter gene [39]. To construct effector plasmids, the cDNAs of OsbZIP23, OsABF2, OsABF4, TRAB1, and OsABI5 were first individually cloned into the pCR8/GW/TOPO Gateway vector (Invitrogen, Waltham, MA, USA), and then cloned upstream of a sequence encoding four copies of the MYC epitope tag in the pGWB17 binary vector [40]. The reporter (4 μg) and effector plasmids (8 μg) were co-transfected into 5 × 104 rice protoplasts by the polyethylene glycol (PEG)-mediated transfection method [41]. The transfected protoplasts were suspended in a protoplast culture medium (0.4 mM mannitol, 4 mM MES buffer, and 15 mM MgCl2 [pH 5.8]), and incubated in the dark for 16 h. Then, LUC activity in each cell lysate was determined using the luciferase assay system kit (Promega, Madison, WI, USA).
2.7. Quantification of Chlorophyll Pigment
Chlorophyll pigments were extracted (using 80% [v/v] ice-cold acetone) from leaf discs homogenized with zirconia beads (Nikkato, Osaka, Japan). The absorbance of extracts was measured at 647 and 664 nm, and chlorophyll contents were calculated as described previously [42].
2.8. Measurements of Agronomic Traits
To measure the agronomic traits of rice plants under normal growth conditions, plants were grown in soil (Honenagri Co., Ltd., Niigata, Japan) in a growth chamber under LD conditions for 150 days. To measure the agronomic traits of plants under drought stress conditions, plants were initially grown in soil in a growth chamber under LD conditions for 90 days, then dehydrated for 7 days, and subsequently rehydrated for 3 days. These processes were repeated six times before measuring the agronomic traits.
2.9. Accession Numbers
Sequence data used in this study can be found in the National Center for Biotechnology Information (NCBI) under the following accession numbers: ONAC054, Os03g0119966; OsABA1, Os04g0448900; OsABA3, Os06g0670000; OsABF1, Os01g0867300; OsABF2, Os06g0211200; OsABF4, Os09g0456200; TRAB1, Os08g0472000; OsCYP707A6, Os08g0472800; OsCYP707A7, Os09g0457100; OsLEA3, Os05g0542500; OsABI1, Os05g0572700; OsABI2, Os01g0513100; OsABI3, Os01g0911700; OsABI4, Os05g0351200; OsABI5, Os01g0859300; OsbZIP23, Os02g0766700; OsRAB16a, Os11g0454300; OsUBQ5, Os01g0328400.
3. Results
3.1. Overexpression of ONAC054 Enhanced Tolerance to Drought Stress in Rice Seedlings
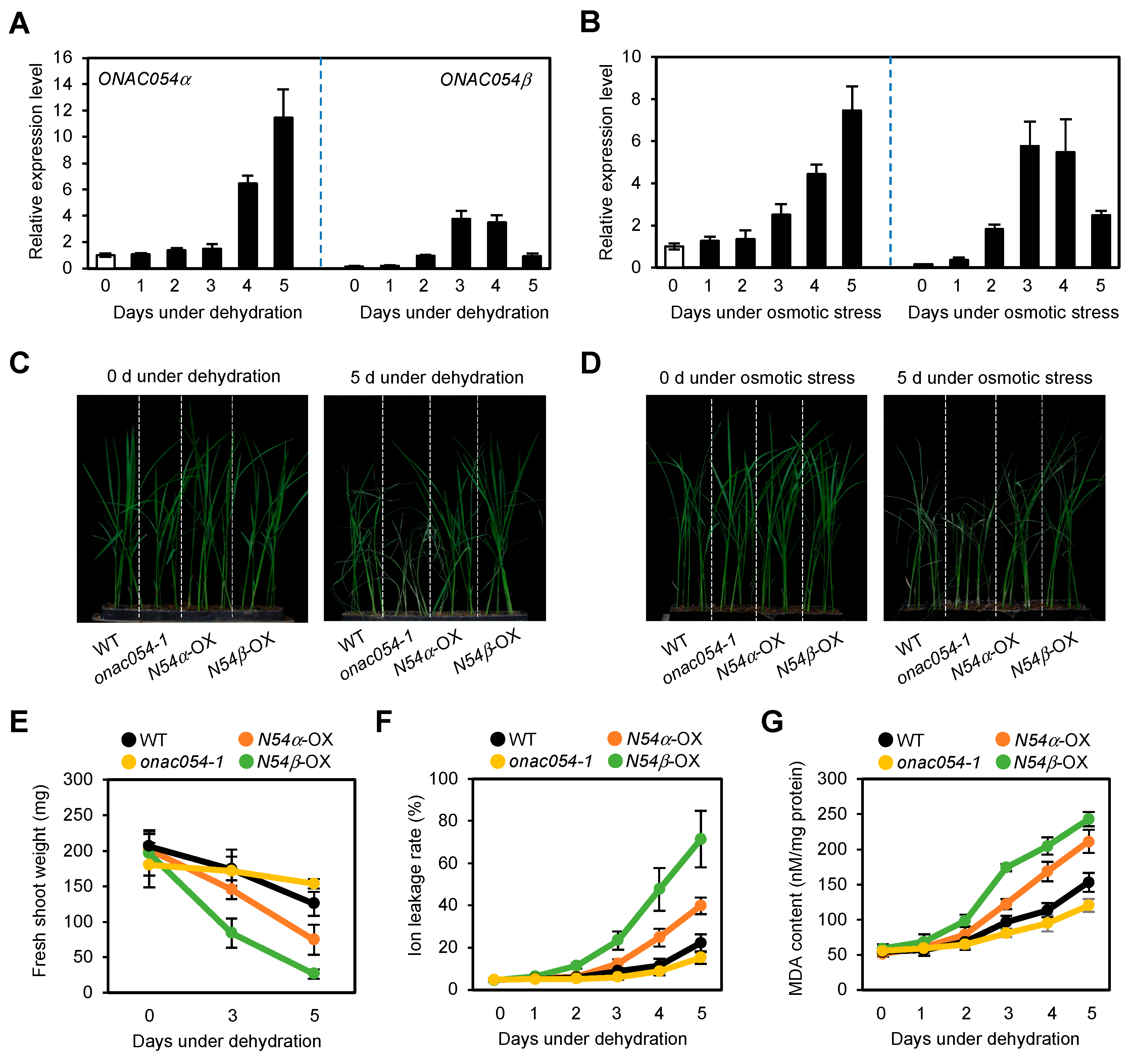
Recently, we showed that rice onac054-KO mutants exhibited accelerated leaf yellowing, while the leaves of ONAC054-OX lines retained greenness during ABA-induced leaf senescence [31]. Since ABA is one of the phytohormones that enhances the tolerance to drought stress [6], it is probable that ONAC054 also functions in drought stress responses. To examine this possibility, we first examined and monitored the expression levels of ONAC054α and ONAC054β in seedling leaves sampled during the dehydration stress treatment. The transcript levels of both ONAC054α and ONAC054β were significantly increased during dehydration stress. However, their expression patterns during dehydration stress were somewhat different: ONAC054β was induced much faster than ONAC054α, while the expression of ONAC054α was much higher than that of ONAC054β in the later stage of dehydration (Figure 1A), consistent with their expression patterns during the ABA treatment [31]. The application of high-molecular-weight PEG is known to disrupt the absorption of water by plant roots, causing drought stress in shoots [43]. Similar to their expression patterns during dehydration stress, ONAC054α and ONAC054β were strongly induced by the PEG treatment (Figure 1B), indicating that both ONAC054 isoforms are associated with drought stress responses.
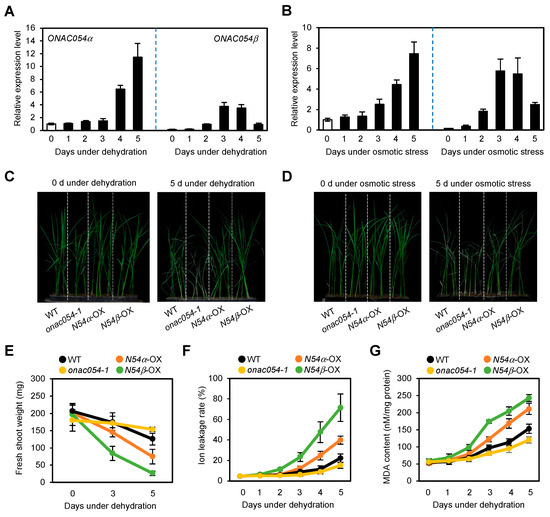
Figure 1.
Overexpression of ONAC054α and ONAC054β enhanced the tolerance to drought stress. (A,B) Expression patterns of ONAC054α and ONAC054β in the leaves of Dongjin (WT) seedlings under dehydration (A) and osmotic (B) stress. Gene expression levels were examined every day for 5 days after the start of each treatment. Transcript levels of ONAC054α and ONAC054β were normalized first against OsUBQ5 transcript levels, and then against the values obtained at time zero. (C,D) Phenotype of Dongjin (WT), onac054-1, ONAC054α-OX (N54α-OX), and ONAC054β-OX (N54β-OX) seedlings under normal growth conditions (C) and drought stress conditions (D). Seedlings were initially grown in soil for 14 days under normal conditions. Then, drought stress was induced for 5 days by either withholding watering or treating the seedlings with 200 mM PEG. (E–G) Changes in fresh shoot weight (E), ion leakage rate (F), and MDA content (G) of Dongjin (WT), onac054-1, ONAC054α-OX (N54α-OX), and ONAC054β-OX (N54β-OX) seedlings under dehydration stress for 5 days. Data represent the mean ± standard deviation (SD) of four biological replicates (A,B) or six biological replicates (E–G).
Next, we examined whether the onac054-KO mutant lines onac054-1 and ONAC054-OX lines (ONAC054α-OX and ONAC054β-OX), which were used in our previous study [31], were susceptible or tolerant to drought stress. Under normal growth conditions, the growth phenotype of onac054-1, ONAC054α-OX, and ONAC054β-OX seedlings was similar to that of the WT (Dongjin). However, after dehydration for 5 days, the onac054-1 seedlings exhibited severe wilting symptoms, whereas ONAC054α-OX and ONAC054β-OX seedlings exhibited strong drought tolerance compared with WT seedlings (Figure 1C). Similar susceptible and tolerant phenotypes of onac054-1 and ONAC054-OX seedlings, respectively, were also observed during the PEG treatment (Figure 1D). Consistent with the growth phenotype data, the fresh shoot weight of onac054-1 dramatically decreased, while ONAC054α-OX and ONAC054β-OX seedlings were unaffected by dehydration compared with the WT (Figure 1E). Furthermore, we measured the ion leakage rate and malondialdehyde (MDA) content, which are used as parameters of membrane damage caused by drought stress. Compared with the WT, the ion leakage rate and MDA content of osnac054-1 seedlings were dramatically higher, while those of ONAC054α-OX and ONAC054β-OX seedlings were much lower under dehydration stress (Figure 1F,G), indicating that the cell membrane of osnac054-1 seedlings was highly unstable. The dehydration susceptible phenotype of the onac054-KO mutant was also confirmed in onac054-2. Similar to onac054-1, the onac054-2 mutant showed a significantly higher ion leakage rate and a faster decline in fresh shoot weight than the WT (Figure S1). To evaluate the effects of the KO mutation and overexpression of ONAC054 on drought stress tolerance, we determined the survival rate of onac054-1 and ONAC054-OX seedlings after 3 days of rehydration. While most of the onac054-1 seedlings failed to recover, the ONAC054α-OX and ONAC054β-OX seedlings showed a significantly higher survival rate than WT seedlings (Figure S2). Taken together, these results suggest that both ONAC054 isoforms act as enhancers of drought stress tolerance in rice.
3.2. ONAC054 Upregulates Genes Associated with Drought Stress Responses
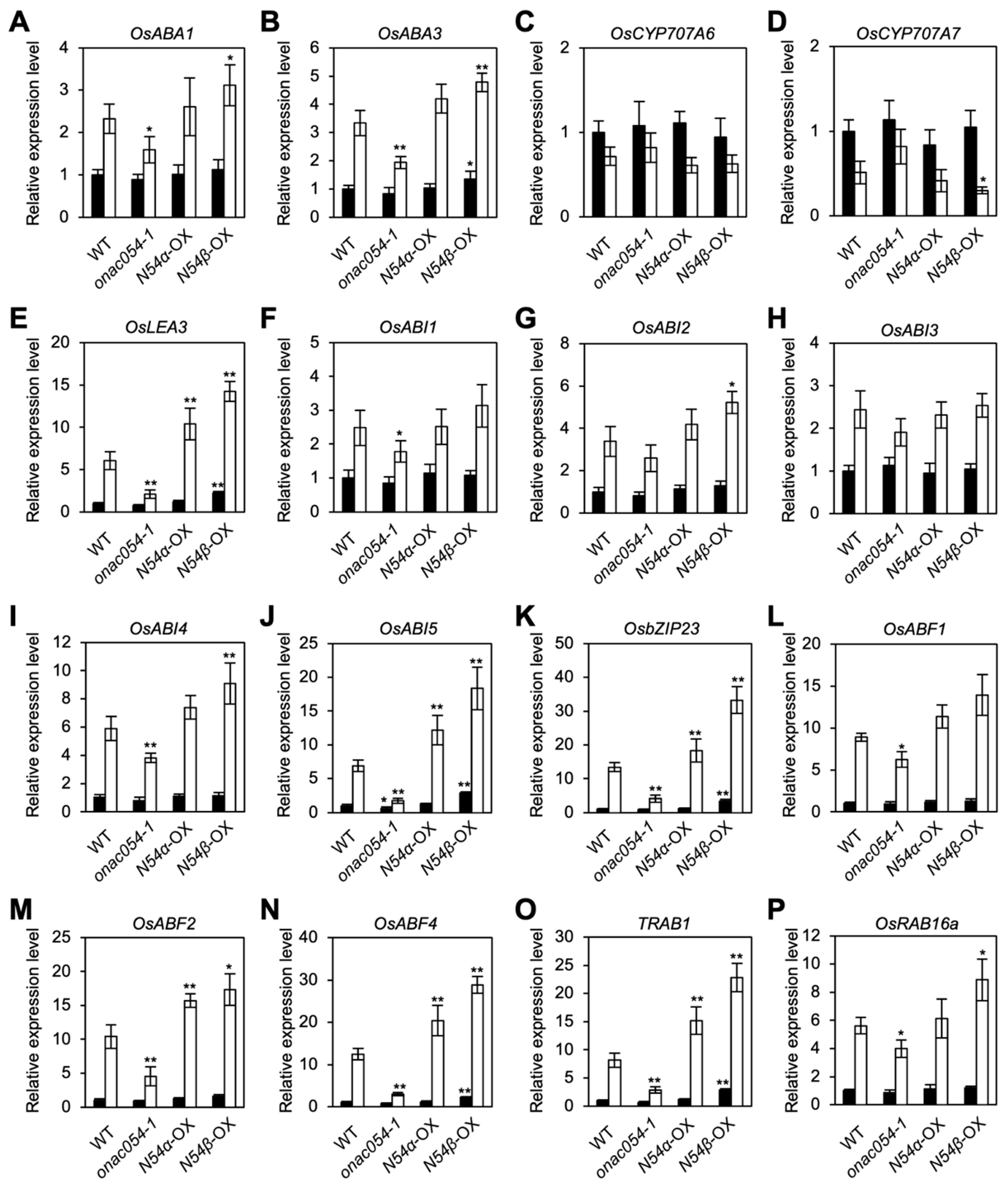
We previously showed that the knockout mutation of ONAC054 modulates the expression levels of a number of genes associated with ABA metabolism and signaling, especially during dark- and ABA-induced leaf senescence [31]. To understand how ONAC054 participates in the drought stress responses, we examined the effects of the KO mutation and overexpression of ONAC054 on the expression of four genes associated with ABA metabolism (OsABA1, OsABA3, CYTOCHROME P450, FAMILY 707, SUBFAMILY A, POLYPEPTIDE 3 [OsCYP707A6], and OsCYP707A7) and twelve genes associated with ABA signaling (OsLEA3, OsABI1–5, OsbZIP23, OsABF1, OsABF2, OsABF4, TRAB1, and RESPONSIVE TO ABA 16A [OsRAB16a]) by real-time quantitative PCR. Most genes examined were highly upregulated after 4 days of dehydration, whereas the expression levels of CYP707A6 and CYP707A7, encoding ABA catabolic enzymes [44], were not significantly changed (Figure 2). Several genes, including OsLEA3, OsABI5, OsbZIP23, OsABF4, and TRAB1, were strongly downregulated in onac054-1 but upregulated in ONAC054α-OX and ONAC054β-OX seedlings after 4 days of dehydration. Moreover, even before the dehydration, these genes were significantly upregulated in ONAC054β-OX but not in ONAC054α-OX, indicating that dehydration stress controls the cleavage of TMD from ONAC054, as observed previously in the ABA treatment [31].
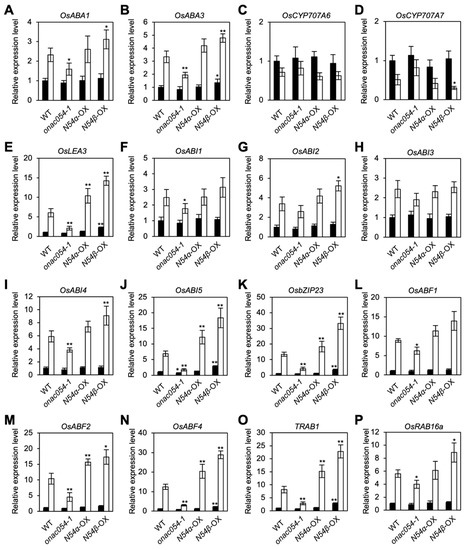
Figure 2.
Effects of the KO mutation and overexpression of ONAC054 on the transcript levels of genes associated with ABA metabolism and signaling. (A–P) Transcript levels of genes associated with ABA metabolism, including OsABA1 (A), OsABI3 (B), OsCYP707A6 (C), and OsCYP707A7 (D), and with ABA signaling, including OsLEA3 (E), OsABI1 (F), OsABI2 (G), OsABI3 (H), OsABI4 (I), OsABI5 (J), OsbZIP23 (K), OsABF1 (L), OsABF2 (M), OsABF4 (N), TRAB1 (O), and OsRAB16a (P), in Dongjin (WT), onac054-1, ONAC054α-OX (N54α-OX), and ONAC054β-OX (N54β-OX) seedlings before (white bars) and after 4 days of dehydration (black bars). The transcript level of each gene was normalized first against that of OsUBQ5, and then against the value obtained in WT seedlings before dehydration. Data represent the mean ± SD of four biological replicates. Asterisks (* p < 0.05; ** p < 0.01) indicate significant differences identified between the WT and other genotypes by Student’s t-test.
3.3. ONAC054 Directly Activates TRAB1 Transcription
We previously showed that ONAC054 specifically interacts with the CTTGXXXXXCA[C/A] sequence, named the mitochondrial dysfunction motif (MDM) [45], in target gene promoters to regulate their expression [31]. ONAC054 binds to the MDM sequence in the promoter of OsABI5 and enhances its expression [31]. However, ONAC054 does not bind to the OsABF4 promoter, which harbors an MDM variant (CTAGAACTTCAAG), indicating that ONAC054 indirectly enhances the expression of OsABF4 [31].
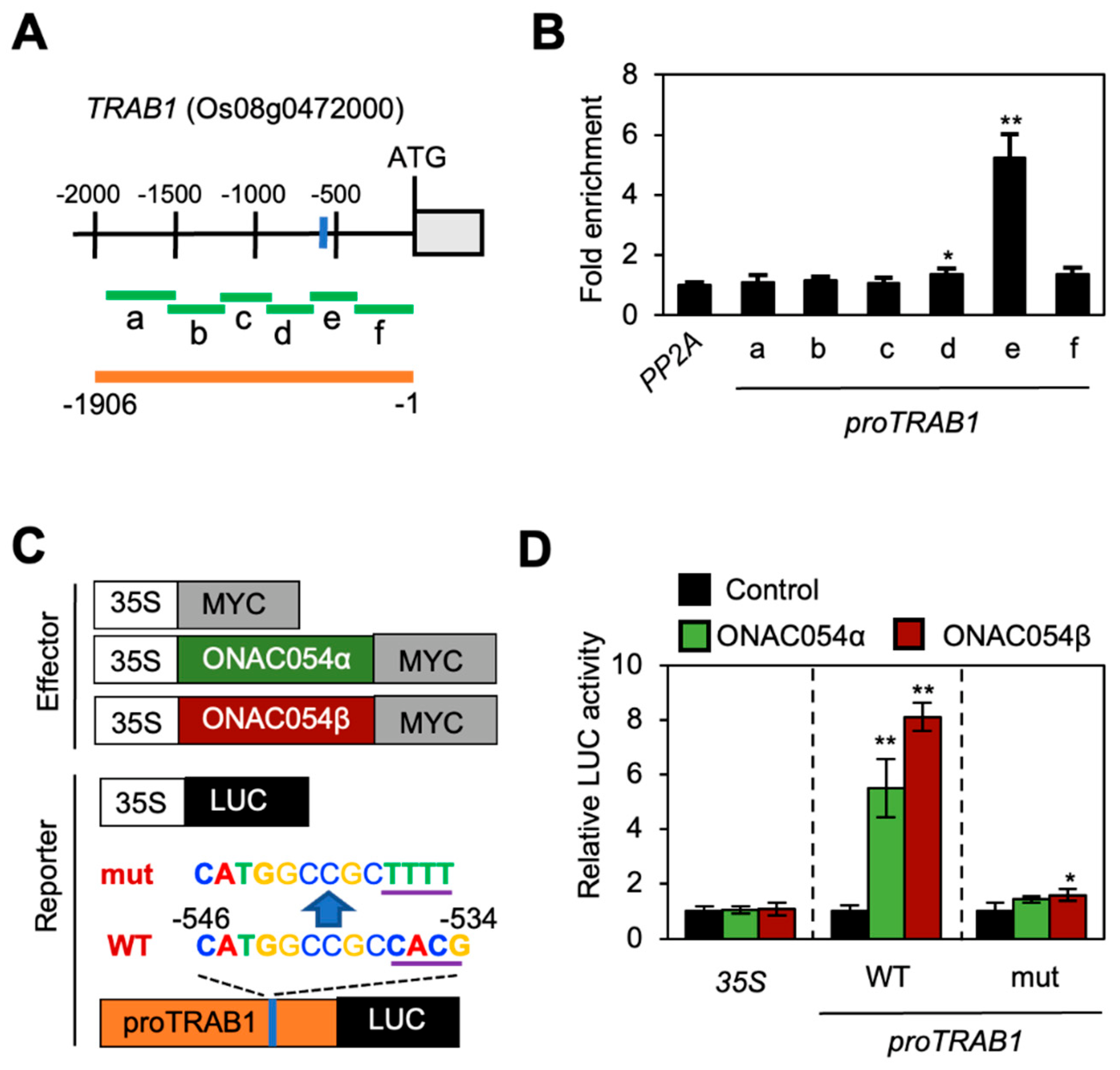
According to the results of RT-qPCR analysis (Figure 2), in addition to OsABI5 and OsABF4, three genes associated with ABA signaling (OsLEA3, OsbZIP23, and TRAB1) were strongly downregulated in onac054-1 seedlings but upregulated in ONAC054-OX lines (Figure 2). To explain this result, we examined the promoter sequences of OsLEA3, OsbZIP23, and TRAB1. An MDM variant (CTAGGCCGCCACG) was found in the promoter of TRAB1 (Os08g0472000; −546 to −534 bp, relative to the translation start site) (Figure 3A); however, OsLEA3 and OsbZIP23 promoters showed neither an MDM nor its variant.
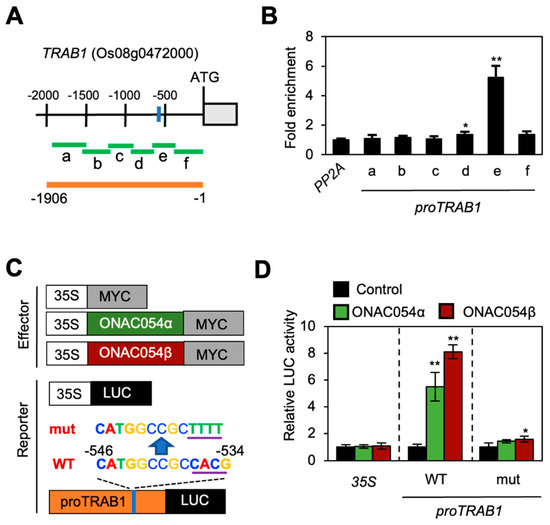
Figure 3.
ONAC054 directly activates TRAB1 expression. (A) Structure of the TRAB1 promoter. Regions amplified in the ChIP assay and used for the protoplast co-transfection assay are indicated using green and orange horizontal bars, respectively. The MDM variant is indicated by a blue vertical bar. (B) Analysis of the binding of ONAC054 (ONAC054β) to the TRAB1 promoter in planta using the ChIP assay. GFP-ONAC054β was transiently expressed in protoplasts isolated from 10-day-old WT seedlings. Fold enrichment of promoter fragments was measured by immunoprecipitation with an anti-GFP antibody (see Materials and Methods). Enrichment of the promoter region of an unrelated gene, PP2A, served as a negative control. Asterisks above each bar indicate significant differences between PP2A and other samples (* p < 0.05; ** p < 0.01; Student’s t-test). (C) Effector and reporter constructs were used to perform the protoplast transient expression assay. Each construct also contained the NOS terminator (not shown). (D) Activation of WT and mutant versions of the TRAB1 promoter (proTRAB1; −1906 to −1 bp, relative to the translation start site) by ONAC054α and ONAC054β in protoplasts. The 35S promoter and empty expression vector (35S:MYC) served as negative controls. Asterisks indicate significant differences compared with the 35S:MYC samples (* p < 0.05; ** p < 0.01; Student’s t-test). Data in (B,D) represent the mean ± SD of four reaction mixtures.
To examine whether ONAC054 binds to the TRAB1 promoter, we performed a ChIP assay using rice protoplasts transiently expressing GFP-ONAC054α. Region e of the TRAB1 promoter, which contained the MDM variant (Figure 3A), was highly enriched in the immunoprecipitate, while other regions showed no enrichment (Figure 3B).
Next, to determine the effect of ONAC054 on TRAB1 expression, we performed a co-transfection assay using rice protoplasts. Both GFP-ONAC054α and GFP-ONAC054β fusions were used to generate the effector constructs. On the other hand, the WT TRAB1 promoter and its mutant version (in which the MDM variant sequence, CTAGGCCGCCACG, was changed to CTAGGCCGCTTTT) were individually fused to the LUC reporter gene to generate the reporter constructs (Figure 3C). Upon protoplast co-transfection, both ONAC054α and ONAC054β activated the WT TRAB1 promoter sequence, although ONAC054β showed a stronger effect (Figure 3D). However, the mutated TRAB1 promoter was hardly activated by GFP-ONAC054α and GFP-ONAC054β co-transfection, indicating that ONAC054 activates the TRAB1 genes by directly binding to the MDM variant in their promoter.
3.4. ONAC054 Expression Is Regulated by Multiple ABA bZIP TFs
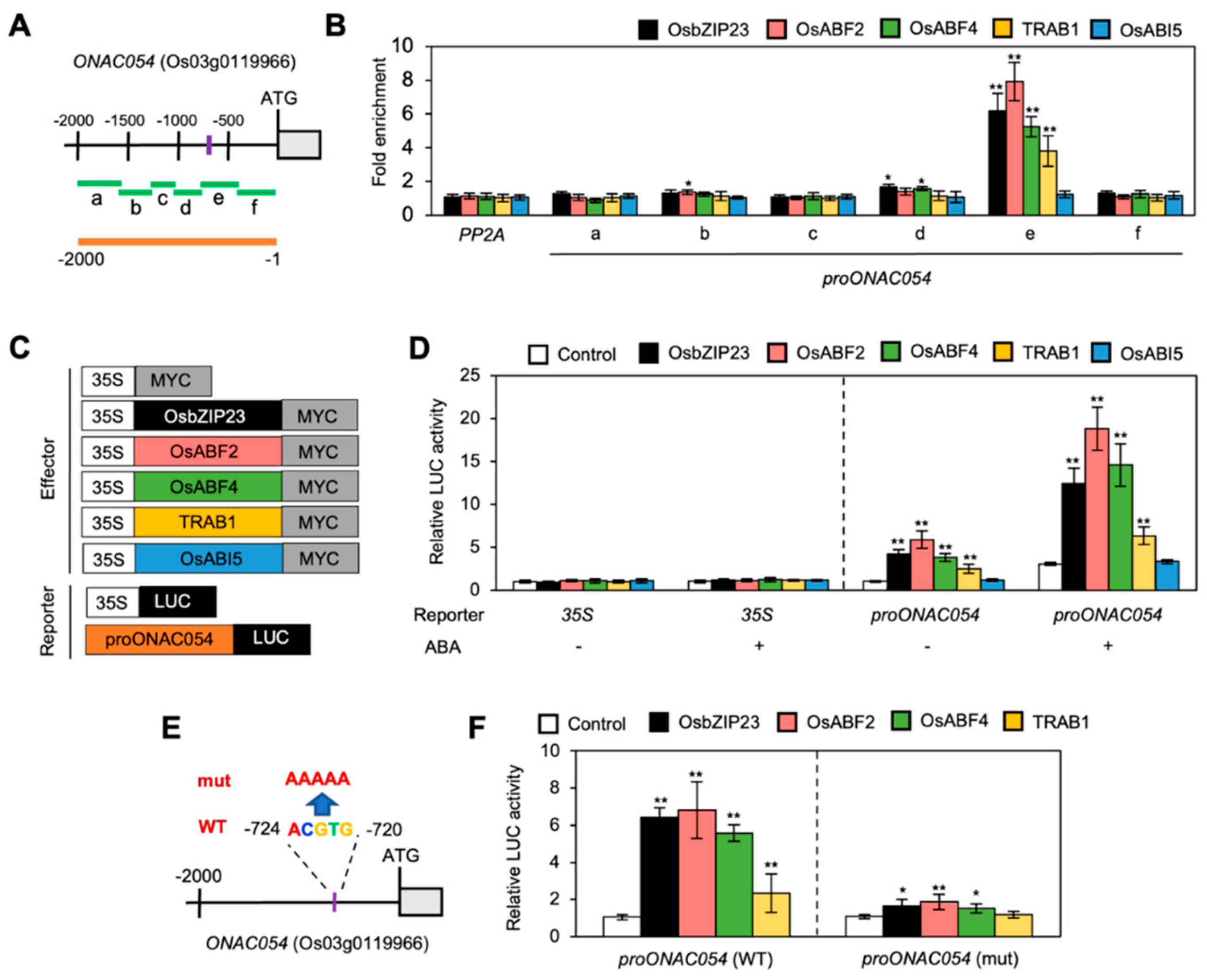
In previous studies and the current study, we showed that ONAC054 modulates the expression of several genes associated with ABA metabolism and signaling as well as those involved in drought stress responses, especially ABA-induced leaf senescence and dehydration [31] (Figure 2). Nonetheless, it remains unclear how the expression of ONAC054 (both ONAC054α and ONAC054β) is induced by ABA and dehydration stress treatments. To understand the regulation of ONAC054 expression, we first checked the promoter sequence of ONAC054 (−2000 to −1 bp, relative to the translation start site). The results revealed the presence of an ABRE core sequence, ACGTG, in the ONAC054 promoter (Figure 4A).
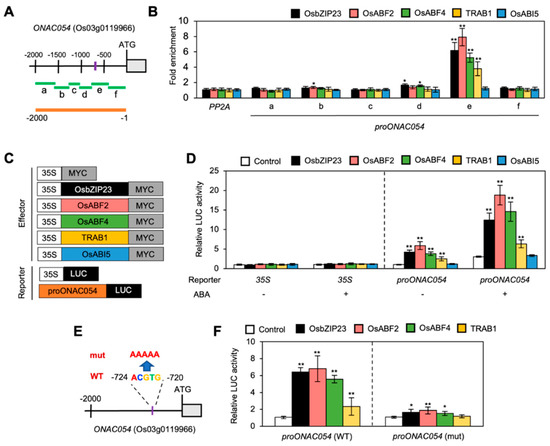
Figure 4.
ABF transcription factors directly activate ONAC054. (A) Structure of the ONAC054 promoter. Regions amplified in the ChIP assay (regions a, b, c, d, e, and f) and used for the protoplast co-transfection assay are indicated using green and orange horizontal bars, respectively. The ABRE motif is indicated by a purple vertical bar. (B) Analysis of the binding of OsbZIP23, OsABF2, OsABF4, TRAB1, and OsABI5 to ONAC054 promoter in planta using ChIP assay. OsbZIP23-MYC, OsABF2-MYC, OsABF4-MYC, TRAB1-MYC, and OsABI5-MYC were transiently expressed in protoplasts isolated from 10-day-old WT seedlings. Fold enrichment of promoter fragments was measured by immunoprecipitation with an anti-MYC antibody (see Materials and Methods). Enrichment of the promoter region of an unrelated gene, PP2A, served as a negative control. Asterisks above each bar indicate significant differences between PP2A and other promoter fragments of ONAC054 (* p < 0.05; ** p < 0.01; Student’s t-test). (C) Effector and reporter constructs are used in the protoplast transient expression assay. Each construct also contained the NOS terminator (not shown). (D) Activation of the ONAC054 promoter (−2000 to −1 bp, relative to the translation start site) by OsbZIP23, OsABF2, OsABF4, TRAB1, and OsABI5 in protoplasts treated with or without 10 μM ABA. The 35S promoter and empty expression vector (35S:MYC) served as negative controls. Asterisks indicate significant differences compared with 35S:MYC samples (** p < 0.01; Student’s t-test). (E) Structure of a mutated ONAC054 promoter. In proNAC054 (mut), the ABRE motif ACGTG (−724 to −1 bp, relative to the translation start site) was changed to AAAAA. (F) Activation of WT and mutated ONAC054 promoters by OsbZIP23, OsABF2, OsABF4, and TRAB1 in protoplasts. Asterisks indicate significant differences compared with 35S:MYC samples (* p < 0.05; ** p < 0.01; Student’s t-test). Data in (B,D,F) represent the mean ± SD values of four biological replicates.
Some ABA-inducible bZIP TFs, including ABFs, interact with the ABRE motif in the promoters of ABA-inducible genes to modulate their expression [9]. Moreover, five rice bZIP TFs, OsbZIP23, OsABF2, OsABF4, TRAB1/OsABF5, and OsABI5, are phylogenetically close to Arabidopsis ABF TFs [46] and are involved in ABA-mediated stress responses [46,47,48,49]. To examine whether these bZIP TFs interact with the ONAC054 promoter, we performed a ChIP assay using rice protoplasts transiently expressing OsbZIP23-MYC, OsABF2-MYC, OsABF4-MYC, TRAB1-MYC, or OsABI5-MYC. The results indicated that OsbZIP23, OsABF2, OsABF4, and TRAB1 bind to the region e of the ONAC054 promoter, which contains an ABRE motif (−724 to −720 bp, relative to the translation start site), but OsABI5 does not (Figure 3B).
We subsequently performed co-transfection experiments using protoplasts isolated from ABA-treated or -untreated WT seedlings, expressing various combinations of reporter and effector plasmids (Figure 3C). OsbZIP23, OsABF2, OsABF4, and TRAB1 enhanced the activity of the ONAC054 promoter, whereas OsABI5 did not (Figure 3D). Notably, the activation of the ONAC054 promoter by these bZIP TFs was much stronger in protoplasts treated with ABA than in those not subjected to the ABA treatment (Figure 3D). Because four bZIP TFs could bind to the ABRE motif-containing region e of the ONAC054 promoter (Figure 3B), we performed protoplast transient assays to examine whether these bZIP TFs could activate the mutated ONAC054 promoter, in which the ABRE sequence (ACGTG) was changed to AAAAA (Figure 3E). We found that the mutated ONAC054 promoter was not strongly activated by co-transfection with OsbZIP23-MYC, OsABF2-MYC, OsABF4-MYC, or TRAB1-MYC (Figure 3F). Taken together, these results suggest that ONAC054 expression is upregulated by several ABFs.
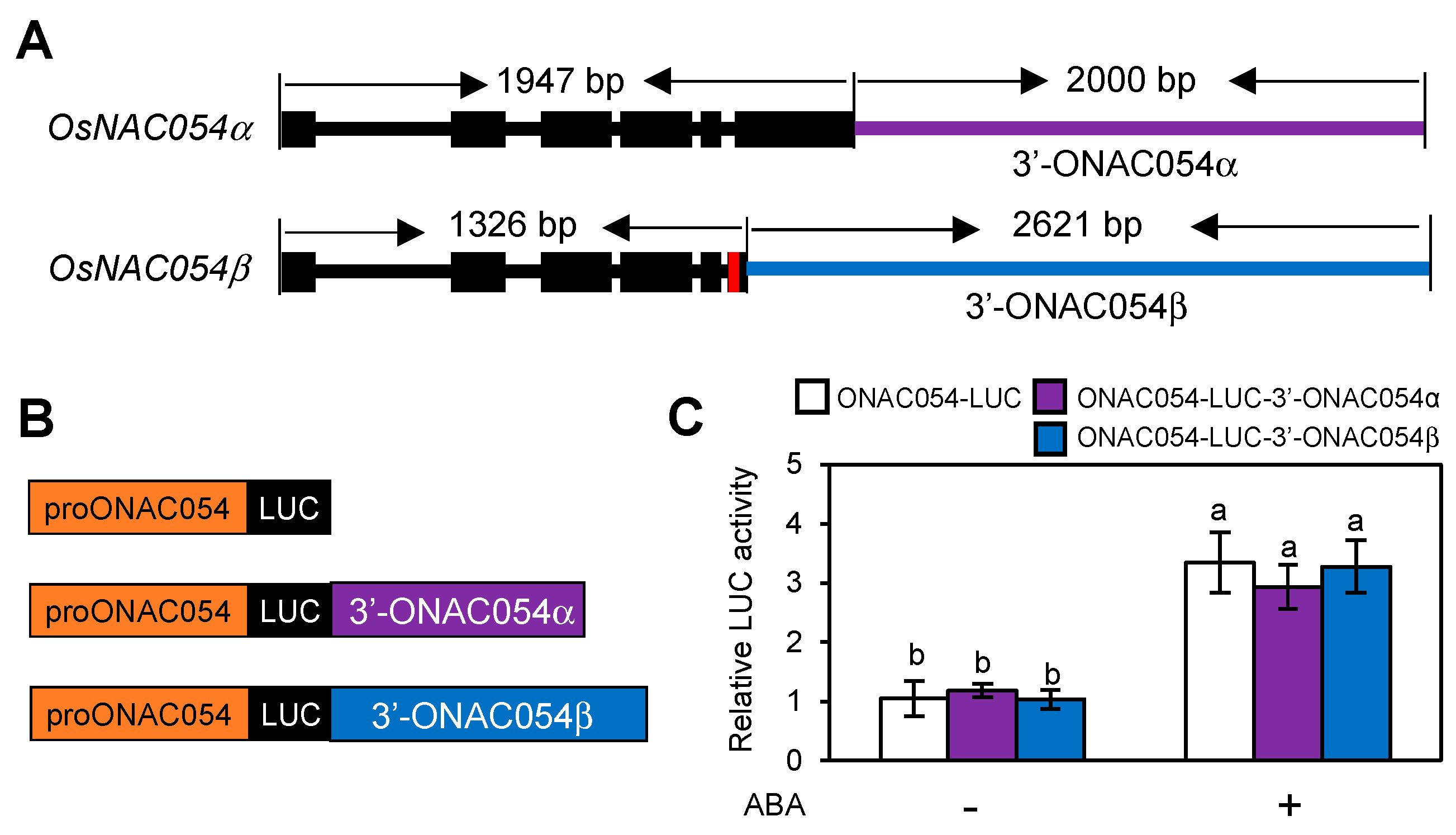
3.5. A Sequence Downstream of the ONAC054 Coding Region Is Not Required for ABA Induction
While we showed that both ONAC054α and ONAC054β were strongly upregulated by dehydration, osmotic stress, and ABA treatment (Figure 1A,B) [31], the timing of their induction was somewhat different; ONAC054β was induced much faster than ONAC054α (Figure 1A,B). This may be because the 3′-untranslated region (3′-UTR) of ONAC054β is different from that of ONAC054α, since ONAC054β carries a 7-nt insertion between intron 5 and exon 6, leading to a frameshift mutation and consequently the insertion of a premature stop codon [31] (Figure 5A). Moreover, the cis elements responsible for gene expression could be located downstream of the coding sequence of ONAC054β, as in the case of Arabidopsis NITRATE REDUCTASE1 (NIA1), which contains nitrate-responsive cis elements downstream of its stop codon [50]. To examine this possibility, we performed protoplast transient expression assays using reporter plasmids in which the 3′-end of ONAC054α or ONAC054β was fused downstream of the LUC gene under the control of the ONAC054 promoter (−2000 to −1, relative to the translation start site) (Figure 5B). While LUC activity was induced in protoplasts transformed with both proONAC054-LUC-3′-ONAC054α and proONAC054-LUC-3′-ONAC054β after the ABA treatment, as in protoplasts transformed with proONAC054-LUC, the LUC activity showed no significant differences among proONAC054-LUC, proONAC054-LUC-3′-ONAC054α, and proONAC054-LUC-3′-ONAC054β protoplasts (Figure 5C), indicating that the difference between the 3′-UTRs of ONAC054α and ONAC054β is not related to the differential expression patterns of the two OsNAC054s under dehydration stress and ABA treatments.
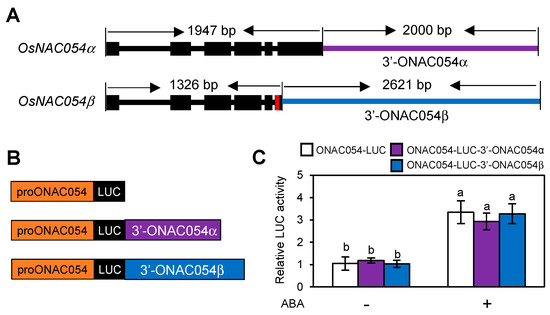
Figure 5.
The 3′-UTRs of ONAC054α and ONAC054β do not affect their expression patterns. (A) Gene structure of ONAC054α and ONAC054β. The insertion of 7 bp in the fifth intron of ONAC054β created an alternative 3′ splice site and a premature stop codon, leading to a difference in the length of the 3′-UTRs of ONAC054α (purple horizontal bar) and ONAC054β (blue horizontal bar). (B) Reporter constructs are used in the protoplast transient expression assay. (C) Effects of 3′-flanking sequences of ONAC054α and ONAC054β on the activity of the ONAC054 promoter (−2000 to −1 bp, relative to the translation initiation site) in protoplasts treated with or without ABA. Data represent the mean ± SD of four biological replicates, and different lowercase letters indicate significant differences (p < 0.05; Tukey’s multiple comparison test).
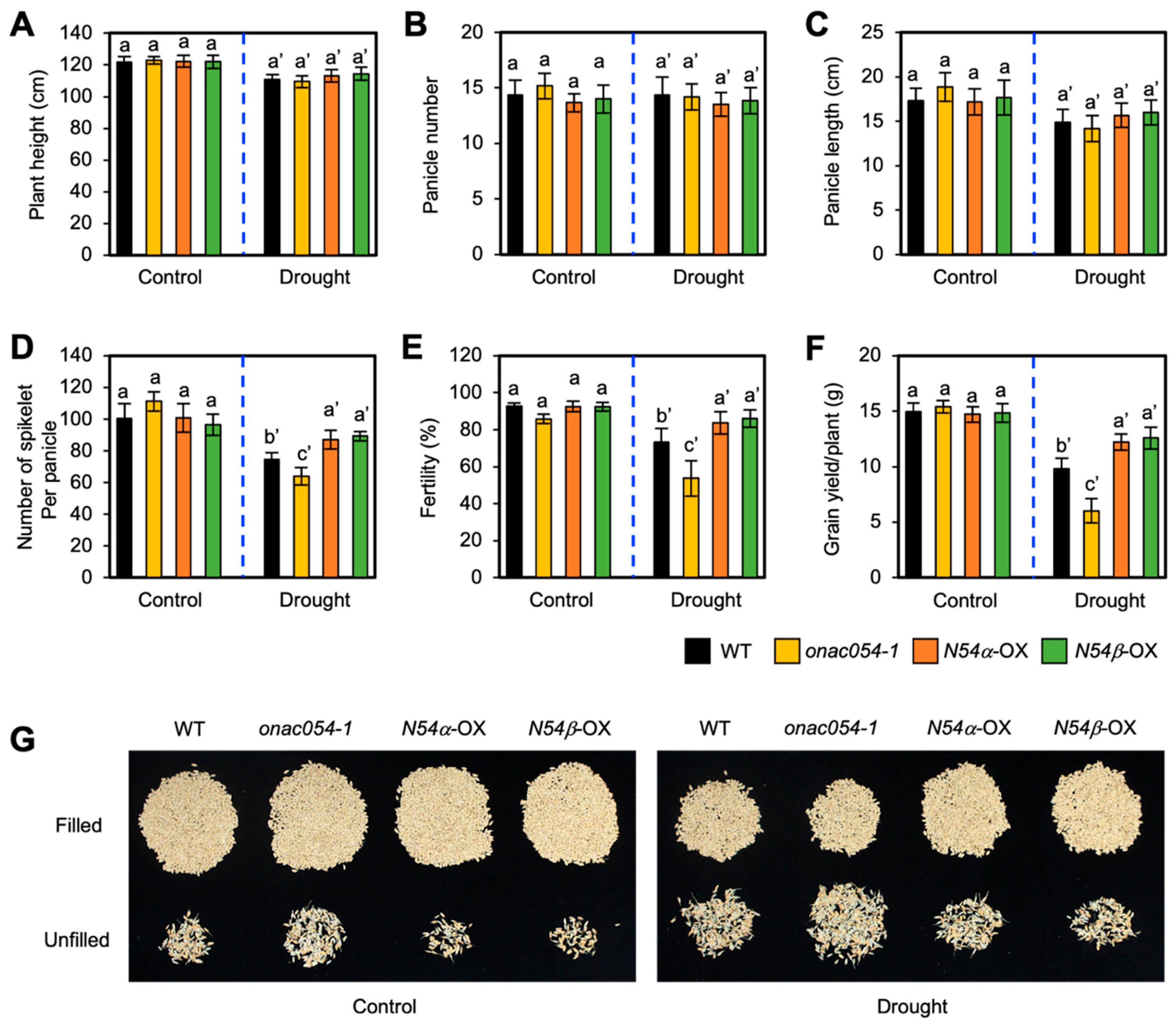
3.6. Overexpression of ONAC054 Increases Grain Yield under Drought Stress Conditions
Drought stress greatly and negatively affects rice grain yield under drought stress conditions [51]. To examine whether the KO mutation and overexpression of ONAC054 affect grain yield under drought stress conditions, we evaluated the agronomic traits of WT, onac054-1, ONAC054α-OX, and ONAC054β-OX plants, including plant height, panicle number per plant, spikelet number per panicle, seed fertility, and grain yield per plant, after sequential drought stress treatments. We previously showed that the KO mutation of ONAC054 did not affect grain yield but slightly decreased seed fertility [31]. Under non-stress conditions, the grain yield and all other traits of onac054-1, ONAC054α-OX, and ONAC054β-OX plants were similar to those of the WT (Figure 6A–G). However, under drought stress conditions, the grain yield of onac054-1 was significantly reduced, while that of ONAC054α-OX and ONAC054β-OX was highly retained compared with the grain yield of the WT (Figure 6F,G). The dramatic decline in the grain yield of onac054-1 was mainly caused by the decrease in two traits: spikelet number per panicle and seed fertility, both of which were highly retained in the two ONAC054-OX lines compared with the WT (Figure 6D,E,G). Overall, ONAC054 overexpression improved the grain yield of rice without causing any growth defects.
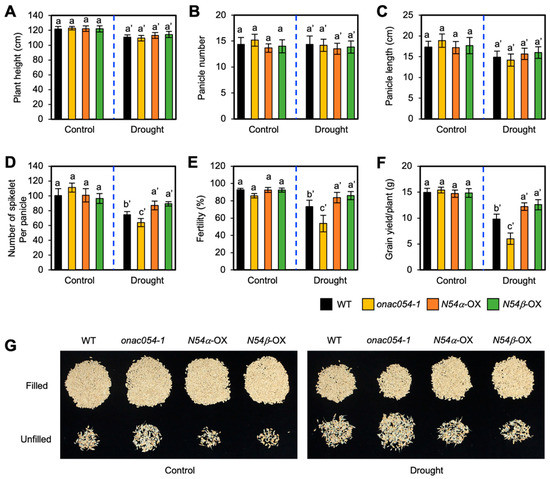
Figure 6.
Agronomic traits of onac054-1 and ONAC054-OX plants under drought stress conditions. (A–F) Agronomic traits, including plant height (A), panicle number per plant (B), panicle length (C), spikelet number per panicle (D), seed fertility (E), and grain yield per plant (F), of WT, onac054-1, ONAC054α-OX (N054α-OX), and ONAC054β-OX (N054β-OX) plants grown under normal growth conditions or drought stress conditions (see Materials and Methods). Data represent the mean ± SD (n = 6 plants), and different lowercase letters (letters with ’ for drought stress conditions) indicate significant differences (p < 0.05; Tukey’s multiple comparison test). (G) Relative abundance of fertile and sterile spikelets in different genotypes.
4. Discussion
4.1. Regulatory Mechanism Underlying the ONAC054-Mediated Drought Stress Responses
In this study, we showed that ONAC054 acts as a key regulator of drought stress responses in rice. After 5 days of dehydration, onac054-KO seedlings exhibited severe wilting, while ONAC054-OX plants showed a tolerant phenotype (Figure 1). These differences in phenotypes may be caused by altered expression of genes associated with ABA metabolism and signaling (Figure 2). Furthermore, we showed that ONAC054 directly or indirectly activates the transcription of several genes associated with ABA signaling, including OsABI5, OsLEA3, OsbZIP23, OsABF4, and TRAB1 [31] (Figure 2 and Figure 3), which have previously been functionally characterized. Overexpression of OsLEA3, OsbZIP23, or OsABF4 enhanced drought tolerance in rice [37,49,52]. The role of TRAB1 in drought stress responses has not yet been elucidated. However, given that TRAB1 is required for the induction of ABA-responsive genes by interacting with the ABRE sequence in their promoters [53], it is likely that TRAB1 plays an important role in the drought stress response. Thus, the upregulation of ABA signaling genes may contribute to the drought tolerance phenotype of ONAC054-OX plants. On the other hand, OsABI5 antisense transgenic rice plants were reported to be tolerant to drought and high salinity stresses [49]. Thus, the role of the ONAC054–OsABI5 regulatory module, which is important for ABA-induced leaf senescence [31], in drought stress responses needs to be elucidated.
In this study, four ABFs, including OsbZIP23, OsABF2, OsABF4, and TRAB1, activated the transcription of ONAC054 (Figure 4). On the other hand, ONAC054 also directly or indirectly activated the transcription of OsbZIP23, OsABF4, and TRAB1 (Figure 3), as shown previously [31]. Thus, our findings suggest that ONAC054 and the four ABFs form a feedforward transcriptional loop for the enhancement of ABA signaling responses (Figure S3). Similar feedforward transcriptional regulatory mechanisms have been previously reported. In Arabidopsis, Dof2.1 directly activates the transcription of MYC2, and the encoded MYC2 TF, a central regulator of the JA response [54], directly activates the transcription of Dof2.1, thus forming a feedforward loop for the enhancement of the JA response [55]. In rice, PHYTOCHROME-INTERACTING FACTOR-LIKE1 (OsPIL1), a basic helix-loop-helix (bHLH) transcription factor, activates chlorophyll biosynthesis genes, OsPORB and OsCAO [56,57], and GARP-type TF genes, OsGLK1 and OsGLK2 [58], while OsGLK1 and OsGLK2 directly activate the transcription of OsPORB and OsCAO, thus forming a feedforward transcriptional loop for the promotion of chlorophyll biosynthesis [59]. Such coherent feedforward loops may increase the robustness of biological signaling processes [60].
Additionally, both ONAC054α and ONAC054β were strongly induced under dehydration and osmotic stress conditions (Figure 1A,B), similar to their expression patterns in ABA-treated plants [31]. However, ONAC054β was induced much faster than ONAC054α (Figure 1) [31]. In this study, we examined whether the difference in the 3′-UTRs of ONAC054α and ONAC054β was responsible for the difference in their expression patterns in the protoplast transient expression assay. However, the 3′-UTRs of ONAC054α and ONAC054β did not affect LUC activity in response to ABA (Figure 5). Thus, the cause of the differential expression patterns of ONAC054α and ONAC054β in response to dehydration and ABA treatments remains unclear. The 3′-UTR is sometimes associated with the regulation of mRNA stability as it contains several sequence elements required for the destabilization of mRNA, which are enriched among short- and long-lived transcripts [61]. Thus, it is necessary to examine whether the stability of ONAC054α and ONAC054β mRNAs is regulated at the post-translational level.
4.2. Advantages of Membrane-Bound NAC TFs in Crop Biotechnology Research
Altering the expression or activity of stress-responsive genes is one of the most useful approaches for generating crops with enhanced environmental stress tolerance [8], although this technique sometimes has a negative impact on plant growth and development. A number of stress-tolerant plants generated by the overexpression of stress-responsive genes exhibit a dwarf phenotype. For instance, the overexpression of DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1A (DREB1A), one of the most extensively studied drought stress-responsive genes, greatly improved drought stress tolerance in Arabidopsis and other crops, including peanut (Arachis hypogaea L.) [62] and wheat (Triticum aestivum L.) [63], but caused severe dwarfism and late flowering in plants [64]. On the other hand, a knockdown mutation of BRASSINOSTEROID INSENSITIVE1 (BRI1) increased tolerance to drought stress but caused severe dwarfism in Brachypodium distachyon [65]. These negative effects on plant growth and development may be caused by the constitutive overexpression or knockout of stress-responsive genes.
Under non-stress conditions, most TMD-containing TFs localize to the ER. Upon stress stimulation, TMD is cleaved through ER-specific proteolytic mechanisms, such as regulated intramembrane proteolysis (RIP) [66], and regulated ubiquitin/proteasome-dependent processing (RUP) [67], which leads to the translocation of TFs to the nucleus, where they induce the expression of target genes [16]. Therefore, the overexpression of TMD-containing TF genes does not affect plant growth and development when plants are grown under moderate growth conditions. Thus, TMD-containing TF genes could be utilized for the development of ideal stress-tolerant crops.
For instance, transgenic Arabidopsis plants overexpressing full-length NTL4, encoding an Arabidopsis TMD-containing NAC TF, were indistinguishable from WT Col-0 plants, while the overexpression of NTL4 lacking the C-terminal TMD caused abnormal growth phenotypes, such as reduced growth and curled leaves with asymmetric leaf axis and serrated margins [30]. Similarly, transgenic Arabidopsis plants overexpressing full-length NAC WITH TRANSMEMBRANE MOTIF1 (NTM1) were indistinguishable from WT plants, while transgenic plants overexpressing a TMD-truncated NTM1 exhibited apparent growth defects such as reduced cell numbers [68]. In the current study, we showed that the overexpression of ONAC054 did not cause any growth defects; rather, ONAC054 overexpression greatly improved the grain yield under drought-stress conditions (Figure 6). Thus, the TMD-containing TF genes show great potential for the development of stress-tolerant crops, although our knowledge of the roles of TMD-containing TFs in crops is still considerably limited.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/crops2040027/s1, Figure S1: Drought stress sensitive phenotypes of two rice onac054 mutants; Figure S2: Survival rate of wild-type (WT), onac054-1 mutants, and ONAC054-OX seedlings after dehydration; Figure S3: Model depicting the ONAC054-mediated drought stress responses in rice; Table S1: Primers used in this study.
Author Contributions
Conceptualization, Y.S. and N.-C.P.; methodology, Y.S.; formal analysis, Y.S.; investigation, Y.S.; resources, Y.S. and N.-C.P.; data curation, Y.S.; writing—original draft preparation, Y.S.; writing—review and editing, Y.S. and N.-C.P.; supervision, Y.S. and N.-C.P.; project administration, Y.S. and N.-C.P.; funding acquisition, Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Japan Society for the Promotion of Science KAKENHI (22K05368).
Data Availability Statement
Not applicable.
Acknowledgments
We thank Gynheung An at Kyung Hee University for donating the seeds of rice T-DNA insertion mutant lines, onac054-1 and onac054-2.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of this study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic erngineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Streeter, J.G.; Lohnes, D.G.; Fioritto, R.J. Patterns of pinitol accumulation in soybean plants and relationships to drought tolerance. Plant Cell Environ. 2001, 24, 429–438. [Google Scholar] [CrossRef]
- Choudhary, M.K.; Basu, D.; Datta, A.; Chakraborty, N.; Chakraborty, S. Dehydration-desponsive nuclear proteome of rice (Oryza Sativa L.) illustrates protein network, novel regulators of cellular adaptation, and evolutionary perspective. Mol. Cell Proteomics 2009, 8, 1579–1598. [Google Scholar] [CrossRef]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118. [Google Scholar] [CrossRef]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef]
- Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 2015, 6, 84. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Choi, H.I.; Hong, J.H.; Ha, J.O.; Kang, J.Y.; Kim, S.Y. ABFs, a Family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Soo, Y.K. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef]
- Gong, W.; Shen, Y.P.; Ma, L.G.; Pan, Y.; Du, Y.L.; Wang, D.H.; Yang, J.Y.; Hu, L.D.; Liu, X.F.; Dong, C.X.; et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004, 135, 773–782. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Seo, P.J.; Kim, S.G.; Park, C.M. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008, 13, 550–556. [Google Scholar] [CrossRef]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; van Aken, O.; de Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia Is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.H.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Kim, S.Y.; Hyeon, D.Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.H.; Kim, S.K.; Hong, J.C.; et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with BZIP-Type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in Rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, Y.S.; Han, S.H.; Lee, B.D.; Paek, N.C. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef]
- Kim, M.J.; Park, M.J.; Seo, P.J.; Song, J.S.; Kim, H.J.; Park, C.M. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 2012, 448, 353–363. [Google Scholar] [CrossRef]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Han, S.H.; Kim, S.H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.C. Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 2020, 32, 630–649. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef]
- Lee, S.H.; Sakuraba, Y.; Lee, T.; Kim, K.W.; An, G.; Lee, H.Y.; Paek, N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006, 45, 616–629. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef]
- Saleh, A.; Alvarez-Venegas, R.; Avramova, Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 2008, 3, 1018–1025. [Google Scholar] [CrossRef]
- Luehrsen, K.R.; de Wet, J.R.; Walbot, V. Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 1992, 216, 397–414. [Google Scholar]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, PGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis Mesophyll Protoplasts: A Versatile Cell System for Transient Gene Expression Analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta. Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Hohl, M.; Schopfer, P. Water relations of growing maize coleoptiles comparison between mannitol and polyethylene glycol 6000 as external osmotica for adjusting turgor pressure. Plant Physiol. 1991, 95, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Choi, D. Characterization of genes encoding ABA 8’-hydroxylase in ethylene-induced stem growth of deepwater rice. Biochem. Biophys. Res. Commun. 2006, 350, 685–690. [Google Scholar] [CrossRef] [PubMed]
- de Clercq, I.; Vermeirssen, V.; van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inzé, A.; Ng, S.; Ivanova, A.; Rombaut, D.; et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013, 25, 3472–3490. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A BZIP Transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, NIA1, in Arabidopsis. Plant Cell Physiol. 2011, 52, 824–836. [Google Scholar] [CrossRef]
- Venuprasad, R.; Lafitte, H.R.; Atlin, G.N. Response to direct selection for grain yield under drought stress in rice. Crop Sci. 2007, 47, 285–293. [Google Scholar] [CrossRef]
- Duan, J.; Cai, W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 2012, 7, e45117. [Google Scholar] [CrossRef]
- Kagaya, Y.; Hobo, T.; Murata, M.; Ban, A.; Hattori, T. Abscisic acid–induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 2002, 14, 3177–3189. [Google Scholar] [CrossRef]
- Dombrecht, B.; Gang, P.X.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Zhuo, M.; Sakuraba, Y.; Yanagisawa, S. A jasmonate-activated MYC2–Dof2.1–MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell 2020, 32, 242–262. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.H.; Kim, Y.S.; Koh, H.J.; Yoo, S.C.; Paek, N.C. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.H.; Eun, S.Y.; Lee, C.H.; Hirochika, H.; An, G. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005, 57, 805–818. [Google Scholar] [CrossRef]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, E.Y.; Han, S.H.; Piao, W.; An, G.; Todaka, D.; Yamaguchi-Shinozaki, K.; Paek, N.C. Rice phytochrome-interacting factor-like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops. J. Exp. Bot. 2017, 68, 4103–4114. [Google Scholar] [CrossRef]
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 2003, 100, 11980–11985. [Google Scholar] [CrossRef]
- Narsai, R.; Howell, K.A.; Millar, A.H.; O’Toole, N.; Small, I.; Whelan, J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis Thaliana. Plant Cell 2007, 19, 3418–3436. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Devi, M.J.; Reddy, D.S.; Lavanya, M.; Vadez, V.; Serraj, R.; Yamaguchi-Shinozaki, K.; Sharma, K.K. Stress-inducible expression of at DREB1A in transgenic peanut (Arachis Hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 2007, 26, 2071–2082. [Google Scholar] [CrossRef]
- Pellegrineschi, A.; Reynolds, M.; Pacheco, M.; Brito, R.M.; Almeraya, R.; Yamaguchi-Shinozaki, K.; Hoisington, D. Stress-induced expression in wheat of the Arabidopsis Thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 2011, 47, 493–500. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Feng, Y.; Yin, Y.; Fei, S. Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium Distachyon. Plant Sci. 2015, 234, 163–173. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Ericsson, J. SREBP in signal transduction: Cholesterol metabolism and beyond. Curr. Opin. Cell Biol. 2007, 19, 215–222. [Google Scholar] [CrossRef]
- Hoppe, T.; Matuschewski, K.; Rape, M.; Schlenker, S.; Ulrich, H.D.; Jentsch, S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 2000, 102, 577–586. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, S.G.; Park, J.E.; Park, H.Y.; Lim, M.H.; Chua, N.H.; Park, C.M. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).