Abstract
Anadenanthera colubrina (Acol) and Anadenanthera peregrina (Aper) (Fabaceae) are two species popularly known as “angicos” that occur in seasonally dry tropical forest (SDTR) remnants in Brazil. Since many of the morphological characteristics of Anadenanthera species are superimposed and species-specific characteristics are difficult to observe, their identification is complex. Therefore, in this research, a set of ISSR (Inter-Simple Sequence Repeat Polymorphic DNA) molecular markers was standardized, aiming to characterize A. colubrina and A. peregrina species and study the genetic diversity of three populations of each species located within a fragmented landscape in São Paulo State, southeastern Brazil. Seven ISSR markers (UBC 2, 820, 851, 858, 864, 866, and 886) that show polymorphism for both species were used. The Bayesian cluster, PCoA and dendrogram analysis show that the total sample divides into two groups corresponding to each species. Also, a genetic divergence (Gst = 0.143) and a high number of migrants per generation (Nm = 3.0) were detected between them. The Acol populations showed significantly higher values for mean genetic diversity (h = 0.30) than Aper (h = 0.25) (p < 0.05). The ISSR marker UBC2250bp showed species-specific electrophoretic fingerprints for both species. The molecular tools generated herein support the conservation of Anadenanthera sp. and the restoration of vegetation where the species naturally occurs.
1. Introduction
Seasonally dry tropical forests (SDTF), including semi-deciduous seasonal forests, occur on fertile soils favorable for agriculture and livestock production and are recognized as one of the most threatened tropical terrestrial ecosystems [1,2]. Fragments of these floras usually include individuals belonging to Anadenanthera (Fabaceae), a tree genus endemic to Latin America and the Caribbean with only two tree species, A. colubrina (Vell.) Brenan, and A. peregrina (L.) Speg. [3,4]. The species are hermaphroditic with abundant small, yellowish-white, fragrant flowers gathered in terminal inflorescences and present a mixed mating system [3,5,6,7]. When adults, the trees reach up to 35 m in height and 100 cm in diameter at breast height [5]. The fruits of both species mature from August to September; and together with their seeds, they have mainly barochoric dispersion [5,7]. The species present medicinal and therapeutic properties [3,8,9], are used in civil construction, and are well suited for reforestation of degraded areas [7,10].
Anadenanthera colubrina and A. peregrina occur in forest remnants in areas of litholic soils or with accentuated stoniness at Ribeirão Preto, a municipality in the interior of São Paulo State—SP (Brazil), where a large part of natural vegetation was converted into monocultures or urban areas [4]. However, since many of the morphological characteristics of Anadenanthera species are superimposed and species-specific characteristics are sometimes difficult to observe, their identification is complex [3]. The natural geographic distribution of these species is also superimposed [3], especially in areas such as Ribeirão Preto, where more than one type of vegetation occurs (mainly semi-deciduous and Cerrado) [4,6], and characteristics such as tree size, leaflet size, and bark vary widely. The trees can be identified most easily on the basis of their pod texture. A. colubrina has nitid, smooth and reticulated pods, while A. peregrina has dull, scurfy, and verrucose pods [3].
Recently, molecular studies with chloroplast simple sequence repeats (cpSSRs) [11], chloroplast DNA (cpDNA) [12], nuclear microsatellite (nSSR) [6,13,14,15], and the Internal Transcribed Spacer region of the rDNA (ITS-rDNA) [16] have been published related to genetic diversity [14], ex situ conservation [13], evolution [11,12,15], and mating systems [6] for Anadenanthera species. However, there has been no genetic study with inter-simple sequence repeat (ISSR) markers in Anadenanthera that may aid in taxonomic distinguishing of species [17].
The ISSR markers, a universal tool of easy access, are based on the amplification of regions between inversely oriented, closely spaced microsatellites (~200–2000 bp) [17]. These markers are highly polymorphic and have been used for evolutionary biology, phylogeny, genetic diversity, genome mapping, and gene tagging [18]. They are especially interesting in differentiating between species and cultivars such as Hordeum vulgare L. (Poaceae) [19], Mangifera indica L. (Anacardiaceae) [20], Brassica carinata A. Braun (Brassicaceae), Arabidopsis thaliana (L.) Heynh. (Brassicaceae), Phaseolus vulgaris L. (Fabaceae), Solanum tuberosum L. (Solanaceae), Nicotiana tabacum L. (Solanaceae), Helianthus annuus L. (Asteraceae) [21], and Stylosanthes species (Fabaceae) [22]. In Brazilian tree species, ISSR molecular markers have been widely used in genetic studies on Sansão-do-Campo (Mimosa caesalpiniifolia Benth., Fabaceae [23]), mangaba (Hancornia speciosa Gomes, Apocynaceae) [24]), carnauba (Copernicia prunifera (Mill.) H.E.Moore, Arecaceae [25]), aroeira (Myracrodruon urundeuva M.Allemão, Anacardiaceae [26]), jacarandá-da-bahia (Dalbergia nigra (Vell.) Allemão ex Benth., Fabaceae [27]), and Carrapateira (Metrodorea nigra A. St. Hil., Rutaceae [28]), among many others.
Herein, we standardized a set of ISSR (Inter-Simple Sequence Repeat Polymorphic DNA) molecular markers aiming to characterize A. colubrina and A. peregrina species and to study the genetic diversity of three populations of each species located within a fragmented landscape in São Paulo State, southeastern Brazil.
2. Materials and Methods
2.1. Identification and Sampling of Anadenanthera Populations
We identify the species using fruit characteristics [3]. Anadenanthera colubrina presents brightly colored fruits, with smooth to cross-linked leaflets, while the fruits of A. peregrina are opaque and verrucous (Figure 1). Samples were collected from three different angicals, or clusters, for each species, located within a fragmented landscape in São Paulo State, Ribeirão Preto Region, southeastern Brazil (Figure 2). The first A. colubrina angical, includes an urban area and is located on the Ribeirão Preto campus of the University of São Paulo—USP/RP (Acol USP; −21.1606609 −47.8618168; Figure 2), we collected leaf samples from 28 adult trees. The second A. colubrina angical (Acol BP; −21.2846649 −47.8142251; Figure 2) is located ~14 km from Acol USP in a rural area with recent deforestation near the district of Bonfim Paulista, SP. From this population, we collected leaf samples from 29 adult trees. In the third angical (Acol BM; −21.1725761 −47.8002551; Figure 2) we collected leaf samples from 30 adult trees. This population is located ~5 km from Acol USP and ~12 km from Acol BP and occurs within one of the last natural vegetation stands in the urban area of the municipality, Morro de São Bento Municipal Park, also known as fragment M103 [4].

Figure 1.
Leaves, trunks, and pods of the species Anadenanthera colubrina (A,B) and A. peregrina (C,D), observed in Acol BP (A,B) and Aper SP255 (C,D) angicals, Bonfim Paulista, district of Ribeirão Preto municipality, São Paulo State, southeastern Brazil. The most easily distinguishable characteristic of the two species is the integumentary surface of the pod. A. colubrina presents brightly colored fruits with smooth to cross-linked leaflets (B), while the fruits of A. peregrina are opaque and verrucous (D).
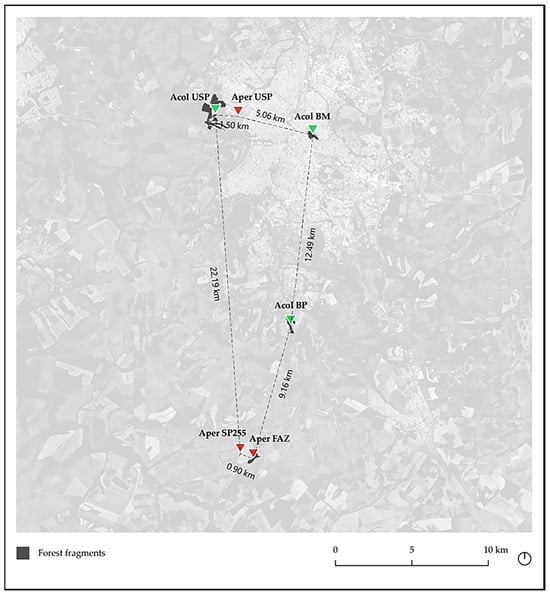
Figure 2.
The graph map of geographic location of the six studied Anadenanthera colubrina (Acol-green) and A. peregrina (Aper-red) populations in forest fragments in Ribeirão Preto, São Paulo, Brazil, made with QGIS3 (Source: Google Earth Satellite).
The first sample area of A. peregrina individuals is also located at USP/RP. This angical (Aper USP; −21.1609271 −47.8474085; Figure 2) consists of 16 isolated individuals, all of which were sampled, with limited association with other species. The second sample area for this species (Aper FAZ; −21.3641896 −47.8383455; Figure 2) is located approximately 22 km from Aper USP, near the district of Bonfim Paulista, from which 17 individuals were sampled. Finally, 0.65 km up the road from Aper Faz, 35 individuals were sampled along the Antonio Machado Sant’Anna highway (Aper SP255; −21.3605059 −47.8460526), from an angical with a prevalence of A. peregrina (approximately 50 individuals). The samples were stored at −20 °C until DNA extraction.
2.2. DNA Extraction, Amplification, and Electrophoresis
We used a modified CTAB protocol for genomic DNA extraction from leaf samples (Alzate-Marin et al. [29]). A total of 10 ISSR primers (University of British Columbia-UBC, Canada, Table 1) that previously showed amplification products in other plant species [22,28] were tested on 23 individuals of each species from the Acol USP and Aper USP populations. ISSR loci were amplified by PCR in a final volume of 12 μL using the GoTaq® Kit Promega (Promega, Madison, WI, USA), which consisted of 5 μL of nuclease-free water, 5 μL of master mix [400 nM of each deoxynucleotide and 3.0 mM of MgCl2], 1 μL of each primer, and 2.0 ng/μL of genomic DNA [28].

Table 1.
Sequence of ISSR (inter-simple sequence repeats) molecular markers tested [N = (A,G,C,T), R = (A,G), Y = (C,T), B = (C,G,T), D = (A,G,T), H = (A,C,T), and V = (A,C,G)]. AT = annealing temperature.
We performed the amplifications with a Mastercycler® pro-S Eppendorf thermocycler (Eppendorf, Hamburg, Germany) under the following conditions: 1 cycle at 95 °C for 10 min; 30 or 35 cycles of denaturation at 95 °C for 1 min, annealing at 50 or 55 °C for 45 s (according to Table 1); 72 °C for 1 min; and final extension at 72 °C for 7 min [22,28]. We separated the PCR products on 8% non-denaturing polyacrylamide gels stained with silver nitrate [30] and estimated allele sizes compared to a 50 bp DNA ladder (GE Healthcare).
2.3. Potential ISSR Species-Specific Markers
Fixed ISSR bands that showed potential species-specific amplification were evaluated in the DNA from 57 and 68 individuals of Acol and Aper, respectively. Amplified DNA samples of both species were run simultaneously run on each gel to facilitate the visualization of polymorphisms.
2.4. Statistical Analyses of Genetic Diversity
The polymorphism obtained through ISSR was tabulated according to the presence (1) or absence (0) of bands. Each polymorphic band was considered a bi-allelic locus, with an amplifiable allele and a null allele. The ancestry between individuals of the two species was generated through the Bayesian approach of the STRUCTURE program [31], assuming two different scenarios, K = 1 and K = 2. The GenAlEx 6.5 (The Australian National University, Acton, Australia) software [32] was employed for the computation of the genetic distance matrix to generate a scatterplot through Principal Coordinates Analysis (PCoA) and to calculate the genetic distances among populations (Nei (1978) [33]) and the Nei’s measures, including the percentage of polymorphic loci (PPL), number of alleles (NA), and h = Nei’s genetic diversity and its standard error (SE) values. Additionally, a genetic dissimilarity dendrogram was constructed using the UPGMA algorithm within the MEGA 11 (Tokyo Metropolitan University, Tokyo, Japan) software [34]. We obtained the parameters coefficient of differentiation (Gst) (Nei 1973 [35]) and the number of migrants per generation (Nm = 0.5 [(1—Gst)/Gst]) using the POPGEN32 (Version 1.31) (University of Alberta, Edmonton, Canada) software [36]. The means were tested for normal distribution with the Shapiro–Wilk test using the PAST (University of Oslo, Oslo, Norway) software [37]. Differences between the indices NA and h between and within species were calculated with ANOVA and a t test (p < 0.05), respectively [24].
3. Results
3.1. Analysis of Ancestry and Clusters
The ancestral analysis suggests that the populations of the two studied species form two genetic groups (Figure 3). Of the evaluated scenarios (K = 1 and K = 2), the model that assumed the existence of two distinct genetic groups (K = 2) presented the highest logarithmic probability (log P (X|K)). This confirms the initial identification of individuals within the species. The average Alpha values suggest a limited mixture between species (Alpha = 0.0518). We found that the species A. colubrina showed that 98% of its genetic composition corresponds to the same genetic structure. A. peregrina showed a greater genetic mix, comprising 91% of the genetic information of this species and 9% of the genetic information from the other studied species (Figure 3). Only one individual of A. peregrina from the population Aper SP255, represented as 90(4), presented a higher level of genetic composition from A. colubrina, suggesting some degree of hybridity, and was excluded from subsequent analyses to mitigate sampling bias. Some individuals from both species also presented the genetic composition of the other species (Figure 3).
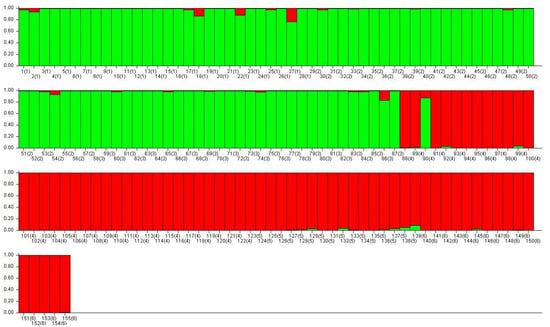
Figure 3.
Ancestry of Anadenanthera colubrina (green) and A. peregrina (red), analyzed with ISSR markers, showing that the species form two genetic groups (Alpha = 0.0518) and that populations within each species form a single group. In parentheses, the number of populations to which each individual belongs (1–3: Acol USP, Acol BP, and Acol BM; 4–6: Aper SP255, Aper USP, and Aper FAZ).
We observed in the Principal Coordinates Analysis (PCoA) based on the genetic distance matrix, two perfectly differentiable groupings containing individuals of each species studied (Figure 4A). The dendrogram shows that individuals in the populations form clusters; however, individuals from the Acol BM population constitute a single cluster while some individuals from the Acol USP and Acol BP populations appear to be part of a larger group (Figure 4B).
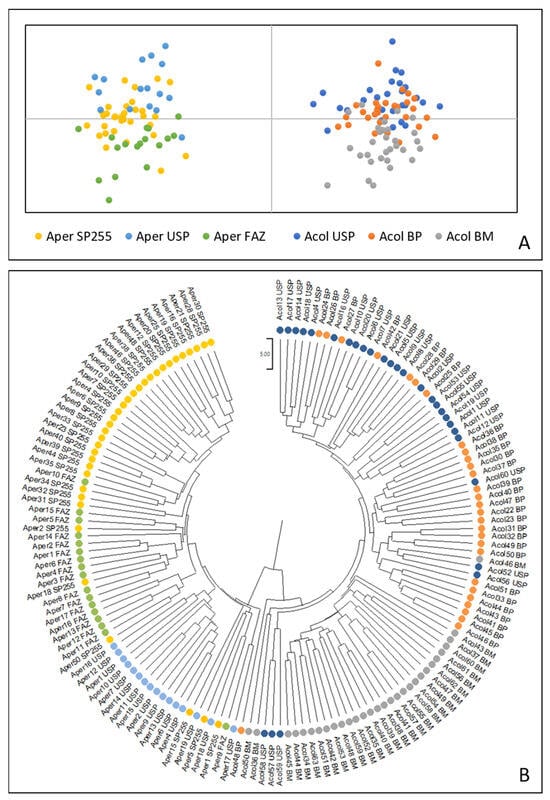
Figure 4.
Principal Coordinates Analysis (PCoA) (A) and UPGMA dendrogram (B) indicating the genetic relationships among individual trees of Anadenanthera colubrina (Acol) and A. peregrina (Aper) for seven ISSR markers. In both analyses, the six populations were grouped into two major clusters: Acol and Aper.
Six Acol individuals from the three populations are isolated but still included in the larger Acol cluster (Figure 4B). Individuals of the Aper populations constitute separate clusters, and some individuals from the Aper SP255 population are dispersed among other Aper populations. Seven individuals from all Aper populations appear to be isolated but included in the larger Aper group. In general, these graphs demonstrate that the two species can be genetically differentiated into two groups and that subpopulations within each species can show greater similarity.
3.2. Gst Analysis and Genetic Distances
We found a genetic divergence (Gst) of 0.143 and a high number of migrants per generation (Nm = 3.0) among species (Table 2), which may be evidence of hybridization, as observed in Figure 3. We also observed a greater genetic divergence within A. peregrina (0.102) than A. colubrina (0.076) populations (Table 2). The number of migrants per generation within populations was higher in Acol (Nm = 6.1) than in Aper (4.4) (Table 2).

Table 2.
Mean values of genetic differentiation between the populations of Anadenanthera colubrina and A. peregrina. Gst = coefficient of population differentiation (Nei 1973); Nm = migrants per generation.
Aper USP/Aper FAZ populations that are located ~22 km apart showing the greatest genetic distance (6%), and Acol USP/Acol BP, located at a distance of ~14 km, presenting the lowest genetic distance (2.4%) (Table 3, Figure 2). Despite the short geographic distance (~5 km) between Acol USP/Acol BM (Figure 2), these populations showed a high genetic distance (5.7%), which is likely related to the urban isolation of the Acol BM population (Figure 2).

Table 3.
Nei’s unbiased measure of genetic distance (Nei 1978).
3.3. Genetic Diversity among Populations of Anadenanthera colubrina and A. peregrina
A similar percentage of polymorphic loci (mean = 82.87%) and number of de alleles (mean = 1.72%) were observed between the species. The Acol populations showed higher values of mean genetic diversity (h = 0.30) than Aper (h = 0.25; p < 0.05) (Table 4). The genetic diversity and number of alleles were significantly different among Aper populations (p < 0.05), with the highest values for Aper USP (h = 0.26) and Aper SP255 (NA = 1.84), respectively (Table 4).

Table 4.
Genetic diversity in three populations of Anadenanthera colubrina and A. peregrina assessed with ISSR molecular markers. N = Number of individuals, LN = loci number, PPL = percentage of polymorphic loci, NA = Number of alleles, and h = Nei’s genetic diversity.
3.4. Species-Specific Molecular Marker Analysis
Among the ISSR molecular markers that showed polymorphism (UBC 2, 820, 851, 858, 864, 866, and 886), UBC 2 amplified two likely species-specific bands of 250 and 370 bp present in A. colubrina and absent in A. peregrina. However, the electrophoretic profiles of the band UBC2250bp were the clearest and easiest to identify across both species (Figure 5). We visually analyzed the reliability of the ISSR UBC2250bp in 57 and 68 individuals from the Acol and Aper populations used in this study, respectively. This band matched A. colubrina (+/presence) and A. peregrina (−/absence) in 98.25 and 97.06% of samples, respectively. Thus, UBC2250bp can potentially identify individuals of both species using direct amplification of their DNA.
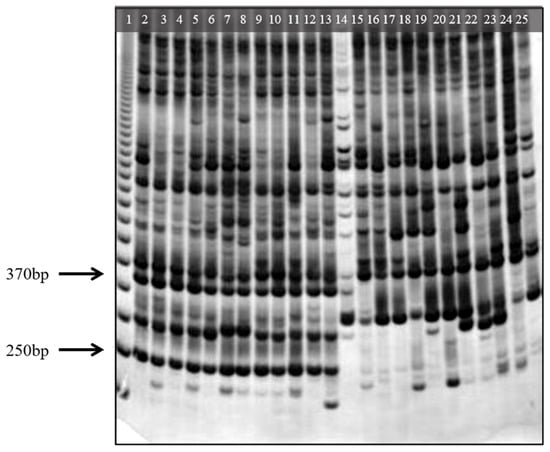
Figure 5.
Electrophoresis amplification products obtained with ISSR UBC 2 on non-denaturing polyacrylamide gel. Channel 1 corresponds to the molecular weight marker 50 pb (GE Healthcare). Channels 2–13 and 14–25 correspond to the amplification products of Anadenanthera colubrina and A. peregrina DNA, respectively. Arrows indicate the potential species-specific ISSR bands present in A. colubrina and absent in A. peregrina.
4. Discussion
Species and populations of trees with vast geographic ranges, outcrossing breeding systems, and seed distribution by animals or wind show higher genetic diversity than species with other combinations of traits [38]. Anadenanthera colubrina and A. peregrina are mixed-mating reproductive species with barochoric seed dispersal, with A. peregrina preferentially allogamous [12]. Its potential pollinators are Apidae bees Apis mellifera, and native Trigona spinipes [39]. Our study shows a degree of hybridization between the studied Anadenanthera species and different levels of genetic distance, lower Gst values, and an increased number of migrants per generation among adult populations. Additionally, A. colubrina exhibits higher genetic diversity (hAcol = 0.30) compared to A. peregrina (hAper = 0.25). These levels may reflect the reproductive characteristics of the species, the degree of isolation of the populations, and their spatial distribution (crossings between nearby trees with moderate pollen dispersal coming from outside the forest fragments) since individuals tend to form dense groups, as studied in the same region [6].
In comparison with other Brazilian forest species, Acol and Aper show higher genetic diversity than both legume Mimosa caesalpiniifolia (h = 0.220 [23]) and the expected by the Fabaceae family (h = 0.18 [40]), and lower than Copernicia prunifera (h = 0.327 [25]), and legume Dalbergia nigra (h = 0.36) [27]. The Aper diversity levels were similar to those of Metrodorea nigra, studied in the same region (h = 0.25 [28]), Hancornia speciosa (h = 0.26 [24]), and Myracrodruon urundeuva (h = 0.27 [26]).
The GST values found in this study (Acol = 0.076, Aper = 0.102) were lower than those expected for family Fabaceae (0.277) and mixed mating species with seed dispersal by gravity (0.25) [40], although these comparisons should be taken with caution [41] if GST was calculated in a different way than that used by Hamrick and Godt [40,41]. The high Nm values of populations of each species resembled some of those expected for tropical trees [42], indicating historical gene exchange between populations despite their current geographic separation.
Furthermore, our results show that the two Anadenanthera species show a low genetic divergence (Gst = 0.143), mirroring the low variability of some phenotypic characteristics, such as flowers and foliage. Nonetheless, the Bayesian cluster, PCoA, and dendrogram analyses discriminated between the two species between the six populations, indicating that the ISSR markers standardized here can be used to identify individuals in the absence of diagnostic traits, such as flowers and fruits, and at any stage of development, such as seedlings. In addition, although it could be linked to the population structure, genetic drift, and isolation of the studied populations, the electrophoretic profile amplified by the ISSR UBC2250bp has the potential to be used as a species-specific marker and should be validated with populations from other areas.
Anadenanthera colubrina and A. peregrina are dominant trees that occur naturally in the Brazilian deciduous and Cerrado forests of Ribeirão Preto and other parts of Brazil and South America [4,5,6,7,11,12,43,44]. These species show resilience in the face of fragmentation, surviving in areas unsuitable for agriculture due to their reproductive characteristics and apparent ease of seed germination and seedling establishment [7,45,46]. The strong dominance of Anadenanthera species, combined with allelopathic effects [44,47,48], inhibits the natural regeneration of other native woody species. Therefore, groups of individuals of these species are frequently found in almost monospecific forest stands [4,6,43,44], playing a role as stepping stones and connections between the few remaining fragments in the region [4,28,49,50]. However, our data show that more than geographic distance, it is urban isolation that most affects the genetic distance between populations, as can be observed in the urban Acol BM and Aper USP clusters (Figure 2 and Table 3). Therefore, it would be necessary to create corridors that connect these populations in the region, which can support the conservation of these angicals. Also, Acol BP and Acol SP255 clusters, with the highest average of alleles, will be an important source of seed for use in the restoration of vegetation where the species naturally occurs.
Author Contributions
Conceptualization, methodology, A.L.A.-M. and F.B.-A.; investigation, formal analysis, F.B.-A.; software, R.M.M.F. and F.B.-A.; resources, L.M.B. and C.A.M.; writing—original draft preparation, A.L.A.-M.; writing—review and editing, A.L.A.-M., F.B.-A., L.M.B., R.M.M.F. and C.A.M.; supervision, project administration, funding acquisition, A.L.A.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by a grant from São Paulo Research Foundation to A.L.A.M.: FAPESP (Grant 2013/01265-8). We also acknowledge the support of CAPES-PROEX grants. F.B.A. was supported by FAPESP (Grant TTIII 2013/18633-0) and CNPq (Grant SI 134699/2012-2, and DSc 141921/2019-6) fellowships. R.M.M.F. was supported by DSc CAPES fellowship (Finance Code 001). A.L.A.M. was supported by research assistantships from CNPq (PD Senior Grant 150277/2009-1, PV Grant 300140/2011-8 and PD Senior Grant 150737/2014-9). C.A.M. is a research fellow from CNPq (Grant 304686/2022-3).
Data Availability Statement
Data are unavailable due to privacy.
Acknowledgments
The authors thank Alexandre Carvalho Gouvêa (Head of Bosque Zoológico Municipal Ribeirão Preto/SP) for providing authorization to collect Anadenanthera samples in BM. Also, we thank Olga Kotchetkoff-Henriques (Prefeitura Municipal Ribeirão Preto/SP), Juliana M. Feres, and Biologists Paulo B. Rodriguez and Rafael P. Campos for collaborating on the sample collections. Access authorization (A380C57) by SISGEN (Sistema de Gestão do Patrimônio Genético e do Conhecimento Tradicional Associado). We thank the architect, Fabio A. Alzate Martinez (TU-Delft Netherlands), for making the graph map of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vieira, D.L.M.; Scariot, A. Principles of Natural Regeneration of Tropical Dry Forests for Restoration. Restaur. Ecol. 2006, 14, 11–20. [Google Scholar] [CrossRef]
- Pennington, R.T.; Lewis, G.P.; Ratter, J.A. An Overview of the Plant Diversity, Biogeography and Conservation of Neotropical Savannas and Seasonally Dry Forests. In Neotropical Savannas and Seasonally Dry Forests: Plant Diversity, Biogeography, and Conservation; Pennington, R.T., Lewis, G.P., Ratter, J.A., Eds.; CRC Press: Boca Raton, FL, USA, 2006; pp. 1–29. [Google Scholar] [CrossRef]
- Altschul, S.R. A taxonomic study of the genus Anadenanthera. Contrib. Gray Herb. Harv. Univ. 1964, 193, 3–65. Available online: http://www.jstor.org/stable/41764816 (accessed on 8 August 2023).
- Kotchetkoff-Henriques, O.; Joly, C.A.; Bernacci, L.C. Soil and floristic composition of native vegetation remnants relationship in the municipality of Ribeirão preto, sp. Braz. J. Bot. 2005, 28, 541–562. [Google Scholar] [CrossRef]
- Carvalho, P.E.R. Angico-Branco. Embrapa—Circular Técnica 56. Colombo. PR Novembro. 2002. Available online: http://www.infoteca.cnptia.embrapa.br/bitstream/doc/306306/1/CT0056.pdf (accessed on 8 August 2023).
- Feres, J.M.; Nazareno, A.G.; Borges, L.M.; Guidugli, M.C.; Bonifacio-Anacleto, F.; Alzate-Marin, A.L. Depicting the mating system and patterns of contemporary pollen flow in trees of the genus Anadenanthera (Fabaceae). PeerJ 2021, 9, e10579. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, H. Árvores Brasileiras—Manual de Identificação e Cultivo de Plantas Arbóreas Nativas no Brasil; Instituto Plantarum: Nova Odessa, Brazil, 2002; Volumes 2–3, p. 368. [Google Scholar]
- Weber, C.R.; Soares, C.M.L.; Lopes, A.B.D.; Silva, T.S.; Nascimento, M.S.; Ximenes, E. Anadenanthera colubrina: A therapeutic potential study. Ver. Bras. Farm. 2011, 92, 235–244. [Google Scholar]
- Chaves, T.L.; Ricardo, L.; Paula-Souza, J.; Brandão, M.G.L. Useful Brazilian plants under the view of the writer-naturalist João Guimarães Rosa. Rev. Bras. Farmacogn. 2015, 25, 437–444. [Google Scholar] [CrossRef][Green Version]
- Durigan, G.; Silveira, E.R. Riparian forest restoration in cerrado, Assis, SP, Brazil. Sci. For. 1999, 56, 135–144. Available online: http://www.ipef.br/publicacoes/scientia/nr56/cap10.pdf (accessed on 29 March 2023).
- Barrandeguy, M.E.; García, M.V.; Prinz, K.; Pomar, R.; Finkeldey, R. Genetic structure of disjunct Argentinean populations of the subtropical tree Anadenanthera colubrina var. cebil (Fabaceae). Plant Syst. Evol. 2014, 300, 1693–1705. [Google Scholar] [CrossRef]
- Barrandeguy, M.E.; Mogni, V.; Zerda, H.; Savino, C.; Dusset, F.; Prado, D.E.; García, M.V. Understanding the spatio-temporal dynamics of Anadenanthera colubrina var. cebil in the biogeographical context of Neotropical seasonally dry forest. Flora 2022, 295, 152149. [Google Scholar] [CrossRef]
- Cortelete, M.A.; Silva Júnior, A.L.; Pereira, M.L.S.; Miranda, F.D.; Caldeira, M.V.W. Molecular characterization as strategy for ex situ conservation of Anadenanthera peregrina (L.) Speg. Sci. For. 2021, 49, e3443. [Google Scholar] [CrossRef]
- García, M.V.; Barrandeguy, M.E.; Prinz, K. Contemporary climate influence on variability patterns of Anadenanthera colubrina var. cebil, a key species in seasonally dry tropical forests. J. For. Res. 2022, 33, 89–101. [Google Scholar] [CrossRef]
- Mangaravite, E.; Silveira, T.C.; Vinson, C.C.; Bueno, M.L.; Silva, R.S.; Carniello, M.A.; Veldman, J.W.; Garcia, M.G.; Oliveira, L.O. Unlocking the secret diversity of Anadenanthera: Insights from molecular genetics of four evolving species. Bot. J. Linn. Soc. 2023, 204, boad037. [Google Scholar] [CrossRef]
- Viana, M.L.; Giamminola, E.; Russo, R.; Ciaccio, M. Morphology and genetics of Anadenanthera colubrina var. cebil (Fabaceae) tree from Salta (Northwestern Argentina). Rev. Biol. Trop. 2014, 62, 757–767. Available online: https://www.redalyc.org/articulo.oa?id=44931383029 (accessed on 8 August 2023). [CrossRef][Green Version]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple se-quence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, M.; Siddiq, E.A. Inter simple sequence repeat (ISSR) poly-morphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Tahir, N.A.-R.; Lateef, D.; Rasul, K.S.; Aziz, R. Assessment of genetic variation and population structure in Iraqi barley accessions using ISSR, CDDP, and SCoT markers. Czech, J. Genet. Plant Breed. 2023, 59, 148–159. [Google Scholar] [CrossRef]
- Uddin, M.S.; Cheng, Q. Recent Application of Biotechniques for the Improvement of Mango Research; Poltronieri, P., Hong, Y., Eds.; Applied Plant Genomics and Biotechnology, Woodhead Publishing: Cambridge, UK, 2015; pp. 195–212. [Google Scholar] [CrossRef]
- Bornet, B.; Branchard, M. Nonanchored Inter Simple Sequence Repeat (ISSR) Markers: Reproducible and Specific Tools for Genome Fingerprinting. Plant Mol. Biol. Report 2001, 19, 209–215. Available online: https://link.springer.com/content/pdf/10.1007/BF02772892.pdf (accessed on 8 August 2023). [CrossRef]
- Alzate-Marin, A.L.; Costa-Silva, C.; Rivas, P.M.S.; Bonifacio-Anacleto, F.; Santos, L.G.; Moraes Filho, R.M.; Martinez, C.A. Diagnostic fingerprints ISSR/SSR for tropical leguminous species Stylosanthes capitata and Stylosanthes macrocephala. Sci. Agric. 2020, 77, e20180252. [Google Scholar] [CrossRef]
- Araújo, F.D.S.; Felix, F.C.; Ferrari, C.D.S.; Vieira, F.D.A.; Pacheco, M.V. Seed quality and genetic diversity of a cultivated population of Mimosa caesalpiniifolia BENTH. Rev. Caatinga 2020, 33, 1000–1006. [Google Scholar] [CrossRef]
- Silva, A.V.C.D.; Amorim, J.A.E.; Vitória, M.F.D.; Ledo, A.D.S.; Rabbani, A.R.C. Characterization of trees, fruits and genetic diversity in natural populations of mangaba. Agric. Sci. 2017, 41, 255–262. [Google Scholar] [CrossRef]
- Fajardo, C.G.; Silva, R.A.R.; Chagas, K.P.T.; Vieira, F.A. Genetic and phenotypic association of the carnauba palm tree evaluated by inter-simple sequence repeat and biometric traits. Genet. Mol. Res. 2018, 17, gmr18018. [Google Scholar] [CrossRef]
- Lopes, J.S.; Costa, M.R.J.; Arriel, D.A.A. Genetic diversity of potential mother trees of Myracrodruon urundeuva Allemão in a remnant population from Brazilian Cerrado using ISSR. Adv. For. Sci. 2020, 7, 1017–1024. [Google Scholar] [CrossRef]
- Santos, A.R.; Gonçalves, E.O.; Silva Júnior, A.L.; Gibson, E.L.; Araújo, E.F.; Mi-randa, F.D.; Caldeira, M.V.W. Diversity and genetic structure in a mini-garden of Dalbergia nigra: A tree threatened with extinction in the Atlantic Forest. Plant Gene 2021, 27, 100304. [Google Scholar] [CrossRef]
- Moraes Filho, R.M.; Bonifacio-Anacleto, F.; Alzate-Marin, A.L. Fragmentation effects and genetic diversity of the key semidecidual forest species Metrodorea nigra in Southwestern Brazil. Genet. Mol. Res. 2015, 14, 3509–3524. [Google Scholar] [CrossRef] [PubMed]
- Alzate-Marin, A.L.; Guidugli, M.C.; Soriani, H.H.; Martinez, C.A.; Mestriner, M.A. An efficient and rapid DNA minipreparation procedure suitable for PCR/SSR and RAPD analyses in tropical forest tree species. Braz. Arch. Biol. Technol. 2009, 52, 1217–1224. [Google Scholar] [CrossRef]
- Sanguinetti, C.J.; Dias, E.N.; Simpson, A.J.G. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 1994, 17, 914–921. [Google Scholar] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. Available online: http://www.genetics.org/content/genetics/155/2/945.full.pdf (accessed on 15 November 2022). [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 586–590. Available online: http://www.genetics.org/content/genetics/89/3/583.full.pdf (accessed on 17 November 2022). [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC427228/ (accessed on 17 November 2022). [CrossRef] [PubMed]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE, Version 1.31. Microsoft Windows-Based Freeware for Population Genetic Analysis. University of Alberta/CIFOR: Edmonton, Alberta, 1999. Available online: https://sites.ualberta.ca/~fyeh/popgene.pdf (accessed on 17 November 2022).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 178kb. Available online: https://palaeo-electronica.org/2001_1/past/past.pdf (accessed on 20 November 2022).
- Hamrick, J.L.; Godt, M.J.W. Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant Species. New For. 1992, 6, 95–124. Available online: https://link.springer.com/content/pdf/10.1007/BF00120641.pdf (accessed on 3 August 2023). [CrossRef]
- Borges, L.A.; Machado, I.C.; Lopes, A.V. Bee pollination and evidence of substitutive nectary in Anadenanthera colubrina (Leguminosae-Mimosoideae). Arthropod-Plant Interact. 2017, 11, 263–271. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Phil. Trans. R. Soc. Lond. 1996, 351, 1291–1298. [Google Scholar] [CrossRef]
- Culley, T.M.; Wallace, L.E.; Gengler-Nowak, K.M.; Crawford, D.J. A comparison of two methods of calculating GST, a genetic measure of population differentiation. Am. J. Bot. 2002, 89, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.S. Dinâmica da movimentação dos alelos: Subsídios para conservação e manejo de populaçõeses naturais em plantas. Braz. J. Genet. 1996, 19, 37–47. [Google Scholar]
- Prado, D.E.; Gibbs, P.E. Patterns of species distribution in the dry seasonal forests of South America. Ann. Miss. Bot. Gard. 1993, 80, 902–927. [Google Scholar] [CrossRef]
- Oliveira-Filho, A.T.; Curi, N.; Vilela, E.A.; Carvalho, D.A. Effects of Canopy Gaps, Topography, and Soils on the Distribution of Woody Species in a Central Brazilian Deciduous Dry Forest. Biotropica 1998, 30, 362–375. Available online: http://www.jstor.org/stable/2389121 (accessed on 20 September 2023). [CrossRef]
- Miranda, C.C.; Souza, D.M.S.; Manhone, P.R.; Oliveira, P.C.; Breier, T.B. Germinação de sementes de Anadenanthera peregrina (L.) Speg. com diferentes substratos em condições laboratoriais. Floresta e Ambiente 2012, 19, 26–31. Available online: http://www.floram.org/files/v19n1/v19n1a4.pdf (accessed on 20 September 2023). [CrossRef]
- Castro, L.E.; Guimarães, C.C.; Faria, J.M.R. Physiological, cellular and molecular aspects of the desiccation tolerance in Anadenanthera colubrina seeds during germination. Braz. J. Biol. 2017, 77, 4–780. [Google Scholar] [CrossRef]
- Silva, L.M.B.; Barbosa, D.C.A. Growth and survival of Anadenanthera macrocarpa (Benth.) Brenan (Leguminosae), in an area of caatinga, Alagoinha, PE. Acta Bot. Bras. 2000, 14, 251–261. [Google Scholar] [CrossRef]
- Silva, R.M.G.; Saraiva, T.S.; Silva, R.B.; Gonçalves, L.A.; Silva, L.P. Potencial alelopático de extrato etanólico de Anadenanthera macrocarpa e Astronium graveolens. Biosci. J. 2010, 26, 632–637. [Google Scholar]
- Alzate-Marin, A.L.; Bonifacio-Anacleto, F.; Moraes Filho, R.M.; Machado, G.P.; Nazareno, A.G. Genetic analysis across life stages of Metrodorea nigra (Rutaceae) in a population located in an urban landscape of Southeastern Brazil using a new set of microsatellite markers. Braz. J. Bot. 2016, 39, 795–799. [Google Scholar] [CrossRef]
- Alzate-Marin, A.L.; Ferreira-Ramos, R.; Guidugli, M.; Martinez, C.A.; Mestriner, M.A. Genetic diversity assessed in individuals of Aspidosperma polyneuron and Cariniana estrellensis used as seed donors in an forest gene bank. BMC Proc. 2011, 5 (Suppl. 7), 8. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).