Abstract
Background/Objectives: Intrachromosomal amplification of chromosome 21 (iAMP21) represents a rare and heterogeneous distinct cytogenetic subgroup of B-cell precursor acute lymphoblastic leukemia (ALL) initially associated with a poor prognosis. Treatment intensification with additional doses of methotrexate and asparaginase was associated with better treatment outcomes. Methods: In this retrospective single-center study, we evaluated the impact of iAMP21 on treatment outcome in a cohort of pediatric patients treated with an intensified asparaginase regimen and describe the genomic landscape of four patients with iAMP21. Results: Four out of 89 patients > 1 year old were classified as iAMP21 positive. Five-year event-free survival (EFS) was inferior in the iAMP21-positive group: 25% versus 85.6% (p = 0.001). The cumulative incidence of relapse and treatment-related mortality were 50% vs. 9.9% and 0% vs. 2.38%, respectively, in the iAMP21-positive and non-iAMP21 groups (p = 0.02 and 0.76, respectively). These results did not translate into a significant difference in overall survival: 100% vs. 93.7% (p = 0.6). The presence of iAMP21 (HR 7.68, 95% CI 2.04–29.05; p = 0.002) and a measurable residual disease ≥1% after induction on day +33 (HR 8.82, 95% CI 2.6–29.91; p = 0.001) retained significant negative impact on EFS in multivariate analysis. Conclusions: We found an independent significant prognostic impact of iAMP21 on EFS among pediatric patients with ALL, and clinical presentation and early treatment response did not classify these patients as HR. Diverse genetic backgrounds among iAMP21-positive patients might influence the treatment response and outcome of this heterogeneous disease.
1. Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignant disease in children, and 85% of cases are from B-cell precursor (BCP) origin [1]. Patient stratification in different risk groups with diverse treatment intensity has largely contributed to the improvement of treatment outcomes of childhood ALL during last decades [2,3]. Patient age, white blood cell count (WBC) at presentation, leukemia immunophenotype, certain disease genetic determinants of outcome, and early response to treatment are factors considered for stratification in different protocols [2,4,5,6]. Reverse transcriptase polymerase chain reaction (PCR), conventional cytogenetics, and fluorescence in situ hybridization (FISH) are considered appropriate techniques for the identification of these genetic abnormalities. Additionally, flow cytometry (FC) for determination of the DNA index is applied for the detection of changes in chromosome numbers [2]. Early treatment response assessment through evaluation of measurable residual disease (MRD) allowed the improvement of results by the adaptation of treatment intensity according the quantification of MRD at different time-points [7,8,9,10].
Intrachromosomal amplification of chromosome 21 (iAMP21) represents a rare genetic alteration recognized as a distinct cytogenetic subgroup of BCP ALL since 2003 [11]. The diagnosis of iAMP21 has largely relied upon the demonstration of multiple signals of the RUNX1 gene on chromosome 21 using FISH [2,3,12,13]. Diverse secondary genetic abnormalities that differ from other BCP ALL subtypes are frequently described in iAMP21-positive ALL cases, defining a high heterogenous disease [3,6,14,15,16].
Early studies associated the presence of iAMP21 with a poor prognosis, with relapse being the most prevalent event [4,5,12,13]. However, treatment intensification with the administration of additional doses of methotrexate and asparaginase was associated with better treatment outcomes [5,6,12,13]. After relapse, iAMP21 has been considered as a high-risk cytogenetic feature by some but not all study groups [17].
We evaluated the impact of iAMP21 on treatment outcome in a cohort of pediatric patients treated with an intensified asparaginase regimen. iAMP21 had a significant impact on event-free survival (EFS) but did not affect overall survival (OS), and all patients who experienced an event could be rescued with second-line treatment.
We further describe the genomic landscape of four patients with this rare genetic ALL subtype.
2. Materials and Methods
In this retrospective single-center study, we aimed to analyze the impact of iAMP21 in the clinical outcome of pediatric patients diagnosed with ALL who underwent treatment according to the Spanish “LAL/SEHOP-PETEHEMA 2013” guidelines regimen. These recommendation guidelines were launched by the Leukemia Working Group of the Spanish Society of Pediatric Hematology and Oncology (SEHOP) and were approved by our local institutional review board in December 2013. Written informed consent before registration and treatment was provided by all patients or legal guardians including consent for subsequent analyses of stored biological samples.
In these guidelines, patients are stratified into three different risk groups, i.e., standard- (SR), intermediate- (IR) and high-risk (HR), according to patient age, WBC, leukemia immunophenotype, central nervous system (CNS) and/or testicular involvement, cytogenetics, and early treatment response (Supplementary Table S1).
Patients younger than 1 year were treated differently and were excluded from this analysis.
Considering results from leukemia immunophenotype and DNA index results provided by FC, conventional cytogenetics, FISH, and PCR analysis, all other patients (≥1 year old) were classified into 10 different biological subtypes according to the World Health Organization (WHO) 2008 classification (Table 1) [18].

Table 1.
Biological classification of patients at primary diagnosis.
The definition of iAMP21-positive cases was based on the identification of ≥5 copies of RUNX1 in an interphase FISH study [3,11,12,14].
In this protocol, the presence of iAMP21 preludes the stratification of patients into the SR group but does not represent a HR criterion itself. Patients with Philadelphia chromosome-positive ALL (BCR/ABL1 rearrangement) were allowed to participate and receive the HR regimen in combination with imatinib (300 mg/m2/day) from day +15 until the end of treatment.
The “LAL/SEHOP-PETHEMA 2013” treatment details are described in the Supplementary Material (Supplementary Tables S2 and S3). In brief, all patients receive a 4-week induction regimen with 4 drugs (induction “IA”) followed by an early intensification (induction “IB”). SR and IR patients are then scheduled to receive the same 8-week extra-compartment chemotherapy consolidation followed by a reinduction phase before maintenance, and IR patients receive an asparaginase-intensified protocol with 10 additional doses of pegylated (PEG-) asparaginase (1000 U/m2 every 2 weeks) during the first 20 weeks of maintenance. HR patients receive three chemotherapy intensification blocks before intensified reinduction chemotherapy and maintenance. Criteria for allogeneic stem cell transplantation (SCT) are restricted to HR patients and poor early MRD response (Supplementary Tables S1–S3).
Early treatment response was evaluated based on morphological assessment of circulating blast count at day +8 and morphological evaluation as well as FC assessment of MRD in bone marrow (BM) at days +15, +33, and +78. HR patients had an additional BM evaluation after the third consolidation block. The indications for SCT in the HR group are provided in Supplementary Table S3.
Our primary objective was to compare the outcome of patients with iAMP21 versus all other patients. We also aimed to analyze and compare clinical presentation as well as early treatment response in these two different groups and to analyze the impact on EFS of the following variables: presence of iAMP21, sex, age and ethnic group, WBC at primary diagnosis, National Cancer Institute (NCI) risk score, CNS involvement, kinetics of early MRD response, as well as initial and final risk-group category.
The Kaplan–Meier method and the log-rank test were used to generate and compare OS and EFS curves. Cumulative incidence of treatment-related mortality (TRM) and relapse (CIR) were compared using the Fine and Grey test. For multivariate analysis, the Cox proportional hazard regression model with estimate of hazard ratios was applied considering any individual risk factors identified in the univariate analysis. Refractoriness to induction was censored as an event at day zero. For other comparisons the Fisher’s exact, Student’s t, or Mann–Whitney tests were applied as appropriate. RStudio version 4.2.2 was used for all statistical analyses.
In order to try to gain insights into possible biological findings affecting outcomes, we retrospectively analyzed stored BM samples from primary diagnosis from four patients with iAMP21-positive ALL using multiplex ligation-dependent probe amplification (MLPA) and next-generation sequencing (NGS).
3. Results
3.1. Patient Characteristics, Risk-Group Assignment, and Early Treatment Response Evaluation
We collected data from 89 pediatric patients diagnosed with ALL at our institution from 27 June 2013 to 21 December 2023, who were included in the “LAL/SEHOP-PETEHEMA 2013” registry. Median age at diagnosis was 7 years (range 1 to 16). Four patients (1 male and 3 females) were classified as iAMP21-positive ALL according FISH analysis (Figure 1), and 85 were classified as non-iAMP21 patients. Table 2 includes clinical characteristics of patients at primary diagnosis, NCI risk stratification, and initial risk-group assignment according to the “LAL/SEHOP-PETHEMA 2013” protocol.
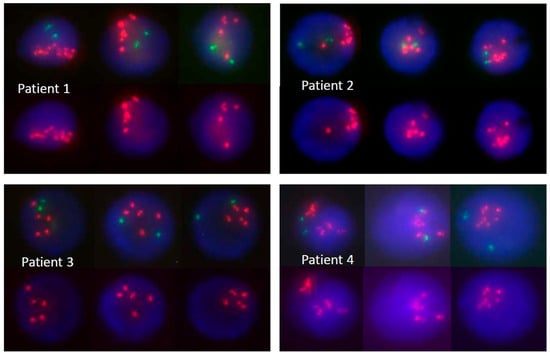
Figure 1.
Bone marrow FISH analysis from four patients diagnosed with iAMP21-positive ALL showing >5 signals of RUNX1 (red). Three nuclei from each patient are presented with upper rows showing both RUNX1 (red) and ETV6 (green) signals and bottom rows depicting only RUNX1 signals.

Table 2.
Clinical and biological characteristics of patients and early treatment response evaluation results.
Although median WBC was significantly lower in the iAMP21-positive group (2.49 × 109/L versus 8.51 × 109/L; p = 0.046), the proportion of high-risk patients according to NCI criteria was not significantly different. The leukemia immunophenotype was common (B-II) in three iAMP21-positive patients and pre-B (B-III) in 1 patient. There were 15 cases (17.6%) of T-lineage immunophenotype in the non-iAMP21 group [19]. CNS involvement did not affect iAMP21-positive patients and was present (CNS3) in four (4.7%) non-iAMP21 patients.
Conventional cytogenetics was non-informative (no available metaphases) in two AMP21-positive cases; however, flow cytometry study informed a DNA index of 1 in all four patients. Three iAMP21-positive patients had additional findings on FISH study, such as CDKN2A deletion, monosomy 11q, and tri-/tetrasomy 4 (Supplementary Table S4).
All patients with iAMP21 were initially stratified within the IR group. According to protocol specifications, all received an intensified PEG-asparaginase schedule with 10 additional doses every 2 week during the first 20 weeks of maintenance. Six out of 85 with non-iAMP21 ALL were stratified within the HR group (Supplementary Table S1).
Results from early treatment response evaluations are presented in Table 2. All four patients with iAMP21-positive ALL had <1 × 109/L circulating blasts in peripheral blood at day +8; however, one patient was refractory to induction chemotherapy (>25% blasts in BM at day +33). The remaining three patients had good early response to treatment with <5% blasts in BM at days +15 and +33 as well as BM MRD < 0.1% at day +15 and at subsequent scheduled evaluations. Accordingly, these three patients remained within the IR group at final stratification. In contrast, 13 patients in the non-iAMP21 group moved from the SR and IR to the HR group after early response evaluation.
3.2. Outcome and Prognostic Factors
One patient in the iAMP21-positive group was refractory to induction chemotherapy and left the protocol. She achieved complete remission (CR) after one cycle of rescue chemotherapy with fludarabine, cytarabine, and idarubicin (“FLAG-Ida”). Then, she received one “extra-compartment” chemotherapy intensification block before SCT from a matched unrelated donor and was alive in remission 59 months after primary diagnosis (Table 3). Two additional patients in the iAMP21-positive group who achieved CR experienced an isolated BM relapse 54 and 24 months after primary diagnosis, respectively. The first one achieved a second CR (CR2) with MRD below 0.1% after standard “ALL REZ BFM 2002” re-induction. A TP53 deletion was later informed to be present at relapse, and he received two consolidation chemotherapy courses followed by one cycle of blinatumomab before SCT and was alive in remission at last contact 89 months after primary diagnosis (35 months after relapse). One patient who relapsed 24 months after primary diagnosis achieved CR2 after one cycle of inotuzumab ozogamycin. The patient then received one chemotherapy consolidation course and one cycle of blinatumomab before SCT and was alive in remission at last contact 33 months after primary diagnosis (9 months after relapse).

Table 3.
Outcome and additional biological information from four iAMP21-positive patients.
Two out of 89 patients with non-iAMP21 ALL died without disease (TRM), and 9 relapsed, yielding a total of 11 events in this group.
After a median follow up of 57 months (range, 1–132 months), 5-year EFS was inferior in the iAMP21-positive group at 25% (95% CI 4.5–100%) versus 85.6% (95% CI 78–93.9%) in the non-iAMP21 group (p = 0.001). CIR was 50% (95% CI 2.3–88.1%) in the iAMP21-positive group and 9.9% (95% CI 4.3–18.3%) in the non-iAMP21 (p = 0.02), and there was no difference in TRM at 0% versus 2.38%, respectively (95% CI 0.5–7.5%) (p = 0.76). These results did not translate into a significant difference in OS: 100% in the iAMP21-positive group versus 93.7% (95% CI 88.5–99.2%) in the no-iAMP21 group (p = 0.6) (Figure 2).
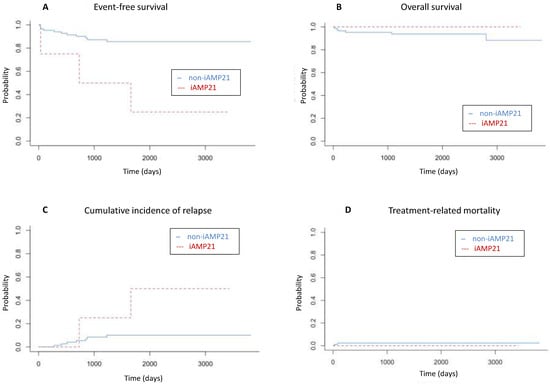
Figure 2.
(A) Five-year event-free survival (25% vs. 85.6%; p = 0.001), (B) overall survival (100% vs. 93.7%; p = 0.6), (C) cumulative incidence of relapse (50% vs. 9.9%; p = 0.02), and (D) treatment-related mortality (0% vs. 2.38%; p = 0.76) among patients with iAMP21-positive (red lines) and non-iAMP21 (blue lines) acute lymphoblastic leukemia.
Apart from the presence of iAMP21, the following factors had a significant negative impact on EFS: Hispanic ethnic group (p = 0.006), ≥5% BM bone marrow blasts on day +15 and +33 evaluations (p = 0.02 and <0.001, respectively), and MRD ≥ 1% on day +33 (p = <0.001; Table 4).

Table 4.
Factors influencing event-free survival: univariate analysis.
In multivariate analysis, the presence of iAMP21 (HR 7.68, 95% CI 2.04–29.05; p = 0.002) and an MRD ≥ 1% after induction on day +33 (HR 8.82, 95% CI 2.6–29.91; p = 0.001) had a significant negative impact on EFS.
All three iAMP21-positive patients who experienced an event had PAX5 deletion/loss on MLPA evaluation. Furthermore, NRAS pathogenic variants were identified in these three patients with >35% variant allele frequencies (VAF) and two patients who relapsed after achieving CR harbored PAX5 and SH2B3 pathogenic or likely pathogenic variants on primary ALL diagnostic samples. None of these alterations were present in the only one case of iAMP21-positive ALL who did not experience any event (Table 3 and Figure 3).
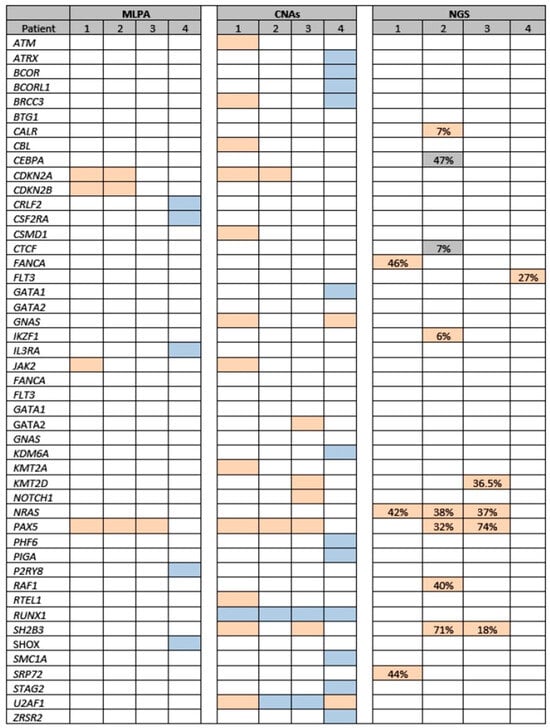
Figure 3.
Genetic alterations in four patients with iAMP21-positive ALL (affected genes are presented in alphabetical order). MLPA: Salmon color, heterozygous deletion; blue, heterozygous duplication. CNAs: Salmon color: copy number < 2 (deletion/loss); blue color: copy number > 2 (amplification). NGS (gene variants): Salmon color, pathogenic or likely pathogenic variants (% VAF); grey, uncertain significance variant (% VAF). Abbreviations; ALL, acute lymphoblastic leukemia; CNAs, copy number alterations; MLPA, multiple ligation probe assay, NGS, next-generation sequencing; VAF, variant allelic frequency.
4. Discussion
Intrachromosomal amplification of chromosome 21 (iAMP21) is currently well recognized as a distinct BCP ALL subtype [18,20]. It is estimated to occur in 1% to 3% of cases of ALL during childhood [5,6,11,12,13].
The presence of iAMP21 is generally exclusive of other primary cytogenetical alterations and is considered a leukemia initiating event. However, a few cases carry other alterations, such as ETV6::RUNX1, high hyperdiploidy, BCR::ABL1, and DUX4-rearrangement, which are currently considered subtype-defining alterations [3,6,15,20,21].
We found the presence of iAMP21 in 4.5% of cases in a series of 89 pediatric ALL patients, excluding those <1 year old at diagnosis. The diagnosis of iAMP21 ALL was based upon FISH analysis results, with ≥5 copies of RUNX1 in interphase. FISH analysis using an ETV6::RUNX1 probe set is considered the standard method for detection of iAMP21 in most reported studies. However, other approaches such as single-nucleotide polymorphism array may be appropriate as well [3,5,13,14,15]. RUNX1 (previously AML1) is commonly amplified in leukemic blasts of iAMP21 ALL. The presence of ≥5 copies of RUNX1 in an interphase FISH study or ≥3 additional copies on a single abnormal chromosome 21 defines the diagnosis of iAMP21 [3,5,6,11,12,13,15,21,22]. The identification of multiple copies of RUNX1 within one metaphase nuclei is also applicable for the definition of iAMP21 [3,5,15,23]. However, extra RUNX1 signals may be found in interphase as the result of additional chromosome 21 in hyperdiploid cases [3]. Moreover, RUNX1 is not always included in the variable region of copy number gain within chromosome 21, amplification of RUNX1 may occur outside chromosome 21 in cases with rearrangements of 21, and these unusual cases of iAMP21-positive ALL would not be detected by FISH [21,22,24]. The definition of iAMP21 will probably be revised according to recent results from gene expression profiles able to recognize variant cases. Copy number profiling (single-nucleotide polymorphism array or whole genome sequencing) has been recommended for adequate diagnosis of these atypical cases [3,15,21]. In a recent comprehensive genomic study from 124 patients with grossly abnormal chromosome 21 in the karyotype and/or ≥5 RUNX1 signals by FISH, the diagnosis of iAMP21 was verified by results from copy number profiles [21].
Children with iAMP21 ALL are frequently diagnosed at older age than non-iAMP21 ALL cases; they commonly present at age older than 10 years, with pre-B or common ALL and WBC below 50,000/µL [4,5,6,12,13,14,15,21,24]. It is reported to be more prevalent among female patients than other cytogenetic subtypes of ALL in children [5,6,12,13,15,21]. In our series, median age was non-significantly higher in the iAMP21-positive group. In addition, three out of four patients were females, and all had WBC below 50,000/µL. Moreover, only one met the NCI high-risk criteria, and none had CNS involvement. None of these presenting findings represented a HR criterion in our protocol.
Early studies associated the presence of iAMP21 with a poor prognosis [4,12,13]. Among 28 patients with iAMP21 ALL collected within the UK Medical Research Council (MRC) ALL97 and ALL97/99 trials, 17 (61%) relapsed, and the 5-year EFS was 29%, which was below that obtained in other poor-risk cytogenetic groups. As non-iAMP21 ALL patients, most relapses (71%) were isolated BM, and almost half of them occurred before the end of first-line treatment. However, an unusual pattern of relapse was observed which appeared constant over time instead of reaching a plateau after 2 years of follow-up. The presence of iAMP21 was an independent prognostic factor for EFS and OS. However, some patients could be rescued after relapse, yielding an OS up to 71%. Only six of these 28 patients were classified and treated as HR according to other risk factors. As a consequence of these results, in the subsequent UK ALL2003 trial, all patients with iAMP21 were stratified and treated as HR patients, they were not randomly allocated to an intensified chemotherapy schedule (“regimen C”) with additional doses of PEG-asparaginase and two courses of escalating doses of methotrexate (Capizzi interim maintenance), and SCT was considered for those with slow early response. CR was achieved in all patients after induction. Half of patients with iAMP21 ALL had an MRD-positive response after induction (≥0.01%), which was not different form the entire cohort, and there was no clear correlation between MRD status after induction and final outcome. Treatment outcome of patients with iAMP21 ALL was significantly better in the UKALL2003 trial compared to that of the previous ALL97/99 trial. Five out of six relapses were classified as late [6,12].
Attarbaschi and colleagues reported a series of 29 patients included in the ALL-BFM 86, 90, 95 and 20 trials. All of them achieved CR after induction. Only one out of 24 patients with available MRD evaluation results by a PCR-based technology (4%) had MRD ≥ 5 × 10−4 at the end of induction. Eleven patients (38%) with iAMP21 relapsed, and the median time to relapse was 2.59 years (3 relapses occurred before the end of front-line therapy). In this series, treatment intensification according to prednisone response and early MRD response seemed to benefit patients with iAMP21. In total 23% of patients allocated to the IR group did relapse, and this proportion increased to 50% among patients with iAMP21-positive ALL. The authors concluded that patients with iAMP21 ALL allocated to the IR group according to early response should receive intensified post-induction therapy as HR patients. After relapse, only five patients survived after a median follow-up of 4.95 years, and four survived after SCT [13].
In the COG studies AALL0331 and AALL0232, patients with iAMP21 had inferior EFS and OS compared with those without iAMP21. In these studies, patients were classified as SR or HR according to other cytogenetic findings and according to early treatment response measured by FC at the end of induction. MRD positivity (≥0.01%) after induction was more frequent among patients with iAMP21. The impact of iAMP21 on treatment outcome significantly affected SR with negative MRD but not HR patients who received intensified post-induction therapy. Accordingly, the presence of iAMP21 affected outcome in patients who received less intensive therapy after induction, and the authors concluded that, in these patients, treatment should be intensified regardless of early response. Subsequent COG trials allocated all iAMP21 ALL patients to HR treatment arms [5].
The introduction of additional doses of PEG-asparaginase and escalating doses of methotrexate in the UK ALL2003 trial were considered as a possible explanation for the better results obtained in this study compared to those reported by previous BFM and COG studies. In the BFM2000 protocol, patients received a high dose of methotrexate but not the escalating (Capizzi) schedule and did not receive PEG-asparaginase. Patients with iAMP21 ALL obtained better results when treated with the AALL0232 instead of the AALL0331 COG regimen, and this different outcome might be explained by the administration of additional doses of methotrexate and asparaginase [5,6,12,13].
The Ponte di Legno International Childhood ALL Group retrospectively analyzed data from 530 pediatric patients with iAMP21-positive ALL from 18 international study groups. Five-year EFS and OS from 283 patients with adequate follow-up time were of 58% and 82%, respectively, and a significant improved outcome was obtained when patients were treated with HR instead of non-HR protocols. However, there was no difference in OS, which indicates that a high proportion could be salvaged after second-line treatment. In the series reported by the Ponte di Legno Childhood ALL Group, only 1% of patients were classified as non-remitters [15]. MRD response might not be able to identify those who benefit from treatment intensification, and, according to the results presented by the Ponte di Legno Childhood ALL Group, all patients with iAMP21-positive ALL should be allocated to HR regimens [5,15].
The following conclusions can be taken from the aforementioned studies: (i) iAMP21 had a significant prognostic impact on treatment outcome that seemed to be independent of early treatment response; (ii) treatment intensification attenuates the negative prognostic impact of this cytogenetic abnormality which should be considered as a HR feature for patient stratification; (iii) PEG-asparaginase intensification and escalating doses of methotrexate might improve treatment outcome; and (iv) early and late relapses were the most prevalent events. In addition, isolated bone marrow was the main site of relapse, but induction failure was also found in some patients.
However, the presence of iAMP21 has not been considered as a HR factor independently from early MRD response by all study groups [3,25]. This is the case of our Spanish “SEHOP-PETHEMA 2013” protocol. In our cohort, apart from one patient who was refractory to induction, the remaining three cases had good early response. Cytological as well as MRD FC evaluations at different early time points did not allow the allocation of these three patients in the HR group; all three were allocated into de IR group and received a PEG-asparaginase-intensified regimen but not escalating doses of methotrexate. In our single-center experience with this regimen, the identification of iAMP21 had an independent significant impact on EFS (25% versus 85.6% in non-iAMP21 patients; p = 0.001). Three out of four patients (75%) experienced an event, including one refractoriness to induction and two isolated BM relapses occurring at 24 and 54 months after primary diagnosis, respectively. iAMP21 and the persistence of ≥1% MRD in BM at day +33 were the only two variables analyzed that retained an independent significantly negative impact on EFS in multivariate analysis.
Since asparaginase activity monitoring was not routinely carried out at our institution until recent years, occult silent inactivation might have contributed to the occurrence of events. This represents a significant drawback of this study since suboptimal exposure to asparaginase has been related to an increased risk of relapse and, as mentioned, asparaginase seems to play an important role in treatment response of iAMP21-positive patients [26]. However, our two patients with iAMP21 who relapsed had asparaginase activity monitoring during second-line treatment without evidence of silent inactivation.
A substantial number of patients with iAMP21-positive ALL can be rescued after relapse [3,4,12]. In the present study, all three patients who failed after frontline therapy achieved CR with second-line treatment, which included conventional chemotherapy, inotuzumab ozogamycin in one patient, blinatumomab in two patients, and SCT in all three patients. All of them were alive in remission 9, 35, and 59 months after the event, respectively. Thus, novel immunotherapy approaches seem to improve outcome of pediatric patients with iAMP21 after relapse.
This is a complex genetic alteration that arises from at least one breakage-fusion-bridge cycle and chromothripsis of chromosome 21 in most cases [14,21]. Overexpression of multiple genes within a common large size region of amplification spanning 5.1 Mb (from 32.8 to 37.9–40.6 Mb) in chromosome 21 occurs, and this remains constant at relapse [3,14,21]. However, one of the four cases presented here was previously reported to show a decreased copy number state of iAMP21 at relapse with only three copies of RUNX1 using FISH. However, SNP array at primary diagnosis and relapse showed similar abnormalities but a reduced copy number in the amplification region of chromosome 21 and the addition of further alterations (including TP53 mutation) at relapse [27].
Certain genes implicated in the pathogenesis of leukemia, such as CHAF1B, DYRK1A, ERG, HMGN1, and RUNX1, are included in this region and are frequently differentially expressed in iAMP21-positive cases [21]. RIPPLY3 has been suggested to be implicated in the pathophysiology of iAMP21 [24]. Secondary genetic abnormalities are frequently described in iAMP21 ALL cases. These include gain of chromosomes X, 10, and 14; monosomy 7 or 7q and 11q deletions; IKZF1, ETV6, RB1, and CDKN2A/B deletions; and P2RY8-CRLF2 rearrangement. These differ from other pre-B ALL subtypes [3,6,14,15]. Alterations of ETV6 and activating mutations of JAK-STAT and the RAS signaling pathway, including SH2B3 and FLT3 aberrations, are particularly frequent in iAMP21 ALL cases, which might offer an opportunity to explore the role of targeted therapy with JAK/STAT inhibitors such as gilterinib and ruxolitinib or MEK inhibitors [16,21,24]. The occurrence of RAS pathway alterations might be related to a higher incidence of relapse through the generation of resistance to chemotherapy [16]. We also found alterations of genes involved in the JAK-STAT signaling pathway (SH2B3, CRLF2, JAK2), RAS signaling (NRAS), B-cell differentiation (PAX5), transcriptional regulation (BTG1), apoptosis (CDKN2A, CDKN2B), and epigenomic signaling (KMT2D, KMT2A, ATM). However, RAS pathway mutations are often described among other BCP ALL subtypes such as high hypediploidy and B-other ALL [16].
Intriguingly, NRAS pathogenic variants were present in all our three patients suffering an event and were absent in the only patient without any event. Moreover, the genomic landscape of this later patient seems to differ from the others (Table 3 and Figure 3). We have not found any previously reported correlation between any particular genetic background and treatment response among patients with iAMP21. The identification of patients with a lower risk of relapse might avoid unnecessary exposure to highly intensive and toxic therapy [14].
We found a pathogenic FANCA gene variant with an allele frequency of 46% in patient 1, which was considered a somatic variant or a possible heterozygous germline alteration provided that there were no clinical findings suggestive of a Fanconi anemia phenotype. This finding did not influence the treatment plan, including the conditioning regimen before stem cell transplantation.
Apart from the lack of data regarding asparaginase activity monitoring during treatment, the retrospective single-center design and the low numbers of patients included in this study are important limitations; however, the comparison of treatment outcome among patients with and without this rare cytogenetic ALL subgroup of homogeneously treated patients may serve to emphasize the need to include it as a HR diagnostic feature.
5. Conclusions
In conclusion, we found an independent significant prognostic impact of iAMP21 on EFS among pediatric patients with ALL. Clinical presentation and early treatment response evaluation did not allow the classification of these patients as HR. Diverse genetic landscapes among iAMP21-positive patients might influence the treatment response and outcome of this heterogeneous disease.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/hemato6030019/s1, Table S1: Risk groups and stratification criteria in “SEHOP-PETHEMA 2013” guidelines; Table S2: “LAL/SEHOP-PETHEMA 2013” treatment protocol for standard- and intermediate-risk patients; Table S3: “LAL/SEHOP-PETHEMA 2013” treatment protocol for high-risk patients; Table S4: Conventional cytogenetics, DNA index, and FISH results from four patients with iAMP21.
Author Contributions
Conceptualization, J.L.F., E.R.-E. and M.H.; methodology, J.L.F., E.R.-E. and M.H.; software, E.R.-E.; validation, J.L.F. and E.R.-E.; formal analysis, J.L.F., E.R.-E. and M.H.; investigation, A.M., J.A.G., H.M.-B. and V.M.-S.; resources, J.L.F., E.R.-E., M.H., M.P., A.M.G., M.E.L., M.B., A.M., J.A.G., H.M.-B. and V.M.-S.; data curation, J.L.F., E.R.-E. and M.H.; writing, J.L.F. and M.H.; writing—review and editing, J.L.F., E.R.-E., I.J. and A.M.; supervision, J.L.F.; project administration, J.L.F., E.R.-E. and M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Patients were included in the Spanish “LAL/SEHOP-PETEHEMA 2013” guidelines regimen. These recommendation guidelines were launched by the Leukemia Working Group of the Spanish Society of Pediatric Hematology and Oncology (SEHOP). The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Ethics Committee of Hospital Clínico Universitario Virgen de la Arrixaca—Instituto Murciano de Investigación Biosanitaria, IMIB, Murcia, Spain (“LAL/SEHOP-PETHEMA 2013”, date of approval 20 December 2013).
Informed Consent Statement
Written informed consent before registration and treatment was provided by all patients or legal guardians including consent for data analysis and analyses of stored biological samples.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Acknowledgments
We would like to acknowledge all patients and families. We want to acknowledge the Asociación Pablo Ugarte (APU) for its support. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ALL | Acute lymphoblastic leukemia |
| BCP | B-cell precursor |
| BM | Bone marrow |
| CIR | Cumulative incidence of relapse |
| CNS | Central nervous system |
| CR | Complete remission |
| CR2 | Second complete remission |
| EFS | Event-free survival |
| FISH | Fluorescence in situ hybridization |
| FLAG-Ida | Fludarabine, cytarabine, and idarubicin |
| FC | Flow cytometry |
| HR | High risk |
| iAMP21 | Intrachromosomal amplification of chromosome 21 |
| IR | Intermediate risk |
| NCI | National Cancer Institute |
| MRD | Measurable residual disease |
| MLPA | Multiplex ligation-dependent probe amplification |
| MRD | Measurable residual disease |
| OS | Overall survival |
| PCR | Polymerase chain reaction |
| NCI | National Cancer Institute |
| NGS | Next-generation sequencing |
| PEG | Pegylated |
| SCT | Allogeneic stem cell transplantation |
| SEHOP | Spanish Society of Pediatric Hematology and Oncology |
| SR | Standard risk |
| TRM | Treatment-related mortality |
| VAF | Variant allele frequencies |
| WBC | White blood cell count |
| WHO | World Health Organization |
References
- Hunger, S.P.; Mullighan, C.G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 2015, 373, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J.; Haas, O.; Harbott, J.; Biondi, A.; Stanulla, M.; Trka, J.; Izraeli, S. Detection of prognostically relevant genetic abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Recommendations from the Biology and Diagnosis Committee of the International Berlin-Frankfurt-Munster study group. Br. J. Haematol. 2010, 151, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J. Blood Spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood 2015, 125, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.V.; Ensor, H.M.; Richards, S.M.; Chilton, L.; Schwab, C.; Kinsey, S.E.; Vora, A.; Mitchell, C.D.; Harrison, C.J. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010, 11, 429–438. [Google Scholar] [CrossRef]
- Heerema, N.A.; Carroll, A.J.; Devidas, M.; Loh, M.L.; Borowitz, M.J.; Gastier-Foster, J.M.; Larsen, E.C.; Mattano, L.A., Jr.; Maloney, K.W.; Willman, C.L.; et al. Intrachromosomal amplification of chromosome 21 is associated with inferior outcomes in children with acute lymphoblastic leukemia treated in contemporary standard-risk children’s oncology group studies: A report from the children’s oncology group. J. Clin. Oncol. 2013, 31, 3397–3402. [Google Scholar] [CrossRef]
- Moorman, A.V.; Robinson, H.; Schwab, C.; Richards, S.M.; Hancock, J.; Mitchell, C.D.; Goulden, N.; Vora, A.; Harrison, C.J. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: A comparison of the MRC ALL97/99 and UKALL2003 trials. J. Clin. Oncol. 2013, 31, 3389–3396. [Google Scholar] [CrossRef]
- Campana, D.; Pui, C.H. Minimal residual disease-guided therapy in childhood acute lymphoblastic leukemia. Blood 2017, 129, 1913–1918. [Google Scholar] [CrossRef]
- Vora, A.; Goulden, N.; Wade, R.; Mitchell, C.; Hancock, J.; Hough, R.; Rowntree, C.; Richards, S. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): A randomised controlled trial. Lancet Oncol. 2013, 14, 199–209. [Google Scholar] [CrossRef]
- Vora, A.; Goulden, N.; Mitchell, C.; Hancock, J.; Hough, R.; Rowntree, C.; Moorman, A.V.; Wade, R. Augmented post-remission therapy for a minimal residual disease-defined high-risk subgroup of children and young people with clinical standard risk and intermediate-risk acute lymphoblastic leukaemia (UKALL 2003): A randomised controlled trial. Lancet Oncol. 2014, 15, 809–818. [Google Scholar] [CrossRef]
- Pieters, R.; de Groot-Kruseman, H.; Van der Velden, V.; Fiocco, M.; van den Berg, H.; de Bont, E.; Egeler, R.M.; Hoogerbrugge, P.; Kaspers, G.; Van der Schoot, E.; et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: Study ALL10 from the Dutch Childhood Oncology Group. J. Clin. Oncol. 2016, 34, 2591–2601. [Google Scholar] [CrossRef]
- Harewood, L.; Robinson, H.; Harris, R.; Al-Obaidi, M.J.; Jalali, G.R.; Martineau, M.; Moorman, A.V.; Sumption, N.; Richards, S.; Mitchell, C.; et al. Amplification of AML1 on a duplicated chromosome 21 in acute lymphoblastic leukemia: A study of 20 cases. Leukemia 2003, 17, 547–553. [Google Scholar] [CrossRef]
- Moorman, A.V.; Richards, S.M.; Robinson, H.M.; Strefford, J.C.; Gibson, B.E.; Kinsey, S.E.; Eden, T.O.B.; Vora, A.J.; Mitchell, C.D.; Harrison, C.J.; et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21). Blood 2007, 109, 2327–2330. [Google Scholar] [CrossRef]
- Attarbaschi, A.; Mann, G.; Panzer-Grumayer, R.; Rottgers, S.; Steiner, M.; Konig, M.; Csinady, E.; Dworzak, M.N.; Seidel, M.; Janousek, D.; et al. Minimal residual disease values discriminate between low and high relapse risk in children with B-cell precursor acute lymphoblastic leukemia and an intrachromosomal amplification of chromosome 21: The Austrian and German acute lymphoblastic leukemia Berlin-Frankfurt-Munster (ALL-BFM) trials. J. Clin. Oncol. 2008, 26, 3046–3050. [Google Scholar] [PubMed]
- Rand, V.; Parker, H.; Russell, L.J.; Schwab, C.; Ensor, H.; Irving, J.; Jones, L.; Masic, D.; Minto, L.; Morrison, H.; et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood 2011, 117, 6848–6855. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J.; Moorman, A.V.; Schwab, C.; Carroll, A.J.; Raetz, E.A.; Devidas, M.; Strehl, S.; Nebral, K.; Harbott, J.; Teigler-Schlegel, A.; et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): Cytogenetic characterization and outcome. Leukemia 2014, 28, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.L.; Matheson, E.; Grossmann, V.; Sinclair, P.; Bashton, M.; Schwab, C.; Towers, W.; Partington, M.; Elliott, A.; Minto, L.; et al. The role of the RAS pathway in iAMP21-ALL. Leukemia 2016, 30, 1824–1831. [Google Scholar] [CrossRef]
- Irving, J.A.E.; Enshaei, A.; Parker, C.A.; Sutton, R.; Kuiper, R.P.; Erhorn, A.; Minto, L.; Venn, N.C.; Law, T.; Yu, J.; et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood 2016, 128, 911–922. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; IARC Press: Lyon, France, 2009; pp. 109–138. [Google Scholar]
- Bene, M.C.; Castoldi, G.; Knapp, W.; Ludwig, W.D.; Matutes, E.; Orfao, A.; van’t Veer, M.B. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia 1995, 9, 1783–1786. [Google Scholar]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Gao, Q.; Ryan, S.L.; Iacobucci, I.; Ghate, P.S.; Cranston, R.E.; Schwab, C.; Elsayed, A.H.; Shi, L.; Pounds, S.B.; Lei, S.; et al. The genomic landscape of acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Blood 2023, 142, 711–723. [Google Scholar] [CrossRef]
- Koleilat, A.; Smadbeck, J.B.; Zepeda-Mendoza, C.J.; Williamson, C.M.; Pitel, B.A.; Golden, C.L.; Xu, X.; Greipp, P.T.; Ketterling, R.P.; Hoppman, N.L.; et al. Characterization of unusual iAMP21 B-lymphoblastic leukemia (iAMP21-ALL) from the Mayo Clinic and Children’s Oncology Group. Genes Chromosomes Cancer 2022, 61, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.D.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: Lymphoid neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Hormann, F.M.; Østergaard, A.; van den Broek, S.; Boeree, A.; van de Ven, C.; Escherich, G.; Sonneveld, E.; Boer, J.M.; den Boer, M.L. Secondary lesions and sensitivity to signaling inhibitors in iAMP21 acute lymphoblastic leukemia. Hemasphere 2025, 9, e70069. [Google Scholar] [CrossRef] [PubMed]
- Izraeli, S. Application of genomics for risk stratification of childhood acute lymphoblastic leukaemia: From bench to bedside? Br. J. Haematol. 2010, 151, 119–131. [Google Scholar] [CrossRef]
- Gottschalk Højfeldt, S.; Grell, K.; Abrahamsson, J.; Lund, B.; Vettenranta, K.; Jónsson, Ó.G.; Frandsen, T.L.; Wolthers, B.O.; Marquart, H.V.H.; Vaitkeviciene, G.; et al. Relapse risk following truncation of pegylated asparaginase in childhood acute lymphoblastic leukemia. Blood 2021, 137, 2373–2382. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Gómez, G.; Minguela, A.; Tazón-Vega, B.; Ribera, J.; Galián, J.A.; Martínez-Banaclocha, H.; García-Garay, M.; Velasco, P.; Fuster-Soler, J.L.; Armengol, G.; et al. Clonal heterogeneity and genomic evolution in intrachromosomal amplification of chromosome 21: A case report Case Report. Br. J. Haematol. 2024, 204, 2512–2515. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).