Abstract
This study addresses challenges in skin cancer detection, particularly issues like class imbalance and the varied appearance of lesions, which complicate segmentation and classification tasks. The research employs deep learning ensemble models for both segmentation (using U-Net, SegNet, and DeepLabV3) and classification (using VGG16, ResNet-50, and Inception-V3). The ISIC dataset is balanced through oversampling in classification, and preprocessing techniques such as data augmentation and post-processing are applied in segmentation to increase robustness. The ensemble model outperformed individual models, achieving a Dice Coefficient of 0.93, an IoU of 0.90, and an accuracy of 0.95 for segmentation, with 90% accuracy on the original dataset and 99% on the balanced dataset for classification. The use of ensemble models and balanced datasets proved highly effective in improving the accuracy and reliability of automated skin lesion analysis, supporting dermatologists in early detection efforts.
1. Introduction
The early detection and accurate classification of skin lesions are critical for effective skin cancer treatment. Dermoscopic analysis has improved diagnostic accuracy, but manual interpretation remains time-consuming and subjective, highlighting the need for automated diagnostic tools [1,2]. Deep learning, particularly with CNNs like VGG16, ResNet-50, and Inception-V3, has proven effective in medical image interpretation [3,4,5]. Skin cancer, especially melanoma, BCC, and SCC, is challenging to diagnose early, and the ISIC dataset provides valuable resources for algorithm development. Deep learning models like U-Net, SegNet, and DeepLabv3 excel in segmentation tasks, capturing spatial details. This study proposes a deep ensemble model combining these architectures to enhance skin lesion segmentation accuracy. For classification, ensemble models of VGG16, ResNet-50, and Inception-V3 are used to improve recognition of BCC, SCC, and melanoma. To address imbalanced datasets, oversampling techniques are employed, aiming to develop a reliable framework for skin cancer diagnosis, contributing to better outcomes and early intervention.
This work [6] focuses on segmenting skin lesions using preprocessed dermoscopic images, removing noise through a fusion of six image-processing techniques. Modified U-Net architectures, particularly U-Net 46, are used, yielding 93% accuracy, 97% specificity, and 91% sensitivity on the ISIC 2018 dataset. Another study [7] employs atrous convolutions in a CNN to improve lesion segmentation by expanding the receptive field without lowering resolution, showing precise lesion region identification. A modified cGAN with Factorized Channel Attention (FCA) [8] enhances segmentation, reducing computational complexity. A multi-task deep neural network [9] achieves high AUCs for lesion classification and segmentation accuracy. A novel CNN architecture [10] uses auxiliary edge prediction and multi-scale feature aggregation, yielding superior segmentation performance. Research [11] reviews 177 deep learning models, highlighting challenges in skin lesion segmentation due to low contrast and fuzzy borders. An attention-based U-Net model with DenseNet [12] improves segmentation via adaptive gamma correction. Another method [13] combines deep and classical learning, achieving high segmentation and classification accuracy using a cubic support vector machine. A deep learning approach [14] with ResUNet++ and post-processing techniques like CRF and TTA improves segmentation, achieving high Jaccard Index scores. Finally, a CNN-based technique [15] recovers lesion locations with enhanced accuracy through pixel-level segmentation and post-processing.
This collection of studies explores various deep learning approaches for skin lesion classification. Study [16] utilized Inception-V3 and DenseNet to categorize seven skin lesion types, with DenseNet outperforming in classification accuracy and using visualizations to enhance interpretability. In study [17], an AI-based system won first place in the ISIC 2019 challenge, addressing challenges like class imbalance by integrating patient data. Study [18] introduced a Deep Focused Sub-network (DFS) and Information-guided Weights Rebalancing (IWR) to highlight significant skin lesion areas and mitigate class disparity, achieving a state-of-the-art performance on ISIC datasets. In study [19], a system using attention mechanisms and SMOTE for data balancing demonstrated improved accuracy on the HAM10000 dataset. The authors [20] presented an advanced deep residual network designed to enhance skin lesion classification accuracy. The model uses multilevel feature extraction and cross-channel correlation to capture complex lesion features and patterns effectively. Additionally, it incorporates an outlier detection mechanism to improve robustness by filtering out data that could negatively impact model performance. This approach demonstrates significant promise for improving diagnostic accuracy in skin lesion classification by capturing subtle lesion characteristics and addressing data anomalies. It introduces a sophisticated residual network that employs multilevel feature extraction and cross-channel correlation, proving effective in capturing complex lesion patterns. However, while Skin-Net offers a robust approach to feature extraction, it may face limitations in handling highly imbalanced datasets without targeted data balancing strategies.
This paper reviews [21] the latest advancements in deep learning and optimization-based techniques for skin lesion segmentation, focusing on the integration of machine learning, neural networks, and optimization algorithms to enhance segmentation accuracy and clinical relevance. It covers prominent deep learning architectures like U-Net, FCN, and GANs, alongside optimization strategies that improve model efficiency and generalization. The review addresses challenges such as class imbalance, variability in lesion appearance, and the need for robust models in real-world applications, providing insights into current trends and future directions for skin lesion segmentation in dermatological diagnostics. It provides a thorough review of segmentation techniques, offering valuable insights into the optimization of segmentation performance. However, as a review, it lacks experimental validation, leaving practical performance considerations in real-world clinical contexts unexplored.
The proposed method enhances skin lesion segmentation and classification by introducing an innovative ensemble model that integrates U-Net, SegNet, and DeepLabv3 for segmentation, and VGG16, ResNet-50, and Inception-V3 for classification. This unique combination leverages the strengths of each model—U-Net’s precision in boundary detection, SegNet’s efficiency in semantic segmentation, and DeepLabv3’s ability to capture multi-scale context—to deliver highly accurate and robust lesion segmentation. For classification, the ensemble of VGG16, ResNet-50, and Inception-V3 captures a broader range of feature representations, boosting diagnostic accuracy for complex lesion patterns. Unlike conventional single-model approaches, our ensemble method addresses critical challenges in differentiating melanoma, BCC, and SCC, especially with a balanced dataset and data augmentation techniques that mitigate the impact of class imbalance. By demonstrating improved precision, recall, and F1-scores across lesion types, this method offers a precise and impactful advancement in automated skin lesion analysis, underscoring its potential for real-world clinical applications. Using the ISIC dataset, the system demonstrates superior diagnostic accuracy, offering a reliable clinical tool for early skin cancer detection.
Our work presents several key contributions aimed at enhancing skin lesion analysis.
- A novel ensemble architecture for segmentation: the proposed method integrates U-Net, SegNet, and DeepLabv3 with augmentation, each contributing unique strengths—boundary detection, semantic segmentation, and multi-scale context capture—for precise and efficient skin lesion segmentation.
- An ensemble of deep models for classification: through combining VGG16, ResNet-50, and Inception-V3, the model captures diverse feature representations, improving classification accuracy for melanoma, basal cell carcinoma (BCC), and squamous cell carcinoma (SCC) lesions.
- Balanced dataset and data augmentation: employing oversampling and augmentation techniques, the model effectively addresses class imbalance, enhancing reliability across lesion categories.
- Enhanced performance metrics: the ensemble method achieves superior precision, recall, and F1-scores compared to single-model approaches, demonstrating its robustness for clinical applications.
- Real-world clinical relevance: the method’s improved diagnostic accuracy and robustness make it a promising tool for real-world skin lesion analysis and potential integration into clinical workflows.
The format of this document is as follows: The proposed deep ensemble model for skin lesion segmentation and the suggested deep ensemble model for skin lesion classification are offered in Section 2. Performance results are discussed in Section 3, Section 4 compares these results with the existing literature, and the last section concludes with future directions for this paper.
2. Materials and Methods
This section outlines proposed skin lesion segmentation and proposed skin lesion classification performance using a dermoscopic image (ISIC) dataset. Algorithm 1 describes the proposed segmentation and classification algorithm.
| Algorithm 1: Proposed Method for Skin Lesion Segmentation and Classification |
| Input: Dermoscopic images of skin lesions Corresponding labels for each image (melanoma, BCC, or SCC) Hyperparameters (e.g., learning rate, batch size, epochs, etc.) Output: Segmentation masks and classification labels |
Step 1: Data Preparation Load the dataset of dermoscopic images and their corresponding labels. Split the dataset into training (80%) and validation (20%) sets for segmentation, and training (75%) and validation (25%) sets for classification. Resize images to appropriate dimensions: Segmentation models: 256 × 256 pixels Classification models: 224 × 224 pixels (VGG16, ResNet50) or 299 × 299 pixels (InceptionV3). Normalize pixel values of images to the range [0, 1]. Apply data augmentation techniques to the training set: Random rotations Horizontal and vertical flips Painting. Step 2: Class Balancing Identify class distribution within the training dataset. Use random oversampling to duplicate instances of minority classes (BCC, SCC) until balanced with the majority class (melanoma). Step 3: Model Training Segmentation Models: Initialize U-Net, DeepLabV3, and SegNet models. Compile each model using an appropriate optimizer (e.g., Adam) and loss function (e.g., Binary Crossentropy, unet3p_hybrid_loss). Train each segmentation model on the augmented and balanced training set for a specified number of epochs. Monitor training and validation loss/accuracy. Classification Models: Initialize VGG16, ResNet50, and InceptionV3 models Compile each model using an appropriate optimizer and loss function (e.g., Categorical Crossentropy). Train each classification model on the augmented and balanced training set for a specified number of epochs. Monitor training and validation metrics (accuracy, precision, recall). Step 4: Ensemble Prediction Generate predictions from each trained segmentation model for the validation set. Combine segmentation outputs using a voting or averaging mechanism to obtain the final segmentation mask. Generate predictions from each trained classification model for the validation set. Combine classification outputs using an ensemble technique (e.g., weighted average or majority voting) to obtain the final class label. Step 5: Model Evaluation Calculate evaluation metrics for segmentation: Intersection over Union (IoU) Dice Coefficient Accuracy. Calculate evaluation metrics for classification: Precision Recall F1-Score AUC Accuracy. End Algorithm |
2.1. Proposed Skin Lesion Segmentation
The system leverages an ensemble of U-Net [22], SegNet [23], and DeepLabV3 [24] to improve segmentation accuracy and robustness. U-Net excels at capturing fine details for precise lesion boundaries, SegNet maintains spatial resolution during upsampling, and DeepLabv3 handles multi-scale lesions with atrous convolutions. This ensemble outperforms individual models on the challenging ISIC dataset, making it applicable in real-world clinical scenarios. The segmentation model’s ability to accurately identify lesion borders aids early detection and the diagnosis of skin malignancies, improving outcomes for patients. Figure 1 describes the segmentation design. The ISIC dataset [25], used for model training, includes 2357 images of various benign and malignant skin conditions, with a focus on three key cancers: melanoma (438 images), basal cell carcinoma (376 images), and squamous cell carcinoma (181 images). The ISIC 2018 dataset is a large and comprehensive dataset used for the analysis of skin lesions, particularly for tasks such as segmentation, classification, and the detection of skin cancer. It was made available as part of the International Skin Imaging Collaboration (ISIC) challenge, aimed at improving the early detection of skin cancer. The dataset contains 10,015 high-resolution dermoscopic images, each labeled with diagnostic information confirmed by expert dermatologists. The images cover various types of skin lesions, with a focus on conditions such as melanoma, BCC, and SCC, among others. The dataset is diverse, including images from different skin types, lesion shapes, sizes, and locations, providing a robust foundation for developing machine learning models that can generalize well to real-world scenarios. Each image is provided in JPG format with varying resolutions, making it essential to preprocess the images before feeding them into models.
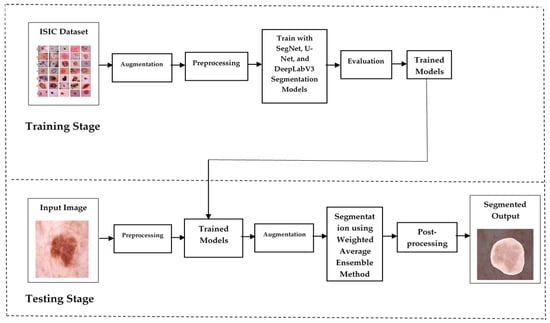
Figure 1.
Skin lesion segmentation design.
The data is split into 80% for training and 20% for validation. Annotation is performed using LabelMe tools, and the annotated JSON files are converted into mask images. Images are resized to 256 × 256 pixels, and pixel intensities are normalized. Data augmentation techniques, such as flipping, in-painting, and rotation, are applied to create five variations of each image to improve model robustness.
This process involves two types of transformations. In flipping (2 images), the original image is flipped to create two additional variations. This might include horizontal and vertical flips. In-painting (2 images) is a technique which involves removing a portion of the image (in this case, one lung) and filling in the missing part with surrounding pixels or predicted values. In medical imaging, this can help models learn to identify structures even when parts of them are missing. Rotation (1 image) involves rotating the original image to create variations. This can involve rotations by angles between −10° to +10°.
Each of the five augmented images is independently fed into the segmentation model, producing a binary or probabilistic mask for each. The masks are averaged pixel-wise across the five augmentations to create a more robust final prediction. The resulting masks from the three models—SegNet, DeepLabv3, and U-Net—are then combined using a weighted average based on each model’s performance (e.g., Dice Coefficient, IoU, and accuracy). This combined mask, containing continuous values representing the probability of each pixel belonging to a skin lesion, is then thresholded at 0.5 to produce a final binary mask, classifying pixels as lesion or non-lesion. This approach leverages the strengths of both augmentations and the model diversity, resulting in a more accurate and reliable skin lesion segmentation.
Binary Mask (i, j) = 1 if Combined Mask (i, j) > 0.5
0 if Combined Mask (i, j) ≤ 0.5
0 if Combined Mask (i, j) ≤ 0.5
In the post-processing stage of skin lesion segmentation, a series of morphological operations is applied to refine the segmentation mask. Morphological Opening removes small noise and smooths lesion boundaries, and is followed by Erosion to reduce small white pixels and shrink the lesion edges. Dilation then restores lesion size while maintaining noise-free regions. Morphological Closing fills small holes within the lesion areas, and an additional Dilation step closes gaps. A final Erosion ensures the lesion regions are continuous and hole-free. These operations enhance the combined mask, leading to more precise and reliable skin lesion segmentation. By applying morphological operations, such as opening and closing, the post-processing stage effectively refines the segmentation masks produced by the ensemble models. Morphological Opening removes small artifacts and noise, leading to smoother contours and more defined boundaries of the lesions. This is particularly important in medical imaging, where the accurate delineation of lesions is critical for diagnosis and treatment planning. Additionally, Morphological Closing fills in gaps and connects fragmented structures, ensuring that the segmented lesions are represented uniformly. These improvements in the quality of segmentation masks facilitate better feature extraction and analysis, enabling subsequent classification tasks to be performed more effectively. Furthermore, post-processing can help mitigate issues arising from class imbalance by ensuring that the detected lesions are consistently represented across different images. Overall, incorporating post-processing techniques significantly enhances the robustness of skin lesion segmentation, ultimately contributing to more reliable and effective medical diagnostics.
2.2. Proposed Skin Lesion Classification
The proposed system combines VGG16 [26], ResNet-50 [27], and Inception-V3 [28] architectures to enhance skin lesion detection and classification by addressing key challenges in dermatological image analysis, such as class imbalance, low intra-class variability, and high inter-class similarity. The ISIC dataset, a comprehensive collection of dermoscopic images, is used for this study. To mitigate class imbalance, oversampling strategies are applied, particularly to underrepresented classes [29]. Figure 2 shows the architecture design for skin lesion classification. The dataset is split into 75% for training and 25% for testing, with images resized to 224 × 224 pixels for VGG16 and ResNet-50, and 299 × 299 pixels for Inception-V3. Image preprocessing includes normalization using the min-max technique [30], scaling pixel values between 0 and 1 to ensure consistent feature contribution. Additional preprocessing steps, such as data augmentation (e.g., rotation, flipping, and zooming), further enhance model performance by increasing data diversity [31]. These include Center Crop to focus on central features, Random Rotate, Grid Distortion, Flips (Horizontal/Vertical), Optical Distortion, and Affine transformations, all applied with a 0.1 probability to introduce controlled distortions and variations. These augmentations helped the model learn robust features by increasing the diversity of the dataset.
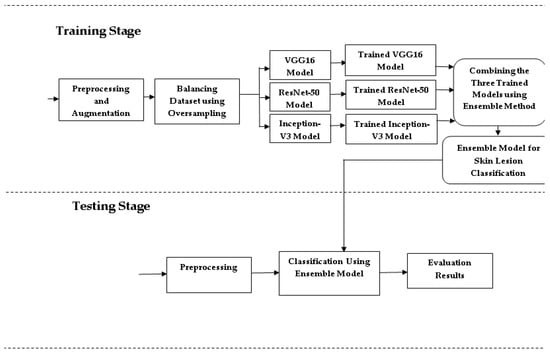
Figure 2.
Skin lesion classification design.
This system classifies skin cancer into three categories: melanoma, BCC, and SCC, using 438 images of melanoma, 376 images of BCC, and 181 images of SCC. The significant disparity in class sizes could bias the model’s performance, making it favor the majority class. To address this imbalance, the dataset was balanced using random oversampling [32]. This technique involves duplicating samples from the minority class, ensuring a more even class distribution. Specifically, simple random sampling with replacement (SRSWR) was employed, allowing the same sample to be chosen multiple times. This method effectively increases the representation of the minority class, mitigating the effects of class imbalance. This technique involves randomly duplicating instances from the minority classes—specifically, basal cell carcinoma (BCC) and squamous cell carcinoma (SCC)—until the number of samples in each class matches that of the majority class, melanoma. The goal of this approach is to prevent the model from being biased toward the overrepresented majority class by ensuring that all classes contribute equally to the training process. In addition to random oversampling, we also applied data augmentation techniques, such as rotation, scaling, and flipping, to introduce more variability into the dataset. This not only enriched the oversampled data but also mitigated the risk of overfitting by providing the model with diverse training samples. Together, random oversampling and data augmentation allowed us to create a balanced and robust dataset for training, improving the model’s ability to classify skin lesions across all categories.
The balanced dataset was then used to train the model over 150 epochs, each representing a full pass through the training data, allowing the model to progressively improve. Once the desired accuracy was reached, the model was saved for potential reuse or further fine-tuning. The final classification system was constructed as a deep ensemble, combining predictions from three models—ResNet-50, VGG16, and Inception-V3—using a weighted average technique.
During testing, input images were first resized according to the model architecture: 224 × 224 pixels for VGG16 and ResNet-50, and 299 × 299 pixels for Inception-V3. These images were then normalized to ensure consistent pixel intensities across the dataset, and converted to tensor format for deep learning compatibility. The classification was performed using the ensemble model, which integrates predictions from multiple architectures to boost accuracy and robustness. Initially, testing was conducted on the original, imbalanced dataset to identify performance issues caused by class disparity. Further testing was carried out on a balanced dataset, achieved through oversampling and data augmentation techniques such as rotations, flips, and color adjustments. Finally, the model was evaluated on both the original and balanced datasets to assess its overall effectiveness and robustness.
3. Results
Performance evaluation for segmentation and performance evaluation for classification are the two sections that make up this section.
3.1. Performance Evaluation for Segmentation
The suggested system is trained and tested using the ISIC dataset, which has an 80% training to 20% testing partition ratio. Using the original dataset, a detailed assessment of the system’s effectiveness and performance is carried out, with a particular focus on important metrics like Dice Coefficient, IoU, and accuracy. In the ensemble model segmentation, performance metrics are evaluated for individual models and the ensemble model. Table 1 describes hyperparameters for segmentation and Table 2 shows the performance results of ensemble model segmentation.

Table 1.
Hyperparameters for segmentation.

Table 2.
Segmentation results.
The segmentation results reveal that the ensemble model of SegNet, U-Net, and DeepLabV3 significantly outperforms the individual models. Specifically, SegNet achieved a Dice Coefficient of 0.82, an IoU of 0.75, and an accuracy of 0.88, while DeepLabV3 delivered higher results with a Dice Coefficient of 0.86, an IoU of 0.79, and an accuracy of 0.90. U-Net also performed well, with a Dice Coefficient of 0.84, an IoU of 0.81, and an accuracy of 0.91. The ensemble model, however, achieved the highest scores with a Dice Coefficient of 0.93, an IoU of 0.90, and an accuracy of 0.95, demonstrating a substantial improvement in segmentation performance by leveraging the strengths of all three models. This enhancement demonstrates how the ensemble approach can effectively improve segmentation performance by utilizing the advantages of many models to produce a more reliable and accurate result.
3.2. Performance Evaluation for Classification
In this section, the performance of the suggested deep ensemble model is compared to that of the three separate models—ResNet-50, Inception-V3, and VGG16—with an emphasis on accuracy metrics. Specifically, the evaluation employs the Kaggle ISIC dataset to classify melanoma, SCC, and BCC using pictures. The initial, unbalanced dataset is used to test the system’s performance. After that, the assessment is carried out once again using a balanced dataset that was produced using oversampling methods. TensorFlow serves as the backend, and Keras is used to build and train these models. In performance evaluation, 25% of the samples are used for testing and 75% are used for training. We use common performance measures including accuracy, recall, precision, and F1-score to assess the effectiveness of our suggested strategy. The model’s ability to distinguish between melanoma, SCC, and BCC lesions may be quantified with the use of these metrics, which provide an extensive assessment of the model’s classification performance across numerous classes. Table 3 describes hyperparameters for classification.

Table 3.
Hyperparameters for classification.
This study compares the performance of VGG16, ResNet-50, Inception-V3, and an ensemble model for skin lesion classification using both balanced and unbalanced datasets, with and without augmentation, as shown in Table 4, Table 5, Table 6 and Table 7. Results show that balanced datasets consistently outperform unbalanced ones, particularly when combined with augmentation. VGG16 and ResNet-50 show significant improvements in precision, recall, F1-Score, and AUC when using balanced datasets with augmentation, while unbalanced datasets yield lower effectiveness. Similarly, Inception-V3 benefits from augmentation and balancing, achieving its highest accuracy on balanced datasets with augmentation. The ensemble model performs best with balanced datasets and augmentation, showing the highest precision and accuracy, underscoring the importance of these techniques in optimizing model performance.

Table 4.
VGG16’s results on balanced and unbalanced datasets.

Table 5.
ResNet-50’s results on balanced and unbalanced datasets.

Table 6.
Inception-V3’s results on balanced and unbalanced datasets.

Table 7.
Deep ensemble model’s results on balanced and unbalanced datasets.
The ensemble model with augmentation significantly outperforms individual models like VGG16, ResNet-50, and Inception-V3 in skin lesion classification for both the original and balanced ISIC datasets. On unbalanced datasets without augmentation, the ensemble model achieves the highest accuracy at 0.78 and, with augmentation, it reaches 0.90. On balanced datasets, the ensemble model’s accuracy improves to 0.92 without augmentation and 0.99 with augmentation, consistently outperforming the individual models. These results highlight the effectiveness of combining multiple models with balanced datasets and augmentation, making skin lesion classification more accurate and reliable. This approach enhances diagnostic precision and holds potential for broader use in medical image analysis. Table 8 describes the confusion matrix. The ROC curves for VGG16, ResNet-50, Inception-V3, and the ensemble model for skin lesion classification using both balanced and unbalanced datasets, with and without augmentation, are shown in Figure 3, Figure 4, Figure 5 and Figure 6.

Table 8.
Confusion matrix.
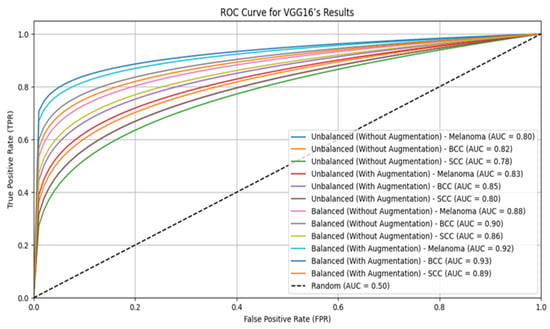
Figure 3.
ROC curve for VGG16.
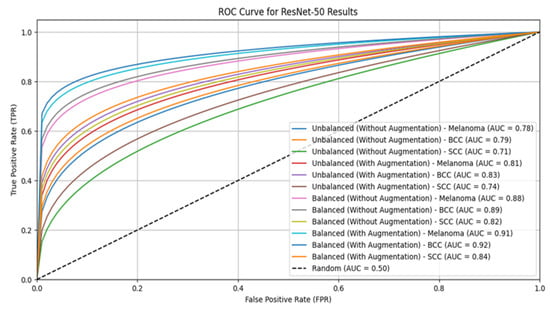
Figure 4.
ROC curve for ResNet-50.
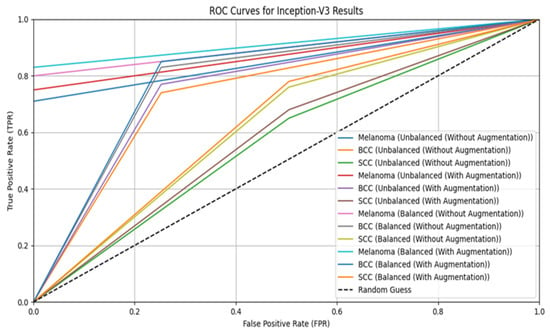
Figure 5.
ROC curve for Inception-V3.
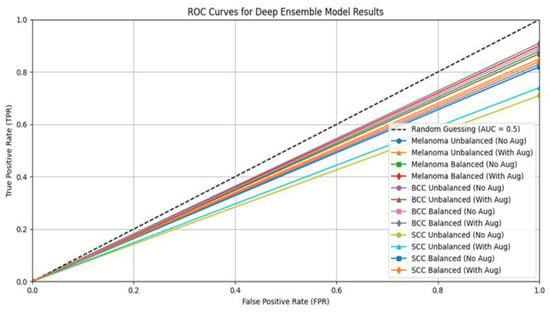
Figure 6.
ROC curve for deep ensemble model.
Then, the system is tested using the ISIC 2019 dataset. The results of the ensemble model for skin lesion classification using both balanced and unbalanced datasets, with and without augmentation, are shown in Table 9. These findings underscore the advantages of integrating various models with balanced datasets and augmentation, improving the accuracy and reliability of skin lesion classification. This strategy increases diagnostic accuracy and shows promise for wider applications in medical image analysis.

Table 9.
Deep ensemble model’s results on balanced and unbalanced datasets of ISIC 2019.
4. Discussion
In this work, we investigated the segmentation and classification of skin lesions using cutting-edge deep learning methods. By leveraging a deep ensemble model combining U-Net, SegNet, and DeepLabv3 for segmentation, and VGG16, ResNet-50, and Inception-V3 for classification, we aimed to enhance the accuracy and reliability of skin lesion detection. The ISIC dataset was balanced using oversampling approaches to address class imbalances and offer a more resilient training process. The results demonstrate that the ensemble models effectively segmented skin lesions and classified them into basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and melanoma, with notable performance improvements when using balanced datasets through oversampling. The segmentation models achieved high accuracy across different types of skin lesions, while the classification models showed significant enhancements in precision, recall, and F1-scores with balanced datasets. These improvements highlight the effectiveness of using ensemble methods and data balancing techniques in medical image analysis.
Comparison with Existing Literature
Table 10 shows comparative results from other existing research in the literature for skin lesion segmentation and classification systems. Our suggested approach yields more accurate findings compared to alternative methods. In comparison with those in the literature, the proposed ensemble model for segmentation outperforms previous methods, achieving a Dice Coefficient of 93 and an IoU of 90 on the ISIC 2018 dataset, surpassing the highest results reported for U-Net (89.3) and DenseUNet (92.23). For classification, the proposed ensemble model also shows superior performance, with an accuracy of 99 on the ISIC 2018 dataset, exceeding the accuracy of other methods such as DenseUNet (97.88) and CNN-based approaches (up to 98.5), and demonstrating its effectiveness in both segmentation and classification tasks.

Table 10.
Comparison with existing literature for segmentation and classification.
5. Conclusions
This study explored advanced deep learning techniques for skin lesion segmentation and classification. Using a deep ensemble approach with U-Net, SegNet, and DeepLabV3 for segmentation and VGG16, ResNet-50, and Inception-V3 for classification, we aimed to improve skin lesion detection. Balancing the ISIC dataset with oversampling addressed class imbalances, leading to enhanced model performance. The significant performance improvements achieved through ensemble methods and data balancing suggest that these techniques are highly effective for skin lesion analysis. The proposed method has several strengths, including improved accuracy for both segmentation and classification through the use of ensemble models, effectively addressing challenges like class imbalance with oversampling. Additionally, data augmentation enhances the model’s robustness, making it more reliable for detecting different types of skin lesions. However, the method has some weaknesses, such as increased computational complexity due to ensemble learning, which demands more resources and time. It also relies heavily on large, well-annotated datasets and requires further clinical validation to assess its practical applicability in real-world settings. Future work could explore additional ensemble configurations or integrate other advanced techniques such as attention mechanisms to further enhance model performance. Additionally, expanding the dataset with more diverse samples could help in validating the robustness of the proposed models across different populations.
Limitations
While our study demonstrated promising results, there are limitations to consider. The oversampling technique, while effective, may introduce some noise in the data, which could potentially impact model generalizability. Future research should address these limitations by exploring alternative balancing techniques or incorporating advanced preprocessing methods. Additionally, we will explore the implications of these limitations in real-world clinical settings, considering factors such as variability in image quality, diversity of skin types, and the integration of our approach into existing clinical workflows. By providing a more nuanced discussion, we aim to better inform readers about the practical considerations of implementing our ensemble models in clinical practice and highlight areas for future research to address these challenges effectively.
Author Contributions
Conceptualization, S.M.T. and H.-S.P.; methodology, S.M.T. and H.-S.P.; software, S.M.T. and H.-S.P.; validation, S.M.T. and H.-S.P.; formal analysis, S.M.T. and H.-S.P.; investigation, S.M.T. and H.-S.P.; resources, H.-S.P. and S.M.T.; data curation, H.-S.P.; writing—original draft preparation, S.M.T.; writing—review and editing, S.M.T. and H.-S.P.; visualization, S.M.T. and H.-S.P.; supervision, H.-S.P.; funding acquisition, H.-S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alkarakatly, T.; Eidhah, S.; Sarawani, M.A.; Sobhi, A.A.; Bilal, M. Skin Lesions Identification Using Deep Convolutional Neural Network. In Proceedings of the 2019 International Conference on Advances in the Emerging Computing Technologies (AECT), Al Madinah Al Munawwarah, Saudi Arabia, 10 February 2020; IEEE: Piscataway, NJ, USA; pp. 209–213. [Google Scholar] [CrossRef]
- Murugan, A.; Nair, S.A.H.; Preethi, A.A.P.; Kumar, K.S. Diagnosis of skin cancer using machine learning techniques. Microprocess. Microsyst. 2021, 81, 103727. [Google Scholar] [CrossRef]
- Salian, A.C.; Vaze, S.; Singh, P. Skin Lesion Classification using Deep Learning Architectures. In Proceedings of the 2020 3rd International Conference on Communication System, Computing and IT Applications (CSCITA), Mumbai, India, 3–4 April 2020; IEEE: Piscataway, NJ, USA; pp. 168–173. [Google Scholar] [CrossRef]
- Ali, M.S.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M.K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Filali, Y.; Khoukhi, H.E.; Sabri, M.A. Texture Classification of skin lesion using convolutional neural network. In Proceedings of the 2019 International Conference on Wireless Technologies, Embedded and Intelligent Systems (WITS), Fez, Morocco, 3–4 April 2019. [Google Scholar] [CrossRef]
- Gouda, W.; Sama, N.U.; Waakid, G.A. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.L.; Rabelo, R.A.; Rodrigues, J.P.C.; Silva, R.V. Automatic Segmentation of Melanoma Skin Cancer Using Deep Learning. In Proceedings of the 2021 IEEE International Conference on E-Health Networking, Application & Services (HEALTHCOM), Shenzhen, China, 1–2 March 2021; IEEE: Piscataway, NJ, USA; pp. 1–6. [Google Scholar] [CrossRef]
- Singh, V.K.; Abdel-Nasser, M.; Rashwan, H.A.; Akram, F.; Pandey, N.; Lalande, A.; Presles, B.; Romani, S.; Puig, D. FCA-Net: Adversarial learning for skin lesion segmentation based on multi-scale features and factorized channel attention. IEEE Access 2019, 7, 130552–130565. [Google Scholar] [CrossRef]
- Yang, X.; Zeng, Z.; Yeo, S.Y.; Tan, C.; Tey, H.L.; Su, Y. A novel multi-task deep learning model for skin lesion segmentation and classification. arXiv 2017, arXiv:1703.01025. [Google Scholar] [CrossRef]
- Liu, L.; Tsui, Y.Y.; Mandal, M. Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. J. Imaging 2021, 7, 67. [Google Scholar] [CrossRef]
- Mirikharaji, Z.; Abhishek, K.; Bissoto, A.; Barata, C.; Avila, S.; Valle, E.; Celebi, M.E.; Hamarneh, G. A survey on deep learning for skin lesion segmentation. Med. Image Anal. 2023, 88, 102863. [Google Scholar] [CrossRef] [PubMed]
- Jimi, A.; Abouche, H.; Zrira, N.; Benmiloud, I. Skin Lesion Segmentation Using Attention-Based DenseUNet. In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023), Lisbon, Portugal, 16–18 February 2023; BIOINFORMATICS. Volume 3, pp. 91–100. [Google Scholar] [CrossRef]
- Bibi, A.; Khan, M.A.; Javed, M.Y.; Tariq, U.; Kang, B.G.; Nam, Y.; Mostafa, R.R.; Sakr, R.H. Skin Lesion Segmentation and Classification Using Conventional and Deep Learning Based Framework. Comput. Mater. Contin. 2022, 71, 2477–2495. [Google Scholar] [CrossRef]
- Ashraf, H.; Waris, A.; Ghafoor, M.F.; Gilani, S.O.; Niazi, I.K. Melanoma segmentation using deep learning with test-time augmentations and conditional random fields. Sci. Rep. 2022, 12, 3948. [Google Scholar] [CrossRef]
- Jafari, M.H.; Karimi, N.; Nasr-Esfahani, E.; Samavi, S.; Soroushmehr, S.M.R.; Ward, K.; Najarian, K. Skin Lesion Segmentation in Clinical Images Using Deep Learning. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancún Center, Cancún, México, 4–8 December 2016. [Google Scholar] [CrossRef]
- Chandra, R.; Hajiarbabi, M. Skin Lesion Detection Using Deep Learning. J. Autom. Mob. Robot. Intell. Syst. 2022, 16, 56–64. [Google Scholar] [CrossRef]
- Gessert, N.; Nielsen, M.; Shaikh, M.; Werner, R.; Schlaefer, A. Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data. MethodsX 2020, 7, 100864. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, Z.; Zheng, Y.; Liu, Z.; Yang, X.X.; Yuan, G.; Xie, J. Deep attention branch networks for skin lesion classification. Comput. Methods Programs Biomed. 2021, 212, 106447. [Google Scholar] [CrossRef] [PubMed]
- Alhudhaif, A.; Almaslukh, B.; Aseeri, A.O.; Guler, O.; Polat, K. A novel nonlinear automated multi-class skin lesion detection system using soft-attention based convolutional neural networks. Chaos Solitons Fractals 2023, 170, 113409. [Google Scholar] [CrossRef]
- Alsahafi, Y.S.; Kassem, M.A.; Hosny, K.M. Skin-Net: A novel deep residual network for skin lesions classification using multilevel feature extraction and cross-channel correlation with detection of outlier. J. Big Data 2023, 10, 105. [Google Scholar] [CrossRef]
- Hosny, K.M.; Elshoura, D.; Mohamed, E.R.; Vrochidou, E.; Papakostas, G.A. Deep Learning and Optimization-Based Methods for Skin Lesions Segmentation: A Review. IEEE Access 2023, 11, 85467–85488. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. DeepLabv3: Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2018, arXiv:1706.05587. [Google Scholar] [CrossRef]
- MNOWAK061. Skin Lesion Dataset. ISIC2018 Kaggle Repository. 2021. Available online: https://www.kaggle.com/datasets/mnowak061/isic2018-and-ph2-384x384-jpg (accessed on 10 April 2022).
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, Hawaii, 21–26 July 2016. [Google Scholar] [CrossRef]
- Sayyad, J.; Patil, P.; Gurav, S. Skin Disease Detection Using VGG16 and InceptionV3. Int. J. Intell. Syst. Appl. Eng. 2024, 12, 148–155. [Google Scholar]
- Barua, S.; Islam, M.M.; Murase, K. A novel synthetic minority oversampling technique for imbalanced data set learning. Lect. Notes Comput. Sci. 2011, 7063, 735–744. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M.; Pei, J. Data Mining: Concepts and Techniques (The Morgan Kaufmann Series in Data Management Systems), 3rd ed.; Elsevier Science Ltd.: Amsterdam, The Netherlands, 2011; pp. 1–703. [Google Scholar] [CrossRef]
- Harangi, B.; Baran, A.; Hajdu, A. Assisted deep learning framework for multi-class skin lesion classification considering a binary classification support. Biomed. Signal Process. Control 2020, 62, 102041. [Google Scholar] [CrossRef]
- Mariani, G.; Scheidegger, F.; Istrate, R.; Bekas, C.; Malossi, C. BAGAN: Data Augmentation with Balancing GAN. arXiv 2018, arXiv:1803.09655. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).