Modeling and Characterizing the Growth of the Texas–New Mexico Measles Outbreak of 2025
Abstract
1. Introduction
2. Materials and Methods
2.1. Exponential Growth Model
2.2. SIR Model
2.3. SEIR Model
3. Results
3.1. Exponential Growth Model Results
3.2. SIR Model Results
3.3. SEIR Model Results
3.4. Sensitivity Analysis of SIR and SEIR Model’s Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Dyer, O. Measles: Texas outbreak spreads to New Mexico. BMJ Br. Med. J. 2025, 388, r357. [Google Scholar] [CrossRef]
- Kuppalli, K.; Perl, T.M. Measles in Texas: Waning vaccination and a stark warning for public health. Lancet Infect. Dis. 2025, 25, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Nowogrodzki, J. Measles is surging in the US: How bad could it get? Nature 2025, 639, 846–847. [Google Scholar] [CrossRef] [PubMed]
- Wikipedia Contributors. 2025 Southwest United States Measles Outbreak—Wikipedia, The Free Encyclopedia. 2025. Available online: https://en.wikipedia.org/w/index.php?title=2025_Southwest_United_States_measles_outbreak&oldid=1286246713 (accessed on 19 April 2025).
- Andreu-Vilarroig, C.; Villanueva, R.J.; González-Parra, G. Mathematical modeling for estimating influenza vaccine efficacy: A case study of the Valencian Community, Spain. Infect. Dis. Model. 2024, 9, 744–762. [Google Scholar] [CrossRef]
- CDC (Center for Disease Control and Prevention). Measles Cases and Outbreaks. 2025. Available online: https://www.cdc.gov/measles/data-research/index.html (accessed on 8 March 2025).
- Johns Hopkins University’s Center for Outbreak Response Innovation (CORI). Measles Cases and Outbreaks. 2025. Available online: https://cori.centerforhealthsecurity.org/resources/measles-outbreak-response (accessed on 6 July 2025).
- CIDRAP. Measles Cases Top 350 in Texas–New Mexico Measles Outbreak. 2025. Available online: https://www.cidrap.umn.edu/measles/measles-cases-top-350-texas-new-mexico-measles-outbreak#:~:text=Steady%20rise%20in%20Texas%20hot,removed%20from%20the%20outbreak%20total (accessed on 15 April 2025).
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Ayodele, O.; Abbasi, A.F.; Prakash, S.; Gosse, J.; Younis, S.; Mangat, J.; Chan, H. Measles outbreak in unvaccinated and partially vaccinated children and adults in the United States and Canada (2018–2019): A narrative review of cases. Inq. J. Health Care Organ. Provision Financ. 2019, 56, 0046958019894098. [Google Scholar]
- Chowell, G.; Viboud, C. Is it growing exponentially fast?—Impact of assuming exponential growth for characterizing and forecasting epidemics with initial near-exponential growth dynamics. Infect. Dis. Model. 2016, 1, 71–78. [Google Scholar] [CrossRef]
- Chowell, G.; Sattenspiel, L.; Bansal, S.; Viboud, C. Mathematical models to characterize early epidemic growth: A review. Phys. Life Rev. 2016, 18, 66–97. [Google Scholar] [CrossRef]
- Chowell, G. Fitting dynamic models to epidemic outbreaks with quantified uncertainty: A primer for parameter uncertainty, identifiability, and forecasts. Infect. Dis. Model. 2017, 2, 379–398. [Google Scholar] [CrossRef]
- Yan, P.; Chowell, G. Quantitative Methods for Investigating Infectious Disease Outbreaks; Springer: Berlin/Heidelberg, Germany, 2019; Volume 70. [Google Scholar]
- Ma, J.; Dushoff, J.; Bolker, B.M.; Earn, D.J. Estimating initial epidemic growth rates. Bull. Math. Biol. 2014, 76, 245–260. [Google Scholar] [CrossRef]
- Hethcote, H.W. The mathematics of infectious diseases. Siam Rev. 2000, 42, 599–653. [Google Scholar] [CrossRef]
- Chowell, G.; Brauer, F. The basic reproduction number of infectious diseases: Computation and estimation using compartmental epidemic models. In Mathematical and Statistical Estimation Approaches in Epidemiology; Springer: Dordrecht, The Netherlands, 2009; pp. 1–30. [Google Scholar]
- Chowell, G.; Viboud, C.; Simonsen, L.; Moghadas, S.M. Characterizing the reproduction number of epidemics with early subexponential growth dynamics. J. R. Soc. Interface 2016, 13, 20160659. [Google Scholar] [CrossRef] [PubMed]
- Duchene, S.; Featherstone, L.; Haritopoulou-Sinanidou, M.; Rambaut, A.; Lemey, P.; Baele, G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020, 6, veaa061. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Bhola, S.; Thakur, P.; Patel, S.K.S.; Kulshrestha, S.; Ratho, R.K.; Kumar, P. Waves and variants of SARS-CoV-2: Understanding the causes and effect of the COVID-19 catastrophe. Infection 2022, 50, 309–325. [Google Scholar] [CrossRef]
- Bugalia, S.; Tripathi, J.P.; Wang, H. Estimating the time-dependent effective reproduction number and vaccination rate for COVID-19 in the USA and India. Math. Biosci. Eng 2023, 20, 4673–4689. [Google Scholar] [CrossRef]
- Córdova-Lepe, F.; Gutiérrez-Jara, J.P.; Chowell, G. Influence of the Effective Reproduction Number on the SIR Model with a Dynamic Transmission Rate. Mathematics 2024, 12, 1793. [Google Scholar] [CrossRef]
- Jorge, D.; Oliveira, J.; Miranda, J.; Andrade, R.; Pinho, S. Estimating the effective reproduction number for heterogeneous models using incidence data. R. Soc. Open Sci. 2022, 9, 220005. [Google Scholar] [CrossRef]
- Koyama, S. Estimating effective reproduction number revisited. Infect. Dis. Model. 2023, 8, 1063–1078. [Google Scholar] [CrossRef]
- Silva, E.P.D.; Lima, A.M. Assessing the Time Evolution of COVID-19 Effective Reproduction Number in Brazil. An. Acad. Bras. CiÊNcias 2024, 96, e20221050. [Google Scholar] [CrossRef]
- Park, S.W.; Champredon, D.; Weitz, J.S.; Dushoff, J. A practical generation-interval-based approach to inferring the strength of epidemics from their speed. Epidemics 2019, 27, 12–18. [Google Scholar] [CrossRef]
- Ma, J. Estimating epidemic exponential growth rate and basic reproduction number. Infect. Dis. Model. 2020, 5, 129–141. [Google Scholar] [CrossRef]
- Park, S.W.; Champredon, D.; Dushoff, J. Inferring generation-interval distributions from contact-tracing data. J. R. Soc. Interface 2020, 17, 20190719. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Bolker, B.M.; Funk, S.; Metcalf, C.J.E.; Weitz, J.S.; Grenfell, B.T.; Dushoff, J. The importance of the generation interval in investigating dynamics and control of new SARS-CoV-2 variants. J. R. Soc. Interface 2022, 19, 20220173. [Google Scholar] [CrossRef]
- Tariq, A.; Undurraga, E.A.; Laborde, C.C.; Vogt-Geisse, K.; Luo, R.; Rothenberg, R.; Chowell, G. Transmission dynamics and control of COVID-19 in Chile, March-October, 2020. PLoS Neglected Trop. Dis. 2021, 15, e0009070. [Google Scholar] [CrossRef]
- Wallinga, J.; Lipsitch, M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc. R. Soc. Biol. Sci. 2007, 274, 599–604. [Google Scholar] [CrossRef]
- Guerra, F.M.; Bolotin, S.; Lim, G.; Heffernan, J.; Deeks, S.L.; Li, Y.; Crowcroft, N.S. The basic reproduction number (R0) of measles: A systematic review. Lancet Infect. Dis. 2017, 17, e420–e428. [Google Scholar] [CrossRef]
- Marinović, A.A.B.; Swaan, C.; Wichmann, O.; van Steenbergen, J.; Kretzschmar, M. Effectiveness and timing of vaccination during school measles outbreak. Emerg. Infect. Dis. 2012, 18, 1405. [Google Scholar] [CrossRef]
- Keeling, M.J.; Grenfell, B.T. Understanding the persistence of measles: Reconciling theory, simulation and observation. Proc. R. Soc. London. Ser. B Biol. Sci. 2002, 269, 335–343. [Google Scholar] [CrossRef]
- Heffernan, J.M.; Keeling, M.J. An in-host model of acute infection: Measles as a case study. Theor. Popul. Biol. 2008, 73, 134–147. [Google Scholar] [CrossRef]
- Fitzgerald, T.L.; Durrheim, D.N.; Merritt, T.D.; Birch, C.; Tran, T. Measles with a possible 23 day incubation period. Commun. Dis. Intell. Q. Rep. 2012, 36, E277–E280. [Google Scholar] [CrossRef]
- O’Donnell, S.; Davies, F.; Vardhan, M.; Nee, P. Could this be measles? Emerg. Med. J. 2019, 36, 310–314. [Google Scholar] [CrossRef]
- Roberts, M.; Tobias, M. Predicting and preventing measles epidemics in New Zealand: Application of a mathematical model. Epidemiol. Infect. 2000, 124, 279–287. [Google Scholar] [CrossRef]
- Klinkenberg, D.; Nishiura, H. The correlation between infectivity and incubation period of measles, estimated from households with two cases. J. Theor. Biol. 2011, 284, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Peter, O.J. Modelling measles transmission dynamics and the impact of control strategies on outbreak Management. J. Biol. Dyn. 2025, 19, 2479448. [Google Scholar] [CrossRef] [PubMed]
- Conlan, A.J.; Grenfell, B.T. Seasonality and the persistence and invasion of measles. Proc. R. Soc. Biol. Sci. 2007, 274, 1133–1141. [Google Scholar] [CrossRef]
- Woelk, C.H.; Jin, L.; Holmes, E.C.; Brown, D.W. Immune and artificial selection in the haemagglutinin (H) glycoprotein of measles virus. J. Gen. Virol. 2001, 82, 2463–2474. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Infectious Diseases of Humans: Dynamics and Control; Oxford University Press: Oxford, UK, 1991. [Google Scholar]
- Cauchemez, S.; Ferguson, N.M. Likelihood-based estimation of continuous-time epidemic models from time-series data: Application to measles transmission in London. J. R. Soc. Interface 2008, 5, 885–897. [Google Scholar] [CrossRef]
- Hempel, K.; Earn, D.J. A century of transitions in New York City’s measles dynamics. J. R. Soc. Interface 2015, 12, 20150024. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Directly transmitted infections diseases: Control by vaccination. Science 1982, 215, 1053–1060. [Google Scholar] [CrossRef]
- Chowell, G.; Nishiura, H.; Bettencourt, L.M. Comparative estimation of the reproduction number for pandemic influenza from daily case notification data. J. R. Soc. Interface 2007, 4, 155–166. [Google Scholar] [CrossRef]
- Viboud, C.; Simonsen, L.; Chowell, G. A generalized-growth model to characterize the early ascending phase of infectious disease outbreaks. Epidemics 2016, 15, 27–37. [Google Scholar] [CrossRef]
- Magal, P.; Webb, G. The parameter identification problem for SIR epidemic models: Identifying unreported cases. J. Math. Biol. 2018, 77, 1629–1648. [Google Scholar] [CrossRef]
- Wallinga, J.; Heijne, J.C.M.; Kretzschmar, M. A measles epidemic threshold in a highly vaccinated population. PLoS Med. 2005, 2, e316. [Google Scholar] [CrossRef]
- Li, M.; Liu, K.; Song, Y.; Wang, M.; Wu, J. Serial interval and generation interval for imported and local infectors, respectively, estimated using reported contact-tracing data of COVID-19 in China. Front. Public Health 2021, 8, 577431. [Google Scholar] [CrossRef]
- Nishiura, H.; Mizumoto, K.; Asai, Y. Assessing the transmission dynamics of measles in Japan, 2016. Epidemics 2017, 20, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, O.; Heesterbeek, J.A.P. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation; John Wiley & Sons: Hoboken, NJ, USA, 2000; Volume 5. [Google Scholar]
- Simon, M.K. Probability Distributions Involving Gaussian Random Variables: A Handbook for Engineers and Scientists; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- He, D.; Ionides, E.L.; King, A.A. Plug-and-play inference for disease dynamics: Measles in large and small populations as a case study. J. R. Soc. Interface 2010, 7, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Gonzàlez-Parra, G.; De Ridder, F.; Huntjens, D.; Roymans, D.; Ispas, G.; Dobrovolny, H.M. A comparison of RSV and influenza in vitro kinetic parameters reveals differences in infecting time. PloS ONE 2018, 13, e0192645. [Google Scholar] [CrossRef] [PubMed]
- González-Parra, G.; Arenas, A.J.; Chen-Charpentier, B.M. A fractional order epidemic model for the simulation of outbreaks of influenza A (H1N1). Math. Methods Appl. Sci. 2014, 37, 2218–2226. [Google Scholar] [CrossRef]
- Trejos, D.Y.; Valverde, J.C.; Venturino, E. Dynamics of infectious diseases: A review of the main biological aspects and their mathematical translation. Appl. Math. Nonlinear Sci. 2022, 7, 1–26. [Google Scholar] [CrossRef]
- Boonpatcharanon, S.; Heffernan, J.M.; Jankowski, H. Estimating the basic reproduction number at the beginning of an outbreak. PLoS ONE 2022, 17, e0269306. [Google Scholar] [CrossRef]
- Davison, A.C.; Hinkley, D.V. Bootstrap Methods and Their Application; Number 1; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Hesterberg, T. Bootstrap. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 497–526. [Google Scholar] [CrossRef]
- Karr, J.; Malik-Sheriff, R.S.; Osborne, J.; Gonzalez-Parra, G.; Forgoston, E.; Bowness, R.; Liu, Y.; Thompson, R.; Garira, W.; Barhak, J.; et al. Model integration in computational biology: The role of reproducibility, credibility and utility. Front. Syst. Biol. 2022, 2, 822606. [Google Scholar] [CrossRef]
- van Boven, M.; Kretzschmar, M.; Wallinga, J.; O’Neill, P.D.; Wichmann, O.; Hahné, S. Estimation of measles vaccine efficacy and critical vaccination coverage in a highly vaccinated population. J. R. Soc. Interface 2010, 7, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Davoudi, B.; Meza, R.; Slim, A.C.; Pourbohloul, B. Epidemics with general generation interval distributions. J. Theor. Biol. 2010, 262, 107–115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Linton, N.; Guha-Sapir, D.; van Loenhout, J.; Keita, M. Outbreak Characteristics and Reproduction Numbers of the 2017 Measles Outbreak in Guinea; Faculté de Santé Publique, Université Catholique de Louvain: Brussels, Belgium, 2018. [Google Scholar][Green Version]
- Grais, R.F.; Ferrari, M.; Dubray, C.; Bjørnstad, O.; Grenfell, B.; Djibo, A.; Fermon, F.; Guerin, P.J. Estimating transmission intensity for a measles epidemic in Niamey, Niger: Lessons for intervention. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 867–873. [Google Scholar] [CrossRef]
- Broutin, H.; Mantilla-Beniers, N.B.; Simondon, F.; Aaby, P.; Grenfell, B.T.; Guégan, J.F.; Rohani, P. Epidemiological impact of vaccination on the dynamics of two childhood diseases in rural Senegal. Microbes Infect. 2005, 7, 593–599. [Google Scholar] [CrossRef]
- Garly, M.L.; Aaby, P. The challenge of improving the efficacy of measles vaccine. Acta Trop. 2003, 85, 1–17. [Google Scholar] [CrossRef]
- Orenstein, W.A.; Bernier, R.H.; Dondero, T.J.; Hinman, A.R.; Marks, J.S.; Bart, K.J.; Sirotkin, B. Field evaluation of vaccine efficacy. Bull. World Health Organ. 1985, 63, 1055. [Google Scholar]
- Kumar, S.S.; Hartner, A.M.; Chandran, A.; Gaythorpe, K.A.; Li, X. Evaluating effective measles vaccine coverage in the Malaysian population accounting for between-dose correlation and vaccine efficacy. BMC Public Health 2023, 23, 2351. [Google Scholar] [CrossRef]
- González-Parra, G.; Pérez, C.L.; Llamazares, M.; Villanueva, R.J.; Villegas-Villanueva, J. Challenges in the mathematical modeling of the spatial diffusion of SARS-CoV-2 in Chile. Math. Biosci. Eng. 2025, 22, 1680–1721. [Google Scholar] [CrossRef]
- Kim, Y.; Barber, A.V.; Lee, S. Modeling influenza transmission dynamics with media coverage data of the 2009 H1N1 outbreak in Korea. PLoS ONE 2020, 15, e0232580. [Google Scholar] [CrossRef]
- Lalwani, S.; Sahni, G.; Mewara, B.; Kumar, R. Predicting optimal lockdown period with parametric approach using three-phase maturation SIRD model for COVID-19 pandemic. Chaos Solitons Fractals 2020, 138, 109939. [Google Scholar] [CrossRef]
- United States Census Bureau. County Population Totals and Components of Change: 2020–2024. 2025. Available online: https://www.census.gov/data/tables/time-series/demo/popest/2020s-counties-total.html (accessed on 14 March 2025).
- London, W.P.; Yorke, J.A. Recurrent outbreaks of measles, chickenpox and mumps: I. Seasonal variation in contact rates. Am. J. Epidemiol. 1973, 98, 453–468. [Google Scholar] [CrossRef]
- Szusz, E.K.; Garrison, L.P.; Bauch, C.T. A review of data needed to parameterize a dynamic model of measles in developing countries. BMC Res. Notes 2010, 3, 1–17. [Google Scholar] [CrossRef]
- Tanaka, H.; Takahashi, Y.; Matsumoto, S.; Matsuyama, M.; Nakayama, K.; Kodaira, H.; Kubota, Y.; Kawakami, J.; Kurata, T.; Kanbayashi, D.; et al. Shorter incubation period in symptomatic measles patients who had no history of measles vaccination. Vaccine 2025, 45, 126652. [Google Scholar] [CrossRef]
- Hulland, E.N.; Charpignon, M.L.; Berkane, T.; Majumder, M.S. Underimmunisation during the 2025 Texas measles outbreak. Lancet Infect. Dis. 2025, 25, 607–609. [Google Scholar] [CrossRef]
- Schweitzer, K. Amid Texas Measles Outbreak, Clinicians Struggle to Offset Increasing Vaccine Hesitancy. JAMA 2025, 333, 1278–1281. [Google Scholar] [CrossRef]
- Liu, G.; Liao, Z.; Xu, X.; Liang, Y.; Xiong, Y.; Ni, J. Accuracy of parent-reported measles-containing vaccination status of children with measles. Public Health 2017, 144, 92–95. [Google Scholar] [CrossRef]
- Dong, E.; Saiyed, S.; Nearchou, A.; Okura, Y.; Gardner, L.M. Trends in county-level MMR vaccination coverage in children in the United States. JAMA 2025, 334, 730–732. [Google Scholar] [CrossRef]
- Ashby, B.; Best, A. Herd immunity. Curr. Biol. 2021, 31, R174–R177. [Google Scholar] [CrossRef]
- Griffith, B.C.; Ulrich, A.K.; Becker, A.B.; Nederhoff, D.; Koch, B.; Awan, F.A.; Basta, N.E. Does education about local vaccination rates and the importance of herd immunity change US parents’ concern about measles? Vaccine 2020, 38, 8040–8048. [Google Scholar] [CrossRef]
- Sanchez, D.J. Measles virus: Continued outbreaks while striving for eradication. Virulence 2024, 15, 2386022. [Google Scholar] [CrossRef]
- Takla, A.; Wichmann, O.; Rieck, T.; Matysiak-Klose, D. Measles incidence and reporting trends in Germany, 2007–2011. Bull. World Health Organ. 2014, 92, 742–749. [Google Scholar] [CrossRef]
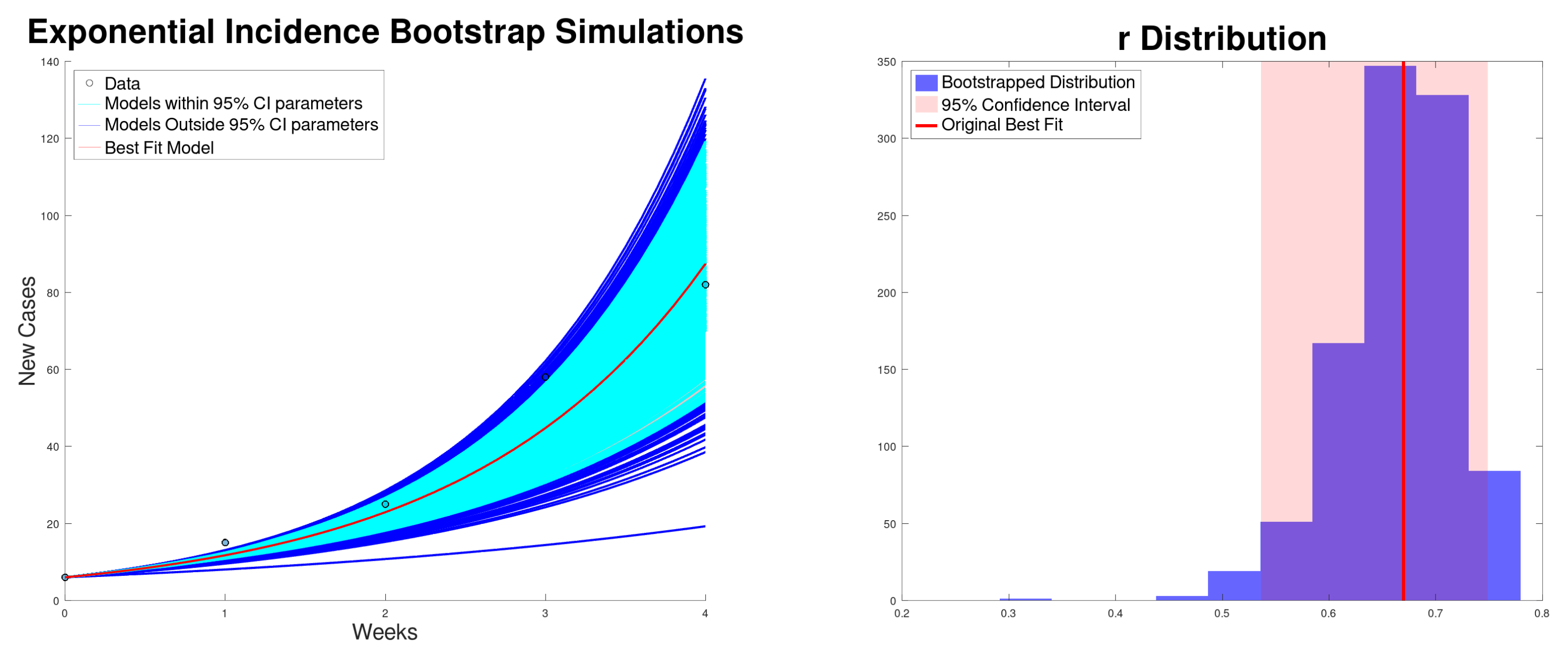

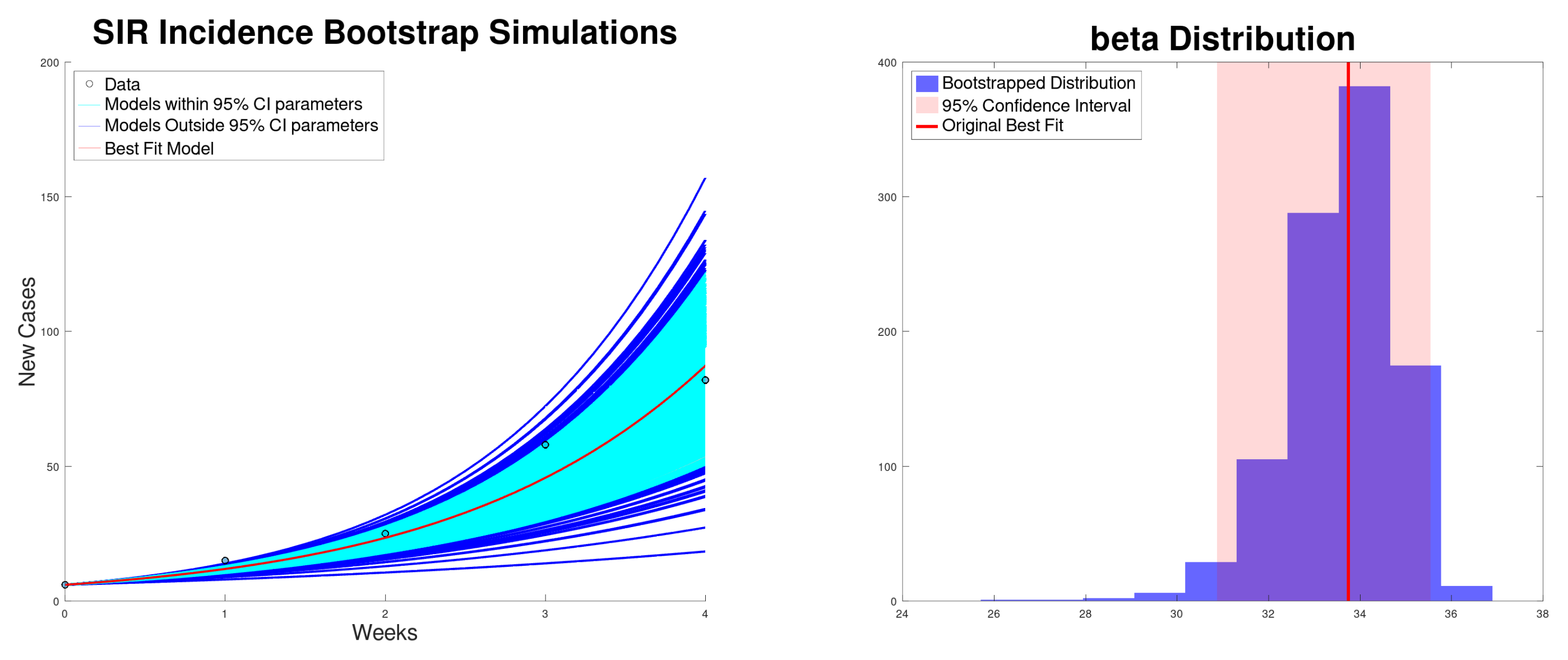

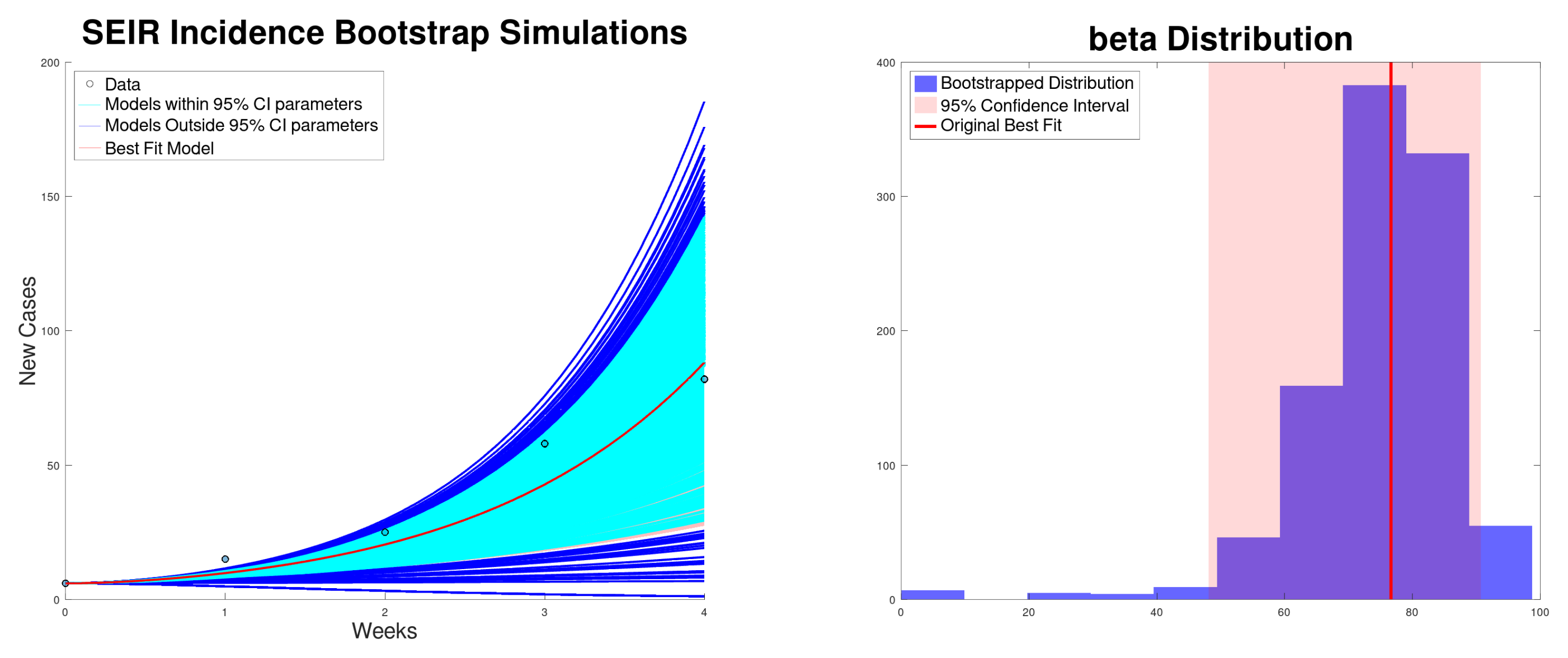

| Growth Rate r (95% CI) | (95% CI) | (95% CI) | SSR | |
|---|---|---|---|---|
| Incidence | 0.669 (0.536–0.748) | 2.51 (2.12–2.75) | 31.93 (27.08–35.06) | 220 |
| Cumulative | 0.869 (0.765–0.936) | 3.16 (2.80–3.41) | 40.28 (35.74–43.39) | 762 |
| Model | (da) (95% CI) | Range of (95% CI) | (95% CI) | SSR |
|---|---|---|---|---|
| SIR incidence | 33.7 (30.9–35.5) | 1.69 (1.54–1.77) | 33.7 (30.9–35.5) | 192 |
| SIR cumulative | 33.1 (32.4–33.7) | 1.65 (1.61–1.68) | 33.1 (32.4–33.7) | 67 |
| SEIR incidence | 76.6 (48.1–90.7) | 3.82 (2.40–4.52) | 76.6 (48.1–90.7) | 318 |
| SEIR cumulative | 74.6 (65.0–82.1) | 3.72 (3.24–4.09) | 74.6 (65.0–82.1) | 243 |
| SIR Sensitivity | Incidence | Cumulative | |||
|---|---|---|---|---|---|
| Original | 33.7 | 1.68 | 33.0 | 1.65 | |
| 92% Vaccinated | 21.0 ▾ | 1.68 | 20.59 ▾ | 1.65 | ← lower |
| 98% Vaccinated | 85.7 ▴ | 1.71 | 83.6 ▴ | 1.67 | ← higher |
| 5 day infectious period | N/A | 28.34 ▾ | 1.42 | ||
| 8.7 day infectious period | N/A | 37.14 ▴ | 1.85 | ||
| SEIR Sensitivity | Incidence | Cumulative | |||
|---|---|---|---|---|---|
| Original | 76.6 | 3.82 | 74.6 | 3.72 | |
| 92% Vaccinated | 47.6 ▾ | 3.80 | 46.4 ▾ | 3.71 | ← lower |
| 98% Vaccinated | 197.3 ▴ | 3.92 | 190.9 ▴ | 3.80 | ← higher |
| 5 day infectious period | N/A | 63.5 ▾ | 3.17 | ← lower | |
| 8.7 day infectious period | N/A | 84.3 ▴ | 4.20 | ||
| 7 day latent period | 60.8 ▾ | 3.04 | 59.3 ▾ | 2.96 | ← lower |
| 15 day latent period | 92.6 ▴ | 4.62 | 90.1 ▴ | 4.49 | ← higher |
| 50% higher E(0) | 71.4 ▾ | 3.56 | 68.3 ▾ | 3.41 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Parra, G.; Vestrand, A.; Mujynya, R. Modeling and Characterizing the Growth of the Texas–New Mexico Measles Outbreak of 2025. Epidemiologia 2025, 6, 60. https://doi.org/10.3390/epidemiologia6040060
González-Parra G, Vestrand A, Mujynya R. Modeling and Characterizing the Growth of the Texas–New Mexico Measles Outbreak of 2025. Epidemiologia. 2025; 6(4):60. https://doi.org/10.3390/epidemiologia6040060
Chicago/Turabian StyleGonzález-Parra, Gilberto, Annika Vestrand, and Remy Mujynya. 2025. "Modeling and Characterizing the Growth of the Texas–New Mexico Measles Outbreak of 2025" Epidemiologia 6, no. 4: 60. https://doi.org/10.3390/epidemiologia6040060
APA StyleGonzález-Parra, G., Vestrand, A., & Mujynya, R. (2025). Modeling and Characterizing the Growth of the Texas–New Mexico Measles Outbreak of 2025. Epidemiologia, 6(4), 60. https://doi.org/10.3390/epidemiologia6040060







