Abstract
Histocompatibility testing is pivotal in any renal transplantation workup, aimed at enhancing prospective donor recipient compatibility and improving transplant outcomes. The evolution and advancement of histocompatibility testing, particularly HLA typing, have significantly improved its precision. This study outlines the historical progression from serologic to DNA-based HLA typing, emphasizing the role of HLA proteins in immune response. Anti-HLA antibodies, targeting HLA proteins, pose challenges in renal transplantation. Monitoring and managing these antibodies are critical for renal transplant success. Complement-dependent cytotoxicity crossmatch and flow cytometry crossmatch are essential techniques for assessing donor–recipient compatibility. Panel-reactive antibody assesses antibodies against a panel of donor antigens, often HLA. Higher PRA levels (percentage) complicate donor matching, requiring specialized protocols. Virtual crossmatch evaluates recipient anti-HLA antibodies against potential donors through synthetic beads. This approach predicts crossmatch outcomes by comparing antibody profiles, offering a valuable tool for the risk assessment of renal transplantation. Despite advancements, a comprehensive understanding of alloreactive immune responses requires a combination of assays, emphasizing the importance of a multifaceted approach in histocompatibility testing. This is an attempt to compile the relevant information, providing a basis for comparison in a clear and foundational format for histocompatibility testing laboratories.
1. Introduction
Histocompatibility testing is an integral part of the organ transplantation workup process, especially in renal transplantation, helping to identify the most compatible donor–recipient pairs and improve the chances of a successful renal transplant outcome [1]. Advances in testing methods continue to enhance the precision and efficiency of histocompatibility assessments. Significant progress in histocompatibility testing has revolutionized the safety of renal transplantation procedures, leading to a notable reduction in incidents of renal rejection [2]. Medawar, Billingham, and others have highlighted the key role of the immune system in recognizing and rejecting foreign tissues through a process termed sensitization. Sensitization occurs when the recipient’s immune system recognizes the transplanted tissue as foreign and mounts an immune response against it. This response involves the activation of immune cells, such as T cells and B cells, and the production of antibodies directed against the transplanted tissue. However, it is important to note that not all transplanted tissues/organs elicit the same degree of sensitization or immune response. Factors such as the degree of histocompatibility between the donor and recipient, the type of tissue or organ transplanted, and the presence of immunosuppressive therapy can influence the outcome of transplantation. Referencing these initial observations and studies is critical for understanding the immunologic basis of transplant rejection and the development of strategies to mitigate it [3,4].
Over the past 55 years, histocompatibility testing in solid organ transplantation has undergone significant changes since its inception by Dr. Paul Terasaki. Despite these changes, the fundamental aim remains consistent: to assess the immunologic risk of a transplant recipient relative to potential donors. Advances in techniques for HLA testing and antibody detection have enhanced the precision and sensitivity of these tests. They aim to predict the likelihood of the recipient’s immune system identifying the transplanted organ as foreign, which could lead to damaging inflammatory responses. The histocompatibility testing laboratory’s assessments complement clinical evaluations by estimating this risk. Importantly, these testing methods are not restricted to pre-transplant evaluations; post-transplant assessments, including antibody evaluation and newer T-cell assays, are increasingly being used to predict acute and chronic alloimmune complications. Therefore, it is crucial for clinical services to grasp the intricate and interconnected nature of available histocompatibility testing methods [5].
Pretransplant histocompatibility testing relies on critical assessments such as human leukocyte antigen (HLA) typing of both the prospective donor and recipient and crossmatches conducted through complement-dependent cytotoxicity (CDC) or flow-cytometer-based methods commonly known as flow cytometry crossmatch (FCXM) [3,4,5]. The evolution of these histocompatibility testing technologies, evolving since the 1960s, has sharpened predictive methodologies for graft (renal) rejection [2,6]. Progression from polymerase chain reaction (PCR)-based HLA typing to sequence-based methods and from cell-based crossmatches to virtual crossmatches using advanced solid-phase platforms has strengthened our comprehension of donor-specific antibodies (DSAs). This has also challenged the conventional belief that the presence of DSA is an absolute bar to renal transplantation [7,8,9]. HLA typing, cell-based crossmatching (CDC and FCXM), panel-reactive antibodies (PRA), and virtual crossmatch (SAB) are some key components related to histocompatibility testing in renal transplantation workup [10]. Despite this progress, pretransplant tests still face practical challenges, particularly in resource-constrained settings like India. There exists a requirement for a comprehensive guide tailored for laboratories, covering all pertinent details regarding histocompatibility testing. The objective here is to furnish such information in a clear and foundational format in the form a concise review.
2. Human Leukocyte Antigen (HLA) Typing
HLAs are proteins on the surface of cells that play a critical role in the immune system. The two main classes of HLA molecules are Class I and II [11]. There exist three primary HLA class I genes, namely HLA-A, HLA-B, and HLA-C. In addition, there are also three major class II genes, HLA-DR (α-chain by HLA-DRA and four β-chains by HLA-DRB1, DRB3, DRB4, and DRB5 loci), DQ (α-chain by HLA-DQA1 and β-chain by HLA-DQB1), and DP (α-chain encoded by HLA-DPA1 locus and β-chain by HLA-DPB1). Both the donor and recipient undergo HLA typing to identify their specific HLA alleles. The goal is to find a donor whose HLA profile closely matches that of the recipient to minimize the risk of rejection. In the past, the HLA phenotype was identified through serologic typing, involving the combination of an individual’s lymphocytes with various sera containing specific HLA antibodies. Nowadays, DNA typing techniques, such as polymerase chain reaction (e.g., sequence-specific primers (SSP), sequence-specific oligonucleotide (SSO), next-generation sequencing (NGS), and third-generation sequencing (MinION, a nanopore sequencing platform)), are employed to determine the HLA phenotype more efficiently [7,8,12]. The major differences between SSP, SSO, and NGS HLA typing are represented in Table 1.

Table 1.
Major differences between SSP, SSO, and NGS HLA typing [7,8].
3. Anti-HLA Antibodies
Anti-HLA antibodies are antibodies that target HLA proteins. HLA proteins are cell surface proteins that play a crucial role in the immune system by presenting antigens to T cells. These proteins are highly polymorphic, meaning they can vary significantly between individuals. Anti-HLA antibodies can be problematic in the context of renal transplantation. When a person receives an organ (renal) transplant, the immune system may recognize the HLA proteins on the transplanted organ (graft) as foreign and mount an immune response against it. This can lead to the rejection of the transplanted organ [10,13]. There are two main types of HLA antibodies: pre-existing (or pre-formed) antibodies and de novo antibodies [14].
Pre-existing antibodies: These antibodies are present in the recipient before transplantation, often due to previous exposure to HLA antigens through blood transfusions, pregnancies, or previous transplants. Pre-existing antibodies can increase the risk of hyperacute or acute rejection of the transplanted organ.
De novo antibodies: These antibodies develop after transplantation as a result of exposure to the new HLA antigens present on the transplanted organ. De novo antibodies can contribute to chronic rejection, a more gradual and ongoing form of rejection that can occur over an extended period.
Monitoring and managing anti-HLA antibodies are crucial in organ transplantation in order to improve transplant outcomes. Transplant centers (TCs) typically assess the recipient’s antibody profile before transplantation to identify the presence of pre-formed antibodies. This information is crucial for developing a strategy to reduce the risk of antibody-mediated rejection (AMR) [15,16].
4. Complement-Dependent Cytotoxicity Crossmatch (CDCXM)
Complement-dependent cytotoxicity crossmatching (CDCXM) is a crucial technique in the field of transplantation, particularly in organ and tissue transplantation [17]. This method is employed to assess the compatibility between the prospective donor and recipient by examining the potential for complement activation and subsequent cell lysis. The procedure involves separating prospective donor lymphocytes (T and B cells). The T and B cells are separately examined against the recipient’s serum [9]. The humoral immunological response operates through the activation of the complement system via the classical pathway [2]. To illustrate the consequences of this sequence, complement (usually from rabbit serum) is introduced to the recipient’s serum mixed with the prospective donor’s lymphocytes, resulting in the observation of cell lysis in the lymphocytes [2]. In 1969, Paul Terasaki proposed the idea that the immediate failure of kidney allografts could be attributed to preexisting allogeneic antibodies in the recipient. He also recommended the utilization of lymphocyte cytotoxicity as a method for identifying and aligning transplantation antigens [5]. The result is expressed as the percentage (%) of lymphocytes within the cell panel that experience lysis due to the activation of the complement system [9]. A negative crossmatch indicates the absence of complement-fixing antibodies against lymphocytes, minimizing the risk of acute graft rejection. Conversely, a positive crossmatch signifies the presence of antibodies against lymphocytes, heightening the risk of graft rejection [11,18,19]. The reliability of the CDCXM assay depends upon the viability of the donor cells. False-positive results may occur due to nonspecific cell death caused by complement sensitivity [2]. In most cases, autoantibodies manifest as IgM rather than IgG antibodies. To determine if autoantibodies are the cause of the positive result, it is essential to conduct an auto-crossmatch. This procedure involves crossmatching the recipient serum with the recipient lymphocytes, as opposed to the donor lymphocytes. Additionally, it is crucial to repeat the initial crossmatch while introducing dithiothreitol (DTT). DTT serves to diminish the disulfide bonds present in IgM, thereby inhibiting the activation of IgM antibodies and preventing false-positive results. IgM antibodies are generally considered to lack significant pathological implications in transplantation scenarios [9]. The test displays a low sensitivity in identifying anti-HLA class II antibodies [11]. Although numerous technical adjustments have been implemented to refine the sensitivity and specificity of the CDCXM test, the incorporation of anti-human globulin (AHG) is one among them. However, the inclusion of AHG does not improve this sensitivity; at times, it may even result in nonspecific binding to fragment crystallizable receptors (FcR) on B cells [11,20]. Complement activation necessitates elevated antibody concentrations, potentially impacting the test accuracy. Additional processing steps, such as B cell isolation and differentiation between anti-HLA class I and class II antibodies, are essential [8]. These factors collectively contribute to a high interlaboratory variability, emphasizing the need for careful consideration when interpreting CDCXM assay results [17].
5. Flow Cytometry Crossmatch (FCXM)
A flow crossmatch is a procedure in which the recipient serum is introduced to prospective donor lymphocytes (T or B), followed by incubation with fluorescein-labeled antibodies targeting human IgG [2]. These labeled antibodies attach to all IgG antibodies present in the recipient serum. When a DSA in the serum binds to the donor lymphocytes, it becomes identifiable through flow cytometry. While the CDCXM relies on assessing cell lysis as a functional readout, the FCXM focuses specifically on detecting the binding of HLA-specific antibodies, without considering their potential for complement fixation or pathogenic effects [9]. In the FCXM, the recipient serum is introduced to the donor lymphocytes (T or B) alongside anti-IgG fluorescein-labeled antibodies [9]. A negative result indicates the absence of donor-specific antibodies, leading to no binding. Conversely, a positive result occurs when donor-specific antibodies attach to the lymphocytes, which are identified through flow cytometry when the tagged lymphocytes are marked by anti-IgG fluorescein-labeled antibodies [1,9,16,21]. A basic comparison of CDCXM and FCXM is presented in Table 2. The CDCXM technique is depicted in Figure 1 below, while Figure 2 illustrates the FCXM. The FCXM, similar to other diagnostic tests, has limitations and may yield inaccurate results, either false positive or false negative. Lymphocytes express various surface antigens, aside from HLA molecules, which can bind antibodies in the recipient’s serum, regardless of their specificity. Several factors contribute to false-negative outcomes, such as a low HLA expression on donor cells, an excessive number of cells, a low serum volume, low DSA levels, lymphocyte impurity, and high background in the negative control serum. False-positive results may occur due to IgG binding to FcR in B cells, a low negative control background, inadequate washing after antibody incubations, the presence of autoantibodies, and the use of therapeutic antibodies like anti-thymocyte globulin, rituximab (anti-CD20), alemtuzumab (anti-CD52), basiliximab (anti-CD25), and daclizumab (anti-CD25) [20]. Pronase treatment has been employed to diminish the attachment of immunoglobulins to Fc receptors and to decrease the responsiveness to therapeutic antibodies targeting CD20 in lymphocytes [21]. It is crucial to correlate the crossmatch results with the patient’s history, sensitizing events, and DSA history to properly interpret potential false-positive or false-negative scenarios. The sensitivity and specificity of CDC crossmatch and flow cytometry crossmatch can vary depending on several factors, including the specific protocol used, the expertise of the laboratory performing the test, and the population being tested. A study published by Ho et al. found a sensitivity of 5% and a specificity of 99% for CDC, compared to a 17% sensitivity and 86% specificity for flow cytometry. Another study from India by Tiwari et al. reported a sensitivity of 12.1% and a specificity of 100% for CDCXM, whereas the sensitivity and specificity for FCXM were 84.8% and 89.6%, respectively. Overall, flow cytometry crossmatch shows a higher sensitivity than CDC crossmatch, whereas CDC crossmatch is found to be more specific than flow cytometry crossmatch [22,23].

Table 2.
Basic comparison of Complement-Dependent Cytotoxicity crossmatch (CDCXM) and Flow Cytometry Crossmatch (FCXM) [16,17,18,19,20,21].
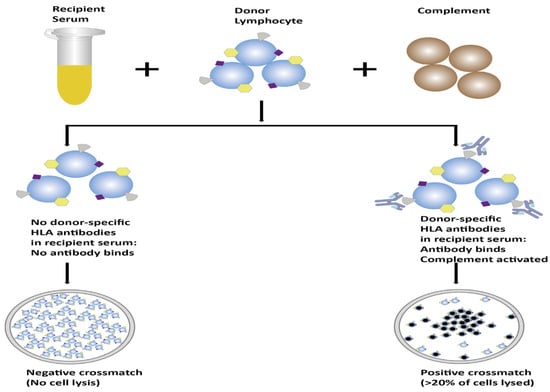
Figure 1.
Schematic representation of the process of CDCXM.
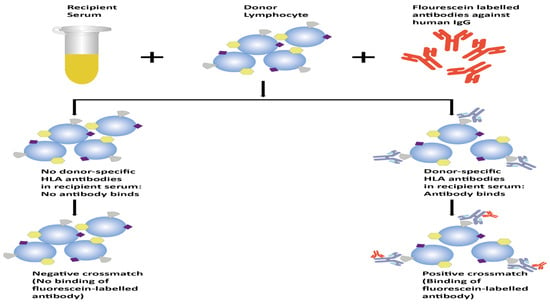
Figure 2.
Schematic representation of the process of FCXM.
6. Panel-Reactive Antibodies (PRA)
The panel-reactive antibodies (PRA) assay is performed to assess the level of antibodies in a patient’s serum that react against a panel of potential donor antigens. In the context of organ (renal) transplantation, these antigens are usually HLAs. The PRA assay involves exposing the patient’s serum to a panel of cells with known HLA types. The test measures the percentage (%) of cells in the panel to which the patient’s antibodies react. A higher PRA percentage (%) indicates a greater likelihood of the patient having antibodies against a wide range of potential organ donors. High PRA levels can complicate the matching process for organ (renal) transplantation. Finding a suitable organ donor with a low risk of rejection becomes challenging when the patient has a high PRA. In such cases, clinicians may consider exploring strategies to mitigate the risk of AMR [20,21,22,23]. The effectiveness of PRA is influenced by both the composition of the panel and the methodology employed for antibody detection. Significant variability in the antigen panel arises when utilizing locally sourced cell panels or diverse commercially available solid-phase immunoassays (SPIs), which may not accurately reflect the potential donor population. To overcome these constraints, the calculated panel-reactive antibody (cPRA) was introduced to standardize the reporting of PRA. It is derived from HLA antigen frequencies that reflect the organ donor population of a particular country or region, resulting in a percentage score. Currently, it stands as the most accurate estimate of the probability of a positive crossmatch with a randomly selected donor [24,25].
7. Virtual Crossmatch (VXM)
Virtual crossmatch (VXM) is a method used to evaluate the immunological compatibility between a recipient and a potential donor. This approach relies on utilizing bead technology to compare the recipient’s anti-HLA antibodies with the donor’s HLA antigens [26,27]. A positive VXM occurs when a DSA is detected. The VXM process involves combining the recipient’s serum with synthetic microspheres (beads) coated with HLA antigens, each distinguished by a unique dye signature. In the presence of anti-HLA antibodies, they selectively attach to the corresponding bead. Subsequently, a detector antibody binds, capturing a reporter dye. Using a laser beam, the beads are examined for the reporter dye presence, establishing an antibody profile in the recipient [6,7]. This profile can then be compared to the HLA makeup of a potential donor, enabling a prediction of the crossmatch outcome. The virtual crossmatch platform, as illustrated in Table 3, provides varied information based on three distinct types of target antigens [28,29]. However, the prozone effect, bead saturation, shared epitopes, and denatured antigen are noteworthy factors that should be considered, as they have the potential to contribute to variability in interpretation. The “prozone effect”, also known as the “hook effect” or “inhibition”, happens when complement interference messes up antibody binding. Diluting the serum can fix this by removing what is blocking the antibodies. Then, the reporter antibody can perform its job properly, attaching to the antigen–antibody complex on the beads [30].
Each of these elements plays a unique role in influencing assay outcomes, and understanding their individual implications is crucial for a comprehensive analysis [27]. A cross-reactive group (CREG) is defined as a set of HLA antigens that share a common public epitope and exhibit a consistent pattern of reactivity. To derive meaningful interpretations as DSAs, it is imperative to analyze the recipient’s antibody profile in relation to the pre-identified CREGs [11]. The key differences between cell-based crossmatch (CDC and FCXM) and VCXM are presented in Table 4. In the extensive evaluation process for renal transplant candidates, following the crucial histocompatibility testing, a series of diverse scenarios may unfold, necessitating a meticulous examination to arrive at a conclusive clinical decision regarding transplantation suitability. These scenarios are intricately linked to the findings obtained from a variety of assays, which encompass both cell-based (CDC and FCXM) and virtual crossmatch single-antigen bead (SAB) techniques [31,32,33,34,35,36,37]. The SAB results are mainly interpreted qualitatively through mean fluorescence intensity (MFI). Converting MFIs into precise antibody titers is complex, so correlations with cell-based assays and patient immunologic history are used. The MFI values are affected by factors like antibody alignment, density, and concentration. The categorization of SAB results as negative or positive relies on the center’s risk tolerance and clinical context, with the MFI cutoff being pivotal. MFI thresholds vary across different organs for transplantation. Lab variability stems from differences in SAB products and internal factors like personnel and equipment. Considering these factors is crucial for accurate SAB result interpretation [28,30].

Table 3.
The representation of varied information in virtual crossmatch tests [32,33].
Table 3.
The representation of varied information in virtual crossmatch tests [32,33].
| Assay | HLA Source | Analysis | Interpretation |
|---|---|---|---|
| Anti HLA antibody (screening) | Most common Antigens (mixed and random) | Positive or negative | Does the patient possess any antibodies? |
| Panel Reactive Antibodies (PRA) | Each bead is a phenotype of an individual | Positive (%) or negative | Extent of sensitization? |
| Single-antigen bead (SAB) | One HLA antigen per bead | Positive or negative (Semi-Quantitative, MFI) | Does patient possess any DSA? |
HLA: Human Leukocyte Antigen; PRA: Panel-Reactive Antibodies; SAB: Single-Antigen Bead, MFI: Mean Fluorescence Intensity; and DSA: Donor-Specific Antibodies.
Despite the advent in technology, the C1q assay, which evaluates the binding of complement component C1q to the Fc region of antibodies bound to antigens, is one approach used to assess complement fixation and antibody-mediated damage. Dr. Stanley Jordan, a renowned figure in transplant immunology, has advocated for the use of C1q testing in assessing the pathogenicity of donor-specific antibodies (DSAs) in kidney transplantation. By assessing C1q binding to antigen–antibody complexes, clinicians can better evaluate the pathogenicity of DSAs and make informed decisions regarding organ transplantation. This approach allows for a more precise assessment of the risk of antibody-mediated rejection and helps to tailor immunosuppressive therapy to mitigate this risk. The use of C1q testing underscores the importance of comprehensive immunological evaluations in transplant medicine to improve patient outcomes and prolong graft survival [38].
To provide a comprehensive understanding of the intricate considerations that guide the transplantation decision-making process, a succinct yet detailed summary of these assessments is systematically presented in tabular format within Table 5 [39]. This visual representation serves as a valuable tool, offering a structured and easily accessible overview of the multifaceted aspects that clinicians or laboratories must consider when determining the feasibility of a renal transplant. The information encapsulated in the table encompasses a spectrum of factors, including immunological compatibility, potential risks, and patient-specific variables, all of which play pivotal roles in shaping the final clinical decision.

Table 4.
Representation of the key differences between cell-based cross match and virtual crossmatch tests [16,17,18,19,20,21,29,30,31].
Table 4.
Representation of the key differences between cell-based cross match and virtual crossmatch tests [16,17,18,19,20,21,29,30,31].
| Characteristic | Cell-Based Crossmatch (CDCXM and FCXM) | Virtual Crossmatch |
|---|---|---|
| Goal | Determine compatibility between prospective donor and recipient by testing for the presence of preformed antibodies. | Assess the percentage of the population to which a patient may be sensitized, indicating the likelihood of finding a compatible donor. |
| Methodology | Involves mixing recipient’s serum with donor lymphocytes (T and B cells) cells to detect any preformed antibodies against donor antigens. | Utilizes HLA typing data to predict potential antibody reactions without physical mixing of donor’s cells. |
| Interpretation | Provides information on the presence of preformed antibodies in the recipient. | Utilizes information about the recipient’s sensitization and the donor’s HLA antigens to predict the likelihood of a positive crossmatch. |
| Assessment of Risk | Identifies immediate risk of rejection due to preformed antibodies. | Predicts the risk of a positive crossmatch based on the recipient’s antibody profile and the donor’s HLA antigen profile. |
CDCXM: Complement-Dependent Cytotoxicity crossmatch; FCXM; Flow Cytometry Crossmatch; HLA: Human Leukocyte Antigen.

Table 5.
Representation of various scenarios following pre-transplant histocompatibility investigations [39].
Table 5.
Representation of various scenarios following pre-transplant histocompatibility investigations [39].
| CDC Crossmatch | Flow Crossmatch | Single-Antigen Bead (SAB) (Virtual Cross Match) | Perspective |
|---|---|---|---|
| Positive | Positive | Positive |
|
| Negative | Positive | Positive |
|
| Negative | Negative | Positive |
|
| Positive | Positive | Negative |
|
| Positive | Negative | Negative |
|
| Negative | Positive | Negative |
|
CDC: Complement-Dependent Cytotoxicity; DSA: Donor-Specific Antibodies; HLA: Human Leukocyte Antigen; SAB: Single-Antigen Bead, and MFI: Mean Fluorescence Intensity.
Pre-transplant risk assessment represents a crucial phase in the comprehensive evaluation of individuals undergoing organ (renal) transplantation [40,41]. This pivotal process serves as the foundation for devising a tailored approach to renal transplant procedures, with subsequent testing strategies being intricately tailored to the specific risk assessment category identified for each patient. This risk assessment encompasses a spectrum of immunological considerations, classifying patients into distinct risk categories that dictate the intensity and nature of the subsequent testing protocols. At the forefront of these categories are individuals deemed to be at high risk, indicating a heightened susceptibility to rejection [1]. This susceptibility is often linked to a significant presence of circulating antibodies specific to mismatched donor antigens detected at the time of transplantation. The identification of such “high-risk profiles” prompts the implementation of more rigorous testing regimens to preemptively address and mitigate potential rejection events. In the “intermediate risk category”, patients exhibit prior donor-reactive sensitization or a moderate level of sensitization to specific mismatched HLA specificities [42,43,44,45]. This nuanced classification necessitates a strategic approach to testing that accounts for the intricacies of sensitization patterns, tailoring interventions to the unique immunological landscape of each patient. This targeted strategy aims to optimize transplant outcomes by addressing specific sensitivities that fall within this intermediate risk spectrum. Conversely, the “low-risk category” characterizes patients as either non-sensitized or minimally sensitized individuals with HLA-mismatched organs [1]. In instances where sensitization is present, it is crucial to assess the absence of known current or historical donor-specific antibodies (DSAs) [46,47,48,49]. This subset of patients requires a more conservative histocompatibility testing approach, with a focus on monitoring and maintaining the delicate balance between the host immune system and the transplanted organ [46,47,48,49]. In summary, the differentiation of pre-transplant risk into high-, intermediate-, and low-risk categories provides a strategic framework for tailoring subsequent testing protocols. This approach allows healthcare professionals to optimize patient care by addressing specific immunological challenges, thereby enhancing the likelihood of successful organ (renal) transplantation while minimizing the risks associated with rejection events.
8. Epitope-Based Matching Algorithms
Nowadays, computational approaches, such as epitope-based matching algorithms, have revolutionized the field of risk stratification for anti-HLA antibodies in transplantation. From machine learning algorithms and bioinformatics tools to network analysis and multi-omics integration, these methodologies provide a multifaceted approach to understanding and predicting the complexities of antibody-mediated immune responses. As technology continues to advance, the synergy between computational approaches and clinical insights will undoubtedly play a pivotal role in optimizing transplant outcomes and improving patient care. Two computational methodologies, namely HLA Matchmaker and PIRCHE-II, have incorporated the epitope-centered HLA matching principle into their algorithms to identify the optimal donor for a recipient [40,41,42,43]. HLA-specific antibodies are like keys that are made to fit specific locks on our cells called epitopes. These epitopes are like small pieces on the surface of our cells, made up of even smaller parts called triplets or eplets. These parts can vary, like the letters in a password. There are two types of epitopes: private and public. Private epitopes are unique to specific cells, like a custom-made key for one lock. Public epitopes, on the other hand, are shared among different cells, like a master key that can open multiple locks. Knowing about these epitopes helps in understanding how specific or cross-reactive these antibodies can be when testing for compatibility, like in organ transplants. HLA Matchmaker is like a smart tool that looks at HLA alleles (important for organ transplants) as special molecular structures. It uses Microsoft Excel to match these structures at a detailed level and also checks for antibody reactions related to specific parts of these structures. This tool helps to match donors with patients for organ and tissue transplants. By entering detailed HLA typing data for both donors and patients, it assesses compatibility. The tool provides detailed analyses of eplet mismatches for both HLA class I and II antigens. This is important for organ and tissue transplants, because eplet mismatches can affect the success of the procedure. By evaluating these mismatches, the tool helps to optimize donor selection, potentially improving transplant outcomes [30,50,51,52].
The HLA Eplet Mismatch Calculator proves to be an invaluable resource, easily accessible via the HLA Eplet Registry (https://www.epregistry.com.br/ (accessed on 12 February 2024). This tool facilitates in-depth analyses of the compatibility between donors and patients, leveraging detailed high-resolution HLA typing information. It is imperative to recognize and incorporate this synergy when selecting a potential donor [30].
Furthermore, while antibodies against recipient grafts are commonly associated with MHC-directed responses, it is important to acknowledge that not all immune responses in transplantation are MHC-mediated. Histocompatibility testing laboratories play a crucial role in the detection of non-MHC antigens, particularly in the context of organ transplantation [49,53,54].
9. Conclusions
The evolution of histocompatibility testing in renal transplantation has significantly contributed to the success and safety of organ transplant procedures. The journey from traditional methods such as complement-dependent cytotoxicity (CDC) and flow-cytometer-based crossmatches (FCXM) to advanced techniques like virtual crossmatches (VXM) using solid-phase platforms has refined our understanding of donor-specific antibodies (DSAs). These advancements challenge the conventional belief that DSAs are an absolute barrier to renal transplantation. The comprehensive understanding provided by histocompatibility testing, as summarized in detailed tables, serves as a valuable tool for clinicians and laboratories involved in renal transplantation decision-making processes. In summary, histocompatibility testing, with its continual evolution and stratified risk assessment, plays a crucial role in ensuring the success of renal transplantation procedures. The ongoing refinement of these testing methods reflects the commitment to improving outcomes and advancing the field of organ transplantation.
10. Future Directions
In the evolving landscape of organ transplantation, the computational-based approach, specifically epitope matching algorithms, is currently in its infancy regarding donor selection. However, the potential for this innovative technique to transform into a pivotal tool for risk assessment and enhanced donor selection is promising. As clinicians increasingly harness the power of these algorithms, the future holds exciting prospects for their widespread application, making this a compelling avenue for further exploration in the field of transplantation.
Author Contributions
V.C.M.: Conceptualization, investigation, resources, writing—original draft preparation, writing—original draft preparation, writing—review and editing, D.C.: data curation, validation, writing—review and editing, V.R.: Conceptualization, visualization, writing—review and editing and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We are grateful to our colleagues at Chimera Labs for their kind support.
Conflicts of Interest
The authors have no conflicts of interest to declare related to the publication of this manuscript.
References
- Nakamura, T.; Shirouzu, T.; Nakata, K.; Yoshimura, N.; Ushigome, H. The role of major histocompatibility complex in organ transplantation-donor specific anti-major histocompatibility complex antibodies analysis goes to the next stage. Int. J. Mol. Sci. 2019, 20, 4544. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mohiuddin, A.; Sharma, A.; El Kosi, M.; Halawa, A. An update on crossmatch techniques in transplantation. J. Kidney 2017, 3, 1–5. [Google Scholar] [CrossRef]
- Billingham, R.E.; Brent, L.; Medawar, P.B. Actively acquired tolerance of foreign cells. Nature 1953, 172, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Billingham, R.E.; Brent, L.; Medawar, P.B. ‘Actively acquired tolerance’ of foreign cells. 1953. Transplantation 2003, 76, 1409–1412. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patel, R.; Terasaki, P.I. Significance of the positive crossmatch test in kidney transplantation. N. Engl. J. Med. 1969, 280, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Tiwari, A.K.; Rajvanshi, C.; Mehra, S.; Aggarwal, G.; Bansal, S.B.; Kher, V. Evaluation of screening tests for pre-transplant compatibility testing in live-related kidney transplants: Single-center report from India—A prospective observational study. Indian J. Transplant. 2021, 15, 99–103. [Google Scholar] [CrossRef]
- Althaf, M.M.; El Kossi, M.; Jin, J.K.; Sharma, A.; Halawa, A.M. Human leukocyte antigen typing and crossmatch: A comprehensive review. World J. Transplant. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Tait, B.D. Detection of HLA antibodies in organ transplant recipients–triumphs and challenges of the solid phase bead assay. Front. Immunol. 2016, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Mulley, W.R.; Kanellis, J. Understanding crossmatch testing in organ transplantation: A case-based guide for the general nephrologist. Nephrology 2011, 16, 125–133. [Google Scholar] [CrossRef]
- Bhaskaran, M.C.; Heidt, S.; Muthukumar, T. Principles of virtual crossmatch testing for kidney transplantation. Kidney Int. Rep. 2022, 7, 1179–1188. [Google Scholar] [CrossRef]
- Aziz, F.; Tiwari, A.K.; Patel, H.V.; Chauhan, R. Pretransplant histocompatibility testing algorithm: Laboratory and clinical approach in the Indian context. Indian J. Transplant. 2021, 15, 4–13. [Google Scholar] [CrossRef]
- Liu, C. A long road/read to rapid high-resolution HLA typing: The nanopore perspective. Hum. Immunol. 2021, 82, 488–495. [Google Scholar] [CrossRef]
- Mishra, V.C.; Chandra, D.; Deshpande, T.; Singh, P.; Anthwal, A.; Raina, V. Human Leukocyte Antigen-A, B, and DRB1 Diversity in Renal Transplant Patients and Donors: A Single-Center Retrospective Observational Study. Indian J. Transplant. 2022, 16, 220–224. [Google Scholar] [CrossRef]
- Alelign, T.; Ahmed, M.M.; Bobosha, K.; Tadesse, Y.; Howe, R.; Petros, B. Kidney transplantation: The challenge of human leukocyte antigen and its therapeutic strategies. J. Immunol. Res. 2018, 2018, 5986740. [Google Scholar] [CrossRef]
- Abu Jawdeh, B.G.; Cuffy, M.C.; Alloway, R.R.; Shields, A.R.; Woodle, E.S. Desensitization in kidney transplantation: Review and future perspectives. Clin. Transplant. 2014, 28, 494–507. [Google Scholar] [CrossRef]
- Gombos, P.; Opelz, G.; Scherer, S.; Morath, C.; Zeier, M.; Schemmer, P.; Süsal, C. Influence of test technique on sensitization status of patients on the kidney transplant waiting list. Am. J. Transplant. 2013, 13, 2075–2082. [Google Scholar] [CrossRef]
- Das, A.; Taner, T.; Kim, J.; Emamaullee, J. Crossmatch, donor-specific antibody testing, and immunosuppression in simultaneous liver and kidney transplantation: A review. Transplantation 2021, 105, e285–e291. [Google Scholar] [CrossRef]
- Salvalaggio, P.R.; Graff, R.J.; Pinsky, B.; Schnitzler, M.A.; Takemoto, S.K.; Burroughs, T.E.; Santos, L.S.; Lentine, K.L. Crossmatch testing in kidney transplantation: Patterns of practice and associations with rejection and graft survival. Saudi J. Kidney Dis. Transplant. 2009, 20, 577–589. [Google Scholar]
- Etta, P.K. Tools for histocompatibility testing and significance of panel reactive antibodies—A narrative review. Indian J. Transplant. 2021, 15, 295–299. [Google Scholar] [CrossRef]
- Rocha, Y.; Jaramillo, A.; Neumann, J.; Hacke, K.; Palou, E.; Torres, J. Crossmatch assays in transplantation: Physical or virtual?: A review. Medicine 2023, 102, e36527. [Google Scholar] [CrossRef]
- Hetrick, S.J.; Schillinger, K.P.; Zachary, A.A.; Jackson, A.M. Impact of pronase on flow cytometric crossmatch outcome. Hum. Immunol. 2011, 72, 330–336. [Google Scholar] [CrossRef]
- Ho, E.K.; Vasilescu, E.R.; Colovai, A.I.; Stokes, M.B.; Hallar, M.; Markowitz, G.S.; D’Agati, V.D.; Cohen, D.J.; Ratner, L.E.; Suciu-Foca, N. Sensitivity, specificity and clinical relevance of different cross-matching assays in deceased-donor renal transplantation. Transpl. Immunol. 2008, 20, 61–67. [Google Scholar] [CrossRef]
- Tiwari, A.K.; Handoo, A.; Choudhary, M.; Mehra, S.; Bacchas, V.; Yadav, A.; Negi, A.; Chopra, R. Prospective multi-centric study to analyze pre-transplant compatibility algorithm for live-related donor kidney transplant in Indian setting: The “Delhi approach”! Transpl. Immunol. 2021, 69, 101487. [Google Scholar] [CrossRef]
- Bray, R.A.; Gebel, H.M.; Ellis, T.M. Flow cytometric assessment of HLA alloantibodies. Curr. Protoc. Cytom. 2004, 27, 6–16. [Google Scholar] [CrossRef]
- Tait, B.D.; Süsal, C.; Gebel, H.M.; Nickerson, P.W.; Zachary, A.A.; Claas, F.H.; Reed, E.F.; Bray, R.A.; Campbell, P.; Chapman, J.R.; et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation 2013, 95, 19–47. [Google Scholar] [CrossRef]
- Alvares, M.; Anwar, S.; Hashmi, S.K.; Zaman, M.B.; Al Mahri, A.; Alvares, C.; Al Katheeri, L.; Purushothaman, A.; Ralonya, M.E.; Sangalang, M.G.; et al. Development of a calculated panel reactive antibody calculator for the United Arab Emirates: A proof of concept study. Sci. Rep. 2023, 13, 8468. [Google Scholar] [CrossRef]
- Cecka, J.M. Calculated PRA (CPRA): The new measure of sensitization for transplant candidates. Am. J. Transplant. 2010, 10, 26–29. [Google Scholar] [CrossRef]
- Schinstock, C.; Tambur, A.; Stegall, M. Current approaches to desensitization in solid organ transplantation. Front. Immunol. 2021, 12, 686271. [Google Scholar] [CrossRef]
- Dunbar, S.A.; Hoffmeyer, M.R. Microsphere-based multiplex immunoassays: Development and applications using Luminex® xMAP® technology. In The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA and Related Techniques; Elsevier: Amsterdam, The Netherlands, 2013; Volume 1, p. 157. [Google Scholar]
- Mishra, V.C.; Raina, V. Enhancing Precision of the Single-antigen Bead (SAB) Assay: Considerations and Challenges. J. Clin. Transl. Pathol. 2024, 4, 12–17. [Google Scholar] [CrossRef]
- Jaramillo, A.; Reddy, K.S.; Heilman, R.L. Using the virtual crossmatch to allow for safer and more efficient kidney transplantation of highly sensitized patients. Transplantation 2020, 104, 1121–1122. [Google Scholar] [CrossRef]
- Chandraker, A.; Sayegh, M.H.; Singh, A.K. Core Concepts in Renal Transplantation; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–242. [Google Scholar] [CrossRef]
- Gebel, H.M.; Bray, R.A. HLA antibody detection with solid phase assays: Great expectations or expectations too great? Am. J. Transplant. 2014, 14, 1964–1975. [Google Scholar] [CrossRef]
- Claas, F.H.; Dankers, M.K.; Oudshoorn, M.; van Rood, J.J.; Mulder, A.; Roelen, D.L.; Duquesnoy, R.J.; Doxiadis, I.I. Differential immunogenicity of HLA mismatches in clinical transplantation. Transpl. Immunol. 2005, 14, 187–191. [Google Scholar] [CrossRef]
- Riethmüller, S.; Ferrari-Lacraz, S.; Müller, M.K.; Raptis, D.A.; Hadaya, K.; Rüsi, B.; Laube, G.; Schneiter, G.; Fehr, T.; Villard, J. Donor-specific antibody levels and three generations of crossmatches to predict antibody-mediated rejection in kidney transplantation. Transplantation 2010, 90, 160–167. [Google Scholar] [CrossRef]
- Batal, I.; Zeevi, A.; Lunz, J.G., III; Aggarwal, N.; Shapiro, R.; Randhawa, P.; Girnita, A. Antihuman leukocyte antigen–specific antibody strength determined by complement-dependent or solid-phase assays can predict positive donor-specific crossmatches. Arch. Pathol. Lab. Med. 2010, 134, 1534–1540. [Google Scholar] [CrossRef]
- Sethi, S.; Choi, J.; Toyoda, M.; Vo, A.; Peng, A.; Jordan, S.C. Desensitization: Overcoming the immunologic barriers to transplantation. J. Immunol. Res. 2017, 2017, 6804678. [Google Scholar] [CrossRef]
- Erdoğmuş, Ş.; Şengül, Ş. Immunologic risk assessment before kidney transplantation: An update. Turk. J. Nephrol. 2019, 28, 216–224. [Google Scholar] [CrossRef]
- Schinstock, C.A.; Gandhi, M.J.; Stegall, M.D. Interpreting anti-HLA antibody testing data: A practical guide for physicians. Transplantation 2016, 100, 1619–1628. [Google Scholar] [CrossRef]
- Bielmann, D.; Hönger, G.; Lutz, D.; Mihatsch, M.J.; Steiger, J.; Schaub, S. Pretransplant risk assessment in renal allograft recipients using virtual crossmatching. Am. J. Transplant. 2007, 7, 626–632. [Google Scholar] [CrossRef]
- Fuggle, S.V.; Martin, S. Tools for human leukocyte antigen antibody detection and their application to transplanting sensitized patients. Transplantation 2008, 86, 384–390. [Google Scholar] [CrossRef]
- Pratschke, J.; Dragun, D.; Hauser, I.A.; Horn, S.; Mueller, T.F.; Schemmer, P.; Thaiss, F. Immunological risk assessment: The key to individualized immunosuppression after kidney transplantation. Transplant. Rev. 2016, 30, 77–84. [Google Scholar] [CrossRef]
- Sugi, M.D.; Joshi, G.; Maddu, K.K.; Dahiya, N.; Menias, C.O. Imaging of renal transplant complications throughout the life of the allograft: Comprehensive multimodality review. Radiographics 2019, 39, 1327–1355. [Google Scholar] [CrossRef]
- Cippà, P.E.; Schiesser, M.; Ekberg, H.; van Gelder, T.; Mueller, N.J.; Cao, C.A.; Fehr, T.; Bernasconi, C. Risk stratification for rejection and infection after kidney transplantation. Clin. J. Am. Soc. Nephrol. 2015, 10, 2213–2220. [Google Scholar] [CrossRef]
- Tambur, A.R.; Campbell, P.; Chong, A.S.; Feng, S.; Ford, M.L.; Gebel, H.; Gill, R.G.; Kelsoe, G.; Kosmoliaptsis, V.; Mannon, R.B.; et al. Sensitization in transplantation: Assessment of risk (STAR) 2019 Working Group Meeting Report. Am. J. Transplant. 2020, 20, 2652–2668. [Google Scholar] [CrossRef]
- Tambur, A.R.; Campbell, P.; Claas, F.H.; Feng, S.; Gebel, H.M.; Jackson, A.M.; Mannon, R.B.; Reed, E.F.; Tinckam, K.; Askar, M.; et al. Sensitization in Transplantation: Assessment of Risk (STAR) 2017 Working Group Meeting Report. Am. J. Transplant. 2018, 18, 1604–1614. [Google Scholar] [CrossRef]
- Lefaucheur, C.; Louis, K.; Morris, A.B.; Taupin, J.L.; Nickerson, P.; Tambur, A.R.; Gebel, H.M.; Reed, E.F.; STAR 2022 Working Group. Clinical recommendations for posttransplant assessment of anti-HLA (Human Leukocyte Antigen) donor-specific antibodies: A Sensitization in Transplantation: Assessment of Risk consensus document. Am. J. Transplant. 2023, 23, 115–132. [Google Scholar] [CrossRef]
- Tambur, A.R.; Bestard, O.; Campbell, P.; Chong, A.S.; Crespo, M.; Ford, M.L.; Gebel, H.M.; Heidt, S.; Hickey, M.; Jackson, A.; et al. Sensitization in transplantation: Assessment of Risk 2022 Working Group Meeting Report. Am. J. Transplant. 2022, 23, 133–149. [Google Scholar] [CrossRef]
- Phanish, M.K.; Hull, R.P.; Andrews, P.A.; Popoola, J.; Kingdon, E.J.; MacPhee, I.A.; South West Thames Renal Transplantation Network. Immunological risk stratification and tailored minimisation of immunosuppression in renal transplant recipients. BMC Nephrol. 2020, 21, 92. [Google Scholar] [CrossRef]
- Duquesnoy, R.J. Reflections on HLA epitope-based matching for transplantation. Front. Immunol. 2016, 7, 469. [Google Scholar] [CrossRef]
- Renaldo, A.; Roa-Bautista, A.; González-López, E.; López-Hoyos, M.; San Segundo, D. Epitope-Level matching—A review of the novel concept of eplets in transplant histocompatibility. Transplantology 2021, 2, 336–347. [Google Scholar] [CrossRef]
- Spitznagel, T.; Matter, L.S.; Kaufmann, Y.L.; Nilsson, J.; von Moos, S.; Schachtner, T. PIRCHE-II scores prove useful as a predictive biomarker among kidney transplant recipients with rejection: An analysis of indication and follow-up biopsies. Front. Immunol. 2022, 13, 949933. [Google Scholar] [CrossRef]
- Lammerts, R.G.; Altulea, D.; Hepkema, B.G.; Sanders, J.S.; Born, J.V.; Berger, S.P. Antigen and cell-based assays for the detection of non-HLA antibodies. Front. Immunol. 2022, 13, 864671. [Google Scholar] [CrossRef] [PubMed]
- Sorohan, B.M.; Baston, C.; Tacu, D.; Bucșa, C.; Țincu, C.; Vizireanu, P.; Sinescu, I.; Constantinescu, I. Non-HLA antibodies in kidney transplantation: Immunity and genetic insights. Biomedicines 2022, 10, 1506. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).