1. Introduction
Sleep is a stable systemic state that, in mammals, is controlled by homeostasis and endogenous circadian rhythms. Ascertaining how sleep is composed makes it easier and faster to evaluate and quantify sleep in research and clinical settings. Nowadays, it is widely known that sleep is composed of two parts, non-rapid eye movement (NREM) sleep and REM sleep. However, it took researchers nearly three decades, from 1924 to 1953, to separate these two types of sleep. In this process, long-term visual judgment by researchers played a key role. Sleep is characterized using an electroencephalogram (EEG), which was first developed by Hans Berger in 1924 [
1]. He called the EEG a “brain mirror,” reflecting the “electrical psychic energy” within cortical tissue. He analyzed the wave phase patterns and described the α and β waves. Over a decade later, Alfred Loomis showed that human EEG patterns dramatically change from the wake to sleep stages [
2]. Loomis initially classified sleep into five stages (A, B, C, D, and E), which are primarily manifested by the characteristic patterns of the α and spindle waves. Initially, the characterization and transition of brainwave frequencies were considered essential features. In his study, Loomis used the six-channel EEG of 30 s because the record sheets were automatically cut by a scissor every 30 s, and this marked the earliest conceptual origin of the classification epoch. These separate epochs were visually judged by researchers in a manner similar to the workflows conducted by modern polysomnography (PSG) technicians. For this reason, even now, most sleep classification algorithms use 30 s as one epoch length to determine the sleep stage.
After the rapid eye movement (REM) sleep stage was discovered by Aserinsky and Kleitman in 1953, electrooculography (EOG) and mentalis muscle electromyography (EMG) were used for sleep classification. Rechtschaffen and Kales then set up the PSG criteria in 1968 [
3], which have been widely used to the present day with only minor modifications. However, without EOG, EMG, or the automatic integral calculation method being used for relative band powers, Loomis’s sleep classification criteria from the 1930s closely resembles the modern one, suggesting that EEG patterns play a more critical role than any other channel. In addition, visual judgments by technicians remain important for classifying sleep stages.
In addition to the human PSG, considerable demand exists for research on rodent sleep data. Rodents also have a sleep–wake cycle consisting of different sleep stages. However, there are some significant differences compared with humans. First, nocturnal rodents tend to get more sleep in the light period compared with humans, who sleep in the dark period. Second, unlike human sleep which is monophasic and repeats the NREM–REM sleep cycle (lasting about 90 min) three to six times successively only during night, the sleep of rodents is polyphasic and occurs both during the day and night time, and does not usually repeat the NREM–REM sleep cycle (lasting for several minutes to longer, irregular duration) successively. Third, the NREM sleep of rodents is not subdivided, unlike humans. Normally, all sleep states, excluding REM, are regarded as NREM. As a result, the rodent sleep cycle is relatively shorter, not continuous, fragmented, and unstable to external environmental changes. Nonetheless, rodents are a great model to understand human sleep, and a simple and reliable method to classify their sleep stage is required. In the specific representation of electrophysiology, the classification criteria of the sleep stages of mice are different from human PSG classification. The murine non-rapid eye movement (NREM) stage shows low EMG amplitude and high EEG δ-wave power, and NREM is classified as one stage without any further subdivision. The REM stage shows a higher θ-wave power than any other frequency band. Thus, three sleep stages, namely wake, NREM, and REM, have typical individual features on the EEG power spectrum. Researchers of murine sleep usually use an automatic scoring commercial software, such as SleepSign (Kissei Comtec Co. Ltd., Matsumoto, Japan) [
4] or a MATLAB advanced toolbox such as EEGLAB [
5]. However, these processing tools may present some obstacles for new researchers due to the cost or the requirement for high-level programming skills. Furthermore, due to the shorter sleep cycle and the relative unstableness of the sleep stage in mice, the one-epoch length is usually set shorter compared with humans, usually being shorter than 30 s.
Thanks to technical advances in machine learning, for the past 10 years we have had the opportunity to utilize artificial neural networks to study the sleep–wake cycle activities generated by natural neural networks. An unsupervised algorithm known as FASTER [
6] (fully automated sleep staging method via EEG/EMG recordings) attained prominence even before the first TensorFlow (Mountain View, CA, USA) beta version was released in 2015. FASTER calculates the power spectrum of both EEG and EMG and performs a clustering of the power spectrum values using principal component analysis. The sensitivity performances of the NREM and wake states are comparatively fine. However, because the clustering of rare events (REM) for “hard” rule classical clustering analysis is complex, the sensitivity of REM is low and unstable in different experimental environments.
After TensorFlow was released, most of the algorithms were aimed at human PSG; however, later, these human-based approaches were found to be instructive for other mammalian sleep studies. In 2017, Guo et al. open-sourced the DeepSleepNet model for EEG single channel-based sleep-stage scoring [
7], which was trained by the Sleep-EDF dataset for humans. Before DeepSleepNet, most classification methods were dependent on complex calculations for extracting band power features. However, the DeepSleepNet model works without utilizing any hand-engineered features by merging the two branches (EEG and EMG) of a convolutional neural network (CNN) and bidirectional long short-term memory (Bi-LSTM) cells.
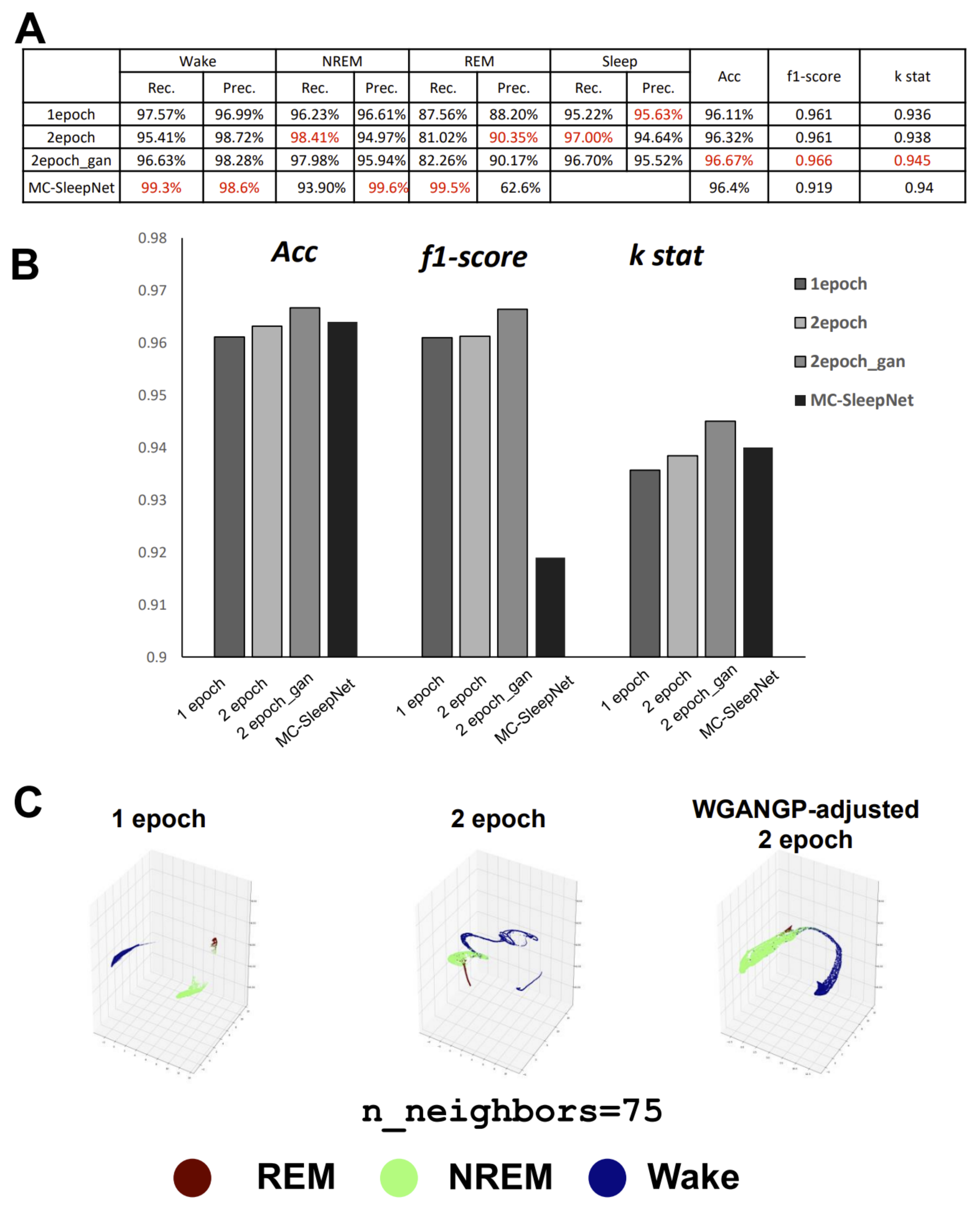
Recently, MC-SleepNet was created for sleep-stage scoring in mice [
8], inspired by DeepSleepNet with the addition of EMG. The performance analysis results, excluding the low precision of REM on small-scale datasets, revealed that MC-SleepNet was superior. However, for laboratory-level studies, particularly for some rare transgenic strains that are not easily propagated, performance on small-scale datasets is also important. This is also true of large-scale datasets, particularly for research related to REM sleep anomalies in mice [
9]. The problem with the one-dimensional CNN is its weakness in outlier detection, especially when applied to sleep studies. This is considered to be a cause of DeepSleepNet’s low sensitivity for N1, as the N1 stage is short and contains various noises. The problem with MC-SleepNet is that it is very difficult for researchers to re-examine the data. This is because one-dimensional data needs to be visualized before examination. This inspired us to potentially visualize the data directly as two-dimensional (2D) to cut out the middle processes. Visualized data can certainly also be analyzed with CNNs.
In fact, the CNN was originally developed to analyze 2D image data. Thus far, 2D image-based CNN-LSTM networks have been applied very successfully in the field of medical diagnostic imaging [
10,
11]. This strategy has also been shown to be successful in the analysis of phenotypic convergence for the taxonomy of species such as butterflies [
12]. An analysis of the features of the images extracted by a 2D CNN even showed that identifying the migration patterns between phases was possible. This type of parsing method can not only classify all discrete data, but it can also provide a visual interpretation of the transformations between various stages and the data relationships within each stage group.
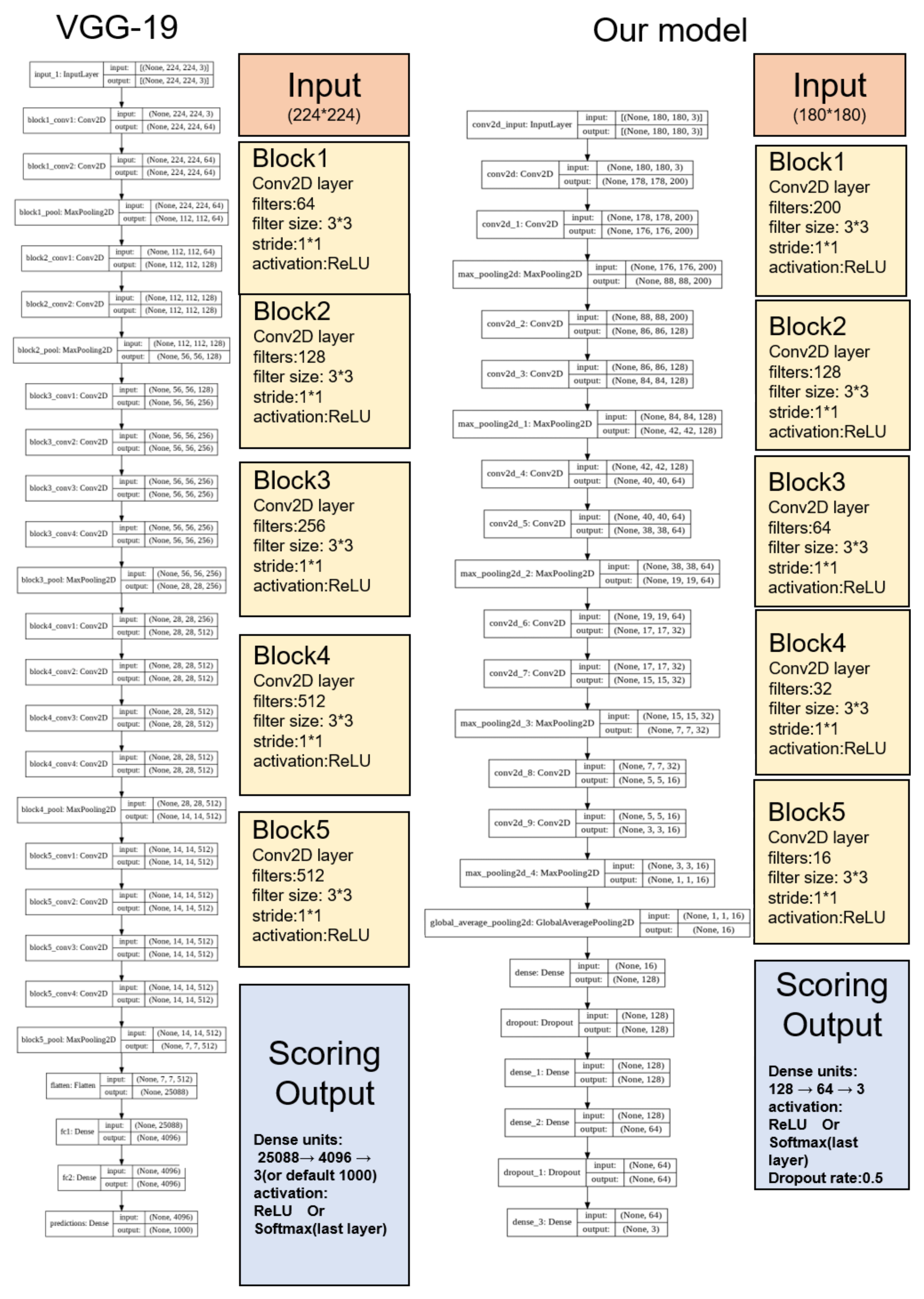
For these reasons, we commenced this project. We hoped that we could develop an algorithm to achieve high accuracy and ease of use for automatic sleep classification in a cost-effective way, allowing researchers to enhance the analysis of sleep as a whole, rather than overly focusing on fragments within a large time scale. In particular, in order to overcome the problems of previous methods that were not easy-to-use, we developed a novel method called GI-SleepNet, a GAN-assisted image-based program, to process EEG and EMG data. As the first step, we produced an image file corresponding to each classification epoch, composed of an EEG power spectrum plot and an EMG raw wave data graph of that epoch. We then manually classified these epoch images into three stages—wake, NREM, and REM. Next, we used a 2D CNN for supervised learning of the designated images. GI-SleepNet precisely follows the logic and workflow of expert technicians, and allows for the easy identification of what the machine is learning. In addition, the accumulated knowledge and numerous methods on the deep learning of images are readily applicable.
3. Discussion
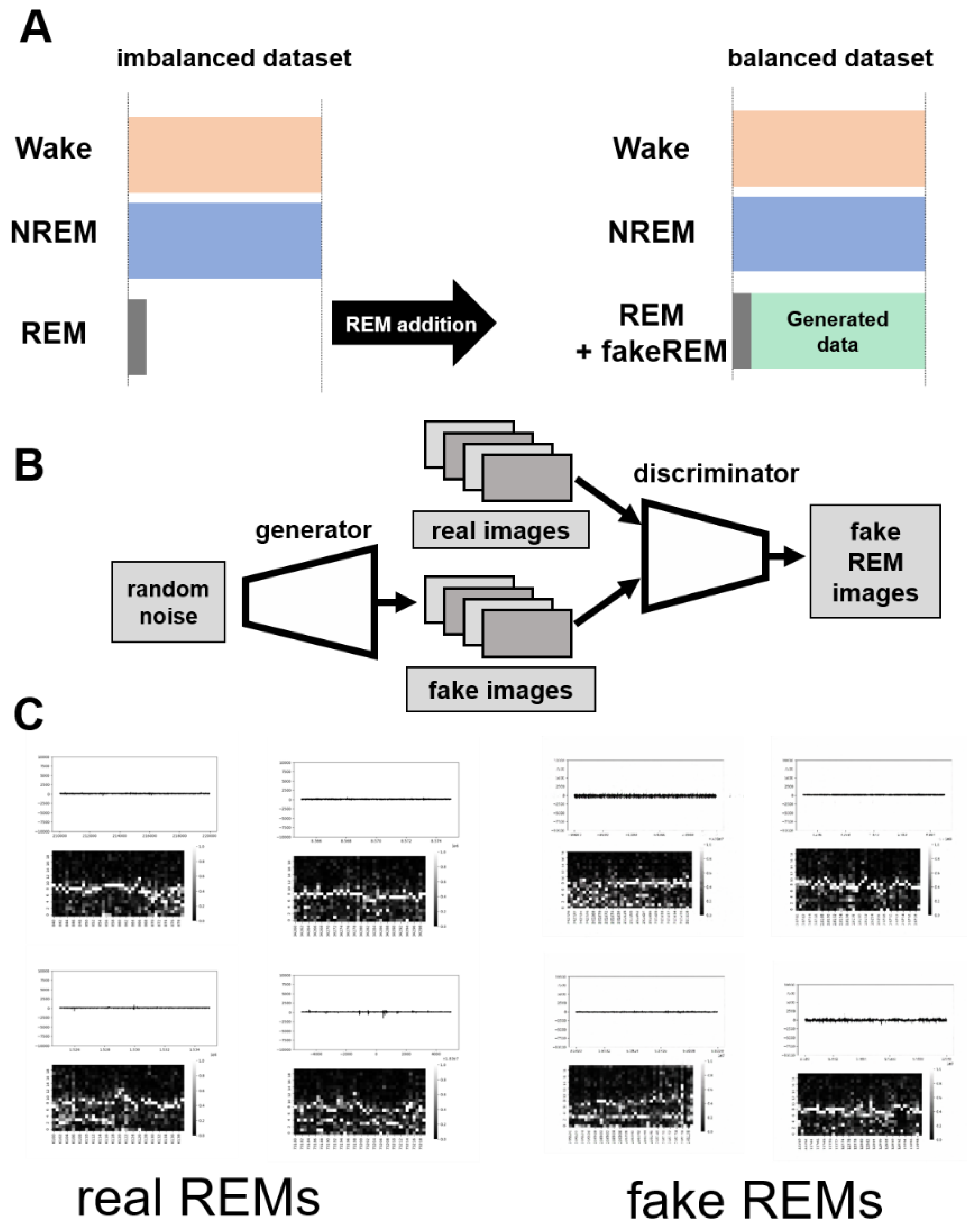
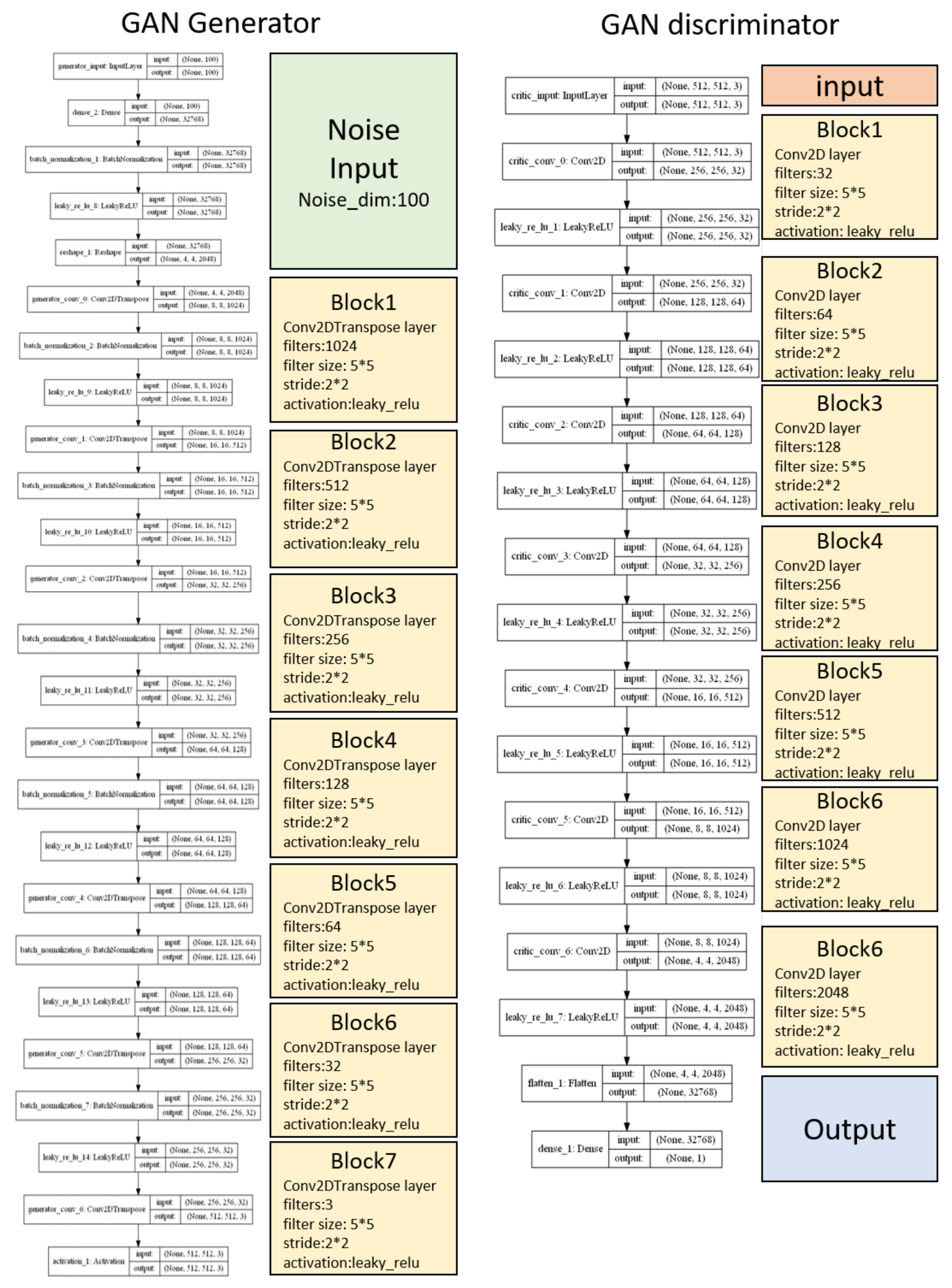
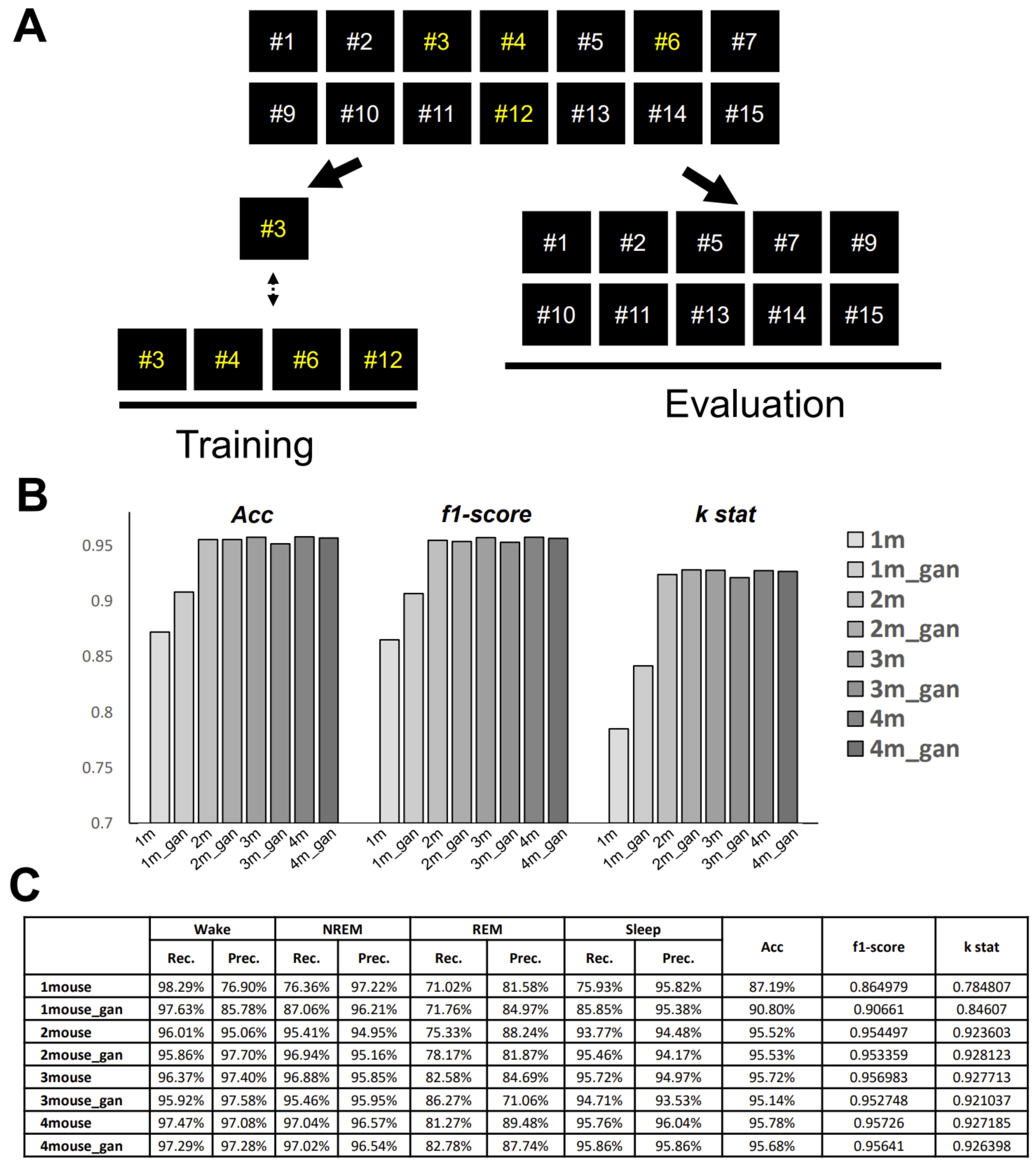
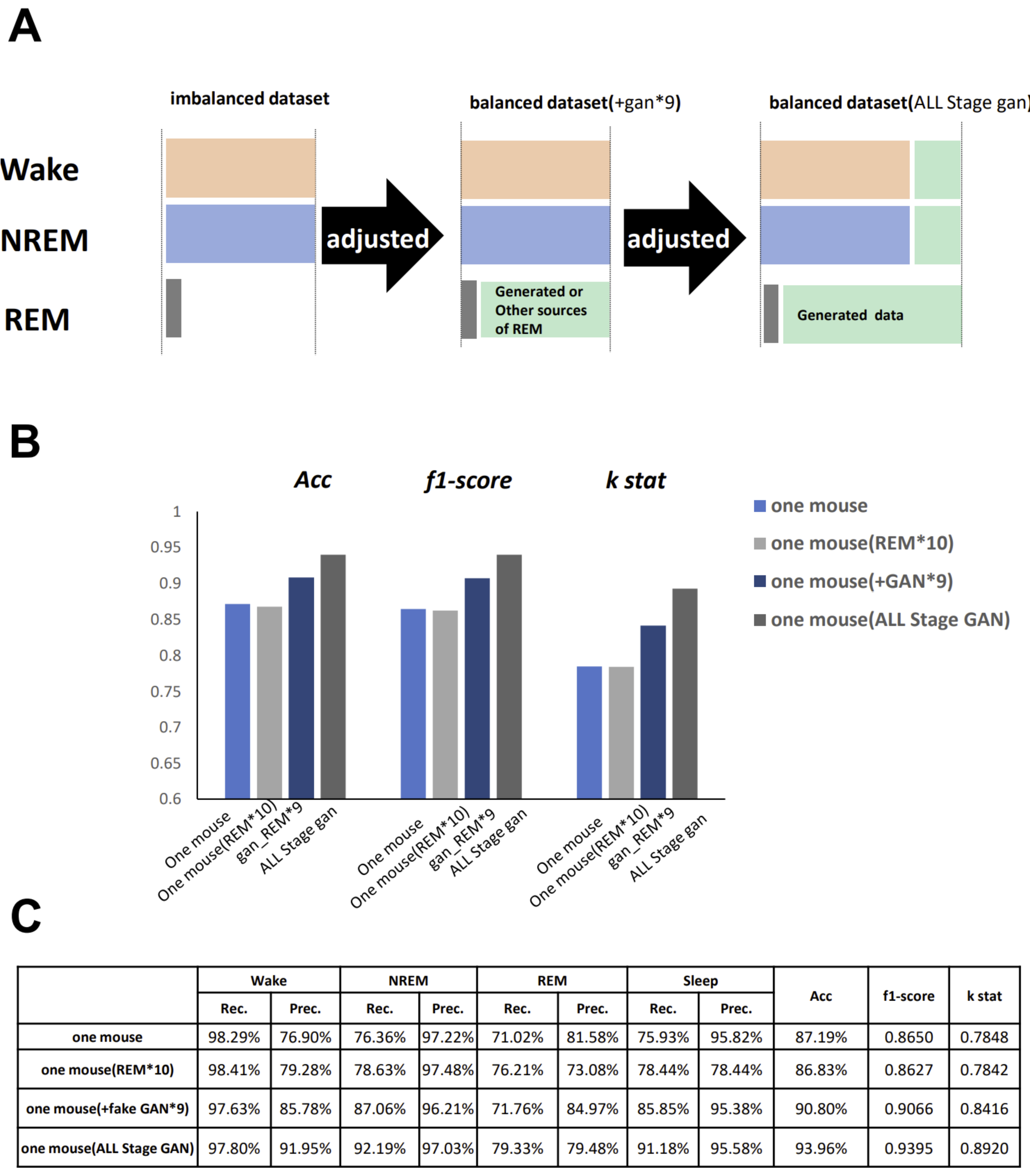
GI-SleepNet, the novel image-based learning algorithm developed in this study, has several advantages over the conventional numerical data-based algorithm. First, the format of the data has excellent flexibility. In our case, one image has both raw EMG data and EEG frequency power spectrum data. EMG and EEG data differ between laboratories with different sampling rates and different data formats. Epoch lengths also differ among animal species. For human PSG, EOG should be included along with EMG and EEG. However, these differences do not matter once all the data are formatted as a single image. Thus, this method is readily applicable to any species and even outside of sleep research. Second, the image data that the machine learns has high interpretability. It is intuitively comprehensive, and each image contains sufficient visual information to classify it into three categories by researchers. Therefore, it is easy to create training datasets manually and to perform post-prediction analysis. Following automatic classification by machine, it is also easy to confirm the results and to find and resolve errors. Third, as image recognition is one of the most advanced fields in AI machine learning, it is easy to find sophisticated algorithms and to find recent progress. Therefore, we included the GAN method to adjust the REM data by producing fake REM images. Fourth, because the size of one set of data is limited to a 2D-image extent, and because the image-processing algorithms are optimized, our method requires relatively low computing power and a short processing time. Fifth, our method exhibits high accuracy with small datasets, making it useful in practice. It performed well on both our own dataset and those from different laboratories, even though these data were recorded using different types of acquisition equipment. This means that it can be easily reused on small datasets of specific strains or transgenic animals that may exhibit atypical EEG patterns. Thus, applying it to precious animal strains from which only a limited amount of data is available can also be advantageous to researchers.
We anticipate that researchers themselves will be able to customize our model. Researchers can use our workflow to create their own dataset and re-train a new model (
Figure 9). To make our image-based scoring system easier to use, we developed a graphical user interface (GUI) for research purposes based on the Python binding GUI toolkit Tkinter (
Supplementary Figure S4). Our GUI includes semi-automatic data-preprocessing, large-scale output of plotting images, neural network training, and prediction functions. We also released several trained h5 files for immediate use without a training dataset. The only action for individual researchers who wish to use our model is to create their own datasets and fit them to our shared network structure. Finally, the DCGAN-created images and forced filters based on our own dataset have also been packaged in the GUI. If this system can be used by a large number of researchers, we hope to collect considerably more data from different devices and further improve the noise resistance of the algorithm.
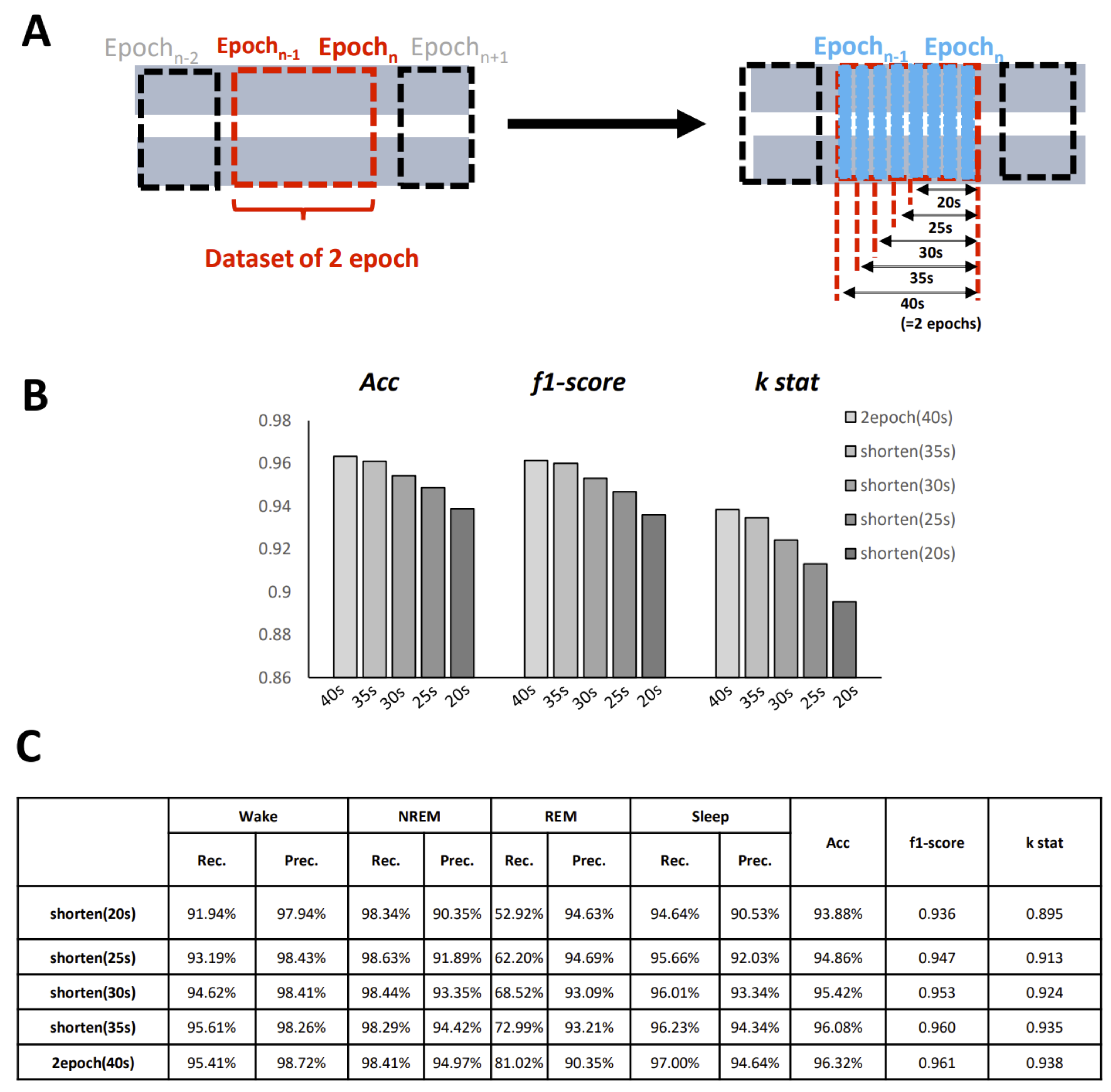
One important setting that should be considered is the length of the epoch. The selection of the epoch length is always difficult in sleep classification files. Conventionally, for humans, the epoch window is set to 30 s, and for mice, it is usually set to 20 s. However, there has been some commentary suggesting that a shorter epoch should be designed to improve the temporal resolution and thus evaluate time more efficiently. As our model is able to create an image for any length of epoch required, we call on others to create a shorter epoch dataset for further study.
Image-based scoring systems can be applied to identify not only sleep activity, but also other physiological and pathological events, such as epileptic seizures or preclinical Alzheimer’s disease symptoms. For example, one study was conducted that showed the differences in the prefrontal cortex EEG power spectrum during Y-maze tests between normal mice and Alzheimer’s disease models. A better understanding and design of experiments may be beneficial if creating image datasets of different performance groups in behavioral tests.
In this study, we only used two channels of biosignal from mice and obtained good results. We therefore expect that the program can be adapted to other systems. For example, small devices equipped with a single or a few channels of EEG are successfully used for many studies, including ours [
16]. Those studies are good targets for our program, and can be used instead of laboratory PSG. It is expected that sleep studies can be facilitated at home and in broader settings [
17]. As the GI-SleepNet is based on the relatively simple VGG-19-based algorithm, we believe that it is able to process the EEG/EMG more easily in order to measure sleep quality in the client terminal. The next step is to apply our model to human clinical PSG data. As the classification of human sleep is more complicated, we may increase the number of biosignals, such as EOG and heart rate, for further research [
18].
Furthermore, in the future, particularly for specific experimental needs where more than three states need to be classified, our model is highly customizable. By modifying the units parameter of the output dense layer at the end of the model, it is possible to apply this model to other wider ranges of classification. In an ongoing study, we are applying this model to human clinical PSG datasets and classifying them into five states (NREM1, NREM2, NREM3, REM, and WAKE). The results so far are promising. Subsequently, for example, there is also the need for research to classify the middle transition state between two pure states. As the transition states are quite small in volume, we assume that the generation of fake data of certain transition states using our GAN model may promote performance if those transition states are recognized and labeled.
Our method does have some limitations. First, a priori knowledge is required to design images. Second, producing images is initially time consuming. In the case of our dataset, we used EMG and EEG signals of almost the same size. However, the ratio of sizes can be changed, which will affect accuracy. Nevertheless, to change the ratio, ensuring that all datasets contain all images for all epochs is required, a process that is time consuming and laborious. Despite these limitations, we believe that our novel algorithm will provide a versatile tool for future research in many fields. Third, a crucial point that needs to be resolved is the unquantified effect of EEG artifacts within the recording. In this study, we used a clear dataset with very few artifacts or noises (Tsukuba-14). Therefore, potential users should be aware of the fact that the performance against noise and artifacts needs to be experimented with in follow-up studies with other datasets containing artifacts. For the next step, we plan to artificially add random noise into the data to examine the artifact condition. As mentioned in the Materials and Methods section, some EEG artifacts featured within the recording could be absorbed by the model with the FFT as many artifacts occur during the wake stage and show an apparent low frequency. We expect that our model will be robust to those artifacts, but this should be examined in each dataset.