Abstract
Neuroblastoma (NB) is the most common extracranial solid tumor in children with incidence rate 10.2 per 1 million children under 15 years old. NB has great diversity in clinical behavior while some tumors are easily curable and spontaneously regressed, the others are resistant to chemotherapies and exhibiting high relapse rates. Despite intensive usage of chemotherapies, still 5-year survival rate is about 40% for advanced stage bad prognostic patient group. Hence, there is need for more effective therapeutics and novel biomarkers for early detection of high-risk patients. In the present study we aimed to study molecular mutation profile of both good and bad prognostic NB patients with whole exome sequencing (WES). The NB patients whose risk group previously was confirmed according to TPOG-2009 protocol. 5 patients with good prognosis (low risk & intermediate with good histology) and 5 patients with bad prognosis (intermediate poor histology & high risk) were chosen. As a result, 23 common mutations were found in both prognostic group. While 7 mutations were found to be specific to bad prognostic group, 2 mutations were only observed in good prognostic group patients. The commonly mutated genes were located commonly in cell membrane and extracellular space and mostly interfering with cell signaling and cell communication pathways.
1. Introduction
Neuroblastoma (NB) is one of the malignant childhood tumors that arisen from primitive neural crest cells. NB is accounting for 7.4% of all childhood cancers in Turkey and approximately 15% of cancer deaths in children [1,2].
Copy number variations (CNV) in patient genome are highly affecting the progression of the disease [3]. MYCN oncogene amplification is one of the biomarker for the disease, levels of MYCN amplification directly correlates with invasive and metastatic behavior of tumor [4]. Not only MYCN, 1p or 11q deletions and 17q addition are commonly observed (especially in poor prognostic groups) and each loss or gain is separately associated with decreased progression free survival among patients [5].
Next generation sequencing (NGS) allows simultaneous sequencing of millions of DNA fragments without previous sequencing knowledge and allowing the sequencing of patient whole genome in low cost and relatively shorter time. Recent developments in NGS technology advances in finding novel mutations in different type of tumors and characterization mutation profiles furthermore revealing insights about tumor evolution and tumor heterogeneity [6]. NGS platforms can provide comprehensive view of DNA aberrations, genetic recombination and other mutations, hence helping clinician to identify specific mutations in each patients and prescribe more accurate and effective therapies, which is now called as precision medicine. Furthermore, NGS technologies enables clinician to monitor disease progression and relapse among the patients. The application of NGS technology in clinics are dramatically increasing since the advent of technology. For biomarker discovery, searching for new therapeutic targets, scientists are benefited strongly from whole genome sequencing or whole exome sequencing, RNA sequencing applications [7].
According to Turkish Pediatric Oncology Group (TPOG) protocol 10 NB patients were classified either in low risk and high risk group based on histological findings, MYCN amplification status, deletion of 1p and 11q arms and DNA ploidy status [8].
In the present study, based on the risk classification we had chosen 5 patients from good prognostic group (including low risk and intermediate risk with good histology) and 5 patients from poor prognosis group (including intermediate risk with poor histology and high risk). It was aimed to analyze the genomic mutations with whole exome sequencing and identify novel mutations and new biomarkers for each group.
2. Material and Methods
The study was conducted in collaboration between Dokuz Eylul University Pediatric Oncology and Basic Oncology Departments and Turkish Pediatric Oncology Group (TPOG). The project was also supported from a grant from Turkish Pediatric Oncology Group in 2016. The ethical committee in Behcet Uz Children Hospital approved the study in 26 September 2013.
Tumor Collection: Ten NB patient biopsy samples were included in the study. The median age for patient was 67.8 months. 6 female and 4 male patients were included in the study. 5 of the patients were in good prognostic group and 5 of the patient were in bad prognostic group. The biopsy samples were taken from transfer medium and directly stored in −80 °C. The DNA isolation were performed from biopsy tissues by using Thermo DNA extraction kit (Thermo Gene Jet, K0721, Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Preparation for Whole Exome Sequencing: 100 ng DNA was subjected to shearing, end repair, phosphorylation and ligation to barcoded sequencing adaptors. The ligated fragments were then selected according to their sizes (~200 bp). Then fragments were amplified using specific primers and sequencing were performed on Illumina platform (Illumina, San Diego, California, USA).
Whole Exome Seqeuncing (WES) analysis: The sequencing results were aligned with BWA MEM and variant detection was made with SAM Tools and variant annotation was done with Variant studio.
3. Results
Clinical Fetures of Patients: The biopsy samples from 10 NB patients were included in the study. Clinical features of patients listed in Table 1.

Table 1.
Clinical Features of Patients.
Good and bad prognostic patients shared many common mutated genes such as ARSD, B3GNT6, CDRT1, MUC4, NBPF14, GLT6D1 as shown in Figure 1. The mutations in A2ML1, CDCP2, CYPA4B1 and FAM182A, ANLN, ADRA2B, PLD1 genes were only exhibiting in patients with bad prognosis, CREB3L1, KIAA1751 mutations were found in patients with good prognosis.
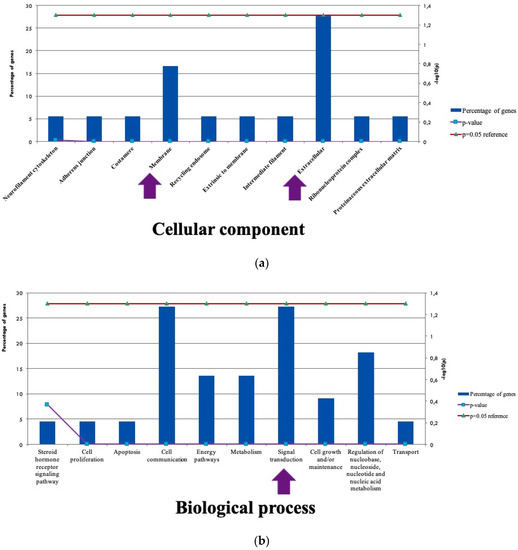
Figure 1.
Location and Functional Profile of Mutated Genes (a) The cellular components associated with mutations (b) The Biological processes related with mutated genes.
4. Discussion
NGS is a highly versatile sequencing technology, used for variety applications in cancer research. In the present study, it was aimed to find novel common and distinctive mutations both in good and bad prognostic patients by whole exome sequencing. We have found commonly mutated genes in both groups, mutations were mostly associated with cell signaling and cellular communication pathways. In bad prognostic group. ANLN was one of the interesting candidate gene, which involved in cytokinesis, cell growth and migration. Another gene was TAF4, which controlled lineage specific differentiation.
5. Conclusions
By whole exome sequencing analysis, we have discovered some novel mutations in both good prognostic and bad prognostic NB patients. Our analyses have shown some infrequent gene mutations that might have an effect NB disease progression and behavior, hence contributing the discovery novel biomarkers in NB.
Acknowledgments
This study was financially supported by TPOG as TPOG project.
References
- Pizzo, P.; Poplack, D.G. Principles and Practice of Pediatric Oncology, 7th ed.; Wolters Kluver: Philadelphia, PA, USA, 2016. [Google Scholar]
- Aksoylar, S.; Varan, A.; Vergin, C.; Hazar, V.; Akici, F.; Dagdemir, A.; Buyukavci, M.; Kebudi, R.; Kurucu, N.; Sevinir, B.; et al. Treatment of high-risk neuroblastoma: National protocol results of the Turkish Pediatric Oncology Group. J. Cancer Res. Ther. 2017, 13, 284–290. [Google Scholar] [PubMed]
- Tonini, G.P. Growth, progression and chromosome instability of Neuroblastoma: A new scenario of tumorigenesis. BMC Cancer 2017, 17, 20. [Google Scholar] [CrossRef]
- Chan, H.S.; Gallie, B.L.; Deboer, G.; Haddad, G.; Ikegaki, N.; Dimitroulakos, J.; Yeger, H.; Ling, V. MYCN protein expression as a predictor of neuroblastoma prognosis. Clin. Cancer Res. 1997, 3, 1699–1706. [Google Scholar]
- Bown, N. Neuroblastoma tumour genetics: Clinical and biological aspects. J. Clin. Pathol. 2001, 54, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Shyr, S.; Liu, Q. Next generation sequencing in cancer research and clinical application. Biol. Proc. Online 2013, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Kamps, R.; Brandao, R.D.; van den Bosh, B.; Paulussen, A.D.; Xanthoulea, S.; Blok, M.J.; Romano, A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int. J. Mol. Sci. 2017, 18, 308. [Google Scholar] [CrossRef] [PubMed]
- TPOG. Turkish Society of Pediatric Oncology Group (TPOG). Neuroblastoma—2009 Treatment Protocol; National Cancer Congress-2009, TURKEY; 2009. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).