Abstract
Subtropical tree species identification is a crucial aspect of forest resource monitoring, and the advancement of deep learning has introduced new opportunities for subtropical tree species identification. But, its performance often relies heavily on the availability of sufficient training samples. In this study, we propose a method for tree species identification via domain generalization with hyperspectral images. The network comprises a generator and a discriminator; the former produces similar samples, and the latter outputs predicted probabilities and classification loss to guide model optimization. The results demonstrate its superiority over traditional CNN-based algorithms.
1. Introduction
As a key part of the global forest ecosystem, subtropical forests, with rich tree species and high productivity, significantly impact regional ecology and help maintain global stability [1]. Understanding the current status, growth, and changes in subtropical forests is crucial for providing data support and forming the foundation for forestry policies, macro planning, and management strategies. Traditional forest surveys mainly use ground-based methods, which, while accurate, are costly, labor-intensive, and time-consuming. They cannot keep pace with the rapid growth in subtropical areas, limiting their ability to provide timely monitoring of forest distribution. Thus, innovative methods are urgently needed to quickly update forest spatial distribution.
Remote sensing technology enables the acquisition of large-scale image data without direct interaction with trees, facilitating species classification across extensive areas without ecological impact. Hyperspectral sensors, a key technology in this field, capture images across dozens to hundreds of continuous spectral bands within specific wavelength ranges, achieving comprehensive spectral coverage [2]. Hyperspectral images are rich in spectral information, and capable of discerning subtle differences between tree species for precise identification and classification. Previous studies [3,4] typically employed traditional machine learning methods such as support vector machines, random forests, and BP neural networks for hyperspectral tree species classification. These methods involve prior feature selection, which can introduce subjectivity and impact classification outcomes. In recent years, deep learning methods have increasingly supplanted traditional approaches in hyperspectral image analysis, offering more objective and accurate classification capabilities.
Deep learning, with its end-to-end architecture, autonomously learns complex nonlinear features from large datasets, avoiding the complexities of traditional feature extraction. Central to this is the Convolutional Neural Network (CNN), known for high classification accuracy through extensive training [5]. Variants like 2D-CNN and 3D-CNN further enhance feature extraction. Researchers have used CNNs for hyperspectral tree species classification, improving accuracy [6]. However, current deep learning methods depend on labeled samples. Limitations in hyperspectral data acquisition hinder feature learning. Self-supervised and transfer learning methods address few-shot classification but struggle with unknown target domains. Leveraging source domain information for sample-free hyperspectral classification is challenging. To tackle this, recent research focuses on domain generalization methods, aiming to create models that generalize effectively to new domains, ensuring reliable classification of hyperspectral images for subtropical tree species identification.
Domain generalization methods address the challenges of domain transfer and target data loss. They aim to train models using data from one or multiple related but different source domains, enabling effective extension to any target domain and enhancing robustness [7]. This approach, similar to human learning patterns, is crucial in real-world applications and has become a key research area in machine learning. In identifying subtropical tree species in hyperspectral imagery, most cross-scene methods depend on labeled source domain data and unlabeled target domain data. However, using only source domain data for cross-scene classification has not been extensively explored.
Based on Zhang Yuxiang’s Single Source Domain Extended Network (SDEnet) [8], we present a classification method for identifying subtropical tree species using domain generalization techniques. This method is trained only in the source domain and tested in the target domain, incorporating adversarial learning generator structures to enhance the use of spectral and spatial information, thereby improving classification performance. This approach offers a novel solution for identifying subtropical tree species without target domain samples using hyperspectral remote sensing images.
2. Related Work
The goal of domain generalization is to learn information from one or several different but related domains and achieve good generalization in untrained regions. In the classification of tree species using HSI, the essential aspect is to solely utilize the source domain dataset during the training process. This approach ensures the attainment of high-quality classification outcomes, as the target domain dataset is not involved in the model training phase. There are currently two main methods for domain generalization: data manipulation and representation learning. In the model proposed in this article, the generator generates additional domains using data operations, while representation learning is used to help the classifier optimize generalization performance. Below are descriptions of two types of methods.
2.1. Data Manipulation
Data manipulation typically involves augmenting or generating data samples that are similar to those in the source domain. These samples are then used as input to train the model, thereby increasing the number of trainable samples and improving the model’s training effectiveness. Data augmentation primarily entails operations such as image flipping, rotation, randomization, scaling, and adding noise to increase the diversity of samples. On the other hand, data generation involves techniques, such as Variational Autoencoders (VAEs) [9] or Generative Adversarial Networks (GANs) [10] to produce diverse samples, thereby facilitating generalization.
2.2. Representation Learning
In the context of domain generalization objectives, representation learning can be described as follows:
In Formula (1), represents the learning function, is the classification function, and is the regularization term. Therefore, it can be seen that improving the performance of representation learning requires a better function for corresponding to . Therefore, the key to representation learning lies in learning domain invariant representations, so that the model can adapt well to different domains.
3. Methodology
3.1. Model Structure
The subtropical tree species classification model based on domain generalization proposed in this article is an improvement of the domain generalization classification model proposed by Zhang Yuxiang [8], mainly divided into two parts: generator and discriminator (Figure 1).

Figure 1.
Composition diagram of subtropical tree species classification model based on domain generalization with HIS.
3.1.1. Generator
To ensure that the additional generated samples (i.e., the generated domain) can effectively contribute to model training, the image style transfer algorithm AdaIN is utilized. It conducts style transfer in the feature space by modulating feature statistics data. And, it not only preserves the content features of the original data in the generated image but also introduces new styles into the generated image through randomization. The feature statistics data primarily encompass the channel mean and variance , which can be expressed as follows:
and represent the image features before and after style transfer, respectively.
For HSI, the original channel mean and variance can be expressed as:
In Formulas (3) and (4), and represents the length and width of the space block, respectively. To ensure the randomness of the generated samples, randomly select the corresponding values before and after the mapping of , related to , and , related to After setting the adaptive linear parameter a, the linear adaptive connection can be obtained by formulas (5) and (6).
By using the style transfer and randomization method, the feature maps of the image are replaced batch by batch and deconvoluted to generate new spatial and spectral features that are consistent with the input feature size.
Due to the high spectral similarity among tree species images, extracting features is challenging, potentially resulting in randomly diverse yet irrelevant samples. Texture information is crucial for depicting image structure and hierarchy. Acting as domain-invariant features, it ensures the reliability of subsequently generated samples. Texture features are extracted using two-dimensional morphological operators, measuring distances between randomly sampled feature points and a central element to determine maximum and minimum values. After two rounds of calculation, these results are concatenated with the original feature points and fed into the AdaIN method to obtain texture features.
Connecting the texture features with spatial and spectral features and inputting them into the model decoder yields the generated domain. However, as previously mentioned, the high spectral similarity of tree species images makes extraction difficult, leading to the potential generation of randomly diverse but useless samples. Hence, this paper introduces a smoothing domain, used as a transition between the source domain and the generated domain when their differences are significant, thereby alleviating the learning and training pressure on the model. The process of generating the smoothing domain is as follows:
represents random weight parameters, represents the smooth domain, represents the source domain, and represents the generated domain.
3.1.2. Discriminator
The discriminator mainly consists of a classification component and a feature extraction component. The classification component primarily outputs predicted probabilities and classification loss, used to guide model optimization. Here, it is defined as:
is the number of samples, is the number of categories, and is the j-th element in the true label vector of the i-th sample.
As the generated domain and smoothing domain are not real samples but are involved in model training, it is also required that they are correctly predicted. Since there is a one-to-one correspondence between the smoothing domain and the generated domain, the classification loss for the smoothing domain and the generated domain is calculated using the labels from the initial domain.
In the feature extraction process, it is necessary to effectively capture the spatial relationships and features in the image. Therefore, in the CNN structure, the convolutional layer, ReLU activation function, and pooling layer are connected. By balancing the impact of image complexity, hardware resources, and performance requirements, a two-stage stacking is selected to gradually extract the features of the image. After feature extraction, the output is used for feature consistency representation to guide the discriminator in learning. Here, InfoNCE is selected to optimize the loss function:
is the number of samples, is the encoding vector of query sample , is the encoding vector of positive samples, and is the encoding vector of negative samples, and uses hyperparameters to adjust the distribution of similarity scores. Based on the above optimization scheme, it is possible to maximize the similarity between the positive sample and the input sample, which can guide the discriminator to learn domain invariant features from the same category. The idea of the generator is opposite to that of the discriminator, but the optimization strategy is the same. Its purpose is to guide the generator to generate samples with greater differences from the initial samples for fully training the network model and extracting deeper features.
3.2. Experiment
3.2.1. Experimental Data
The two research areas selected in this article are both from Gaofeng Forest Farm in Guangxi. Region 1 consists of 1368 × 1101 pixels, while Region 2 consists of 1913 × 1639 pixels. Both of them contain 125 bands with 1m spatial resolution. There are three categories of the same tree species in Region 1 and Region 2, namely eucalyptus, Masson pine, and logging sites. To facilitate model input, Region 2 is cropped to the same size as Region 1, which is 1368 × 1101 pixels, based on the number of common category samples. Figure 2 displays the hyperspectral images of both regions alongside their corresponding labeled category images. Table 1 shows the number of pixels for each class of labeled samples in the two cropped regions.
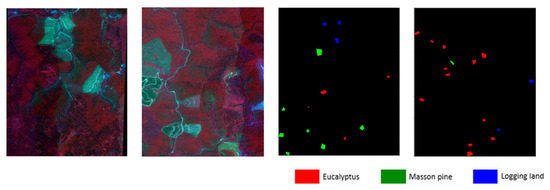
Figure 2.
Hyper spectral images and label maps of two regions.

Table 1.
Number of Labeled Samples for Each Category in the Two Regions.
3.2.2. Parameter Settings
In the experiment, the batch input network sample size of the dataset was set to 13 × 13, and the default value for initial weight decay l2 norm regularization was set to 1 × 10−4. Optimizer Adam uses the adaptive learning rate to accelerate convergence speed, trains the model for 200 epochs, and adjusts the learning rate at equal intervals.
The computer hardware environment used in the experiment is AMD Ryzen9 5900HX processor, NVIDIA GeForce RTX 3080 Laptop graphics card, 3200MHz@32GDDR4 Memory, compiled in Python 3.8.10+Python: 1.11.0 environment.
4. Results
After 200 epochs, the confusion matrix obtained by training on Region 1 and classifying Region 2 is shown in Figure 3. The classification accuracy for the three common categories, namely eucalyptus, Pinus massoniana, and Logging Land, reached 78.81%, 96.69%, and 97.27%, respectively, with an overall accuracy of 81.86%.
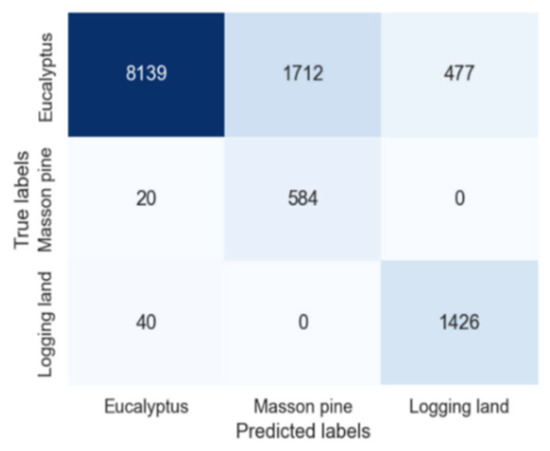
Figure 3.
Classification confusion matrix.
To validate the superiority of the proposed network model (SCM-DG), its classification performance is compared with several traditional classification models directly applied to the target domain, as well as with the PDEN method [10,11], a single-source domain generalization approach in computer vision. Traditional models use 80% of samples for training and the remaining 20% for testing. Both PDEN and the proposed method utilize 80% samples from the source domain for training, 20% for validation, and then test on the target domain. All methods maintain consistent input parameters and network configurations. For clarity in classification accuracy, each class is simplified into a binary problem focusing on correct classification. Table 2 summarizes the classification performance on the tree species dataset, while Figure 4 illustrates the classification results from these methods.

Table 2.
Comparison of Classification Performance of Different Methods.
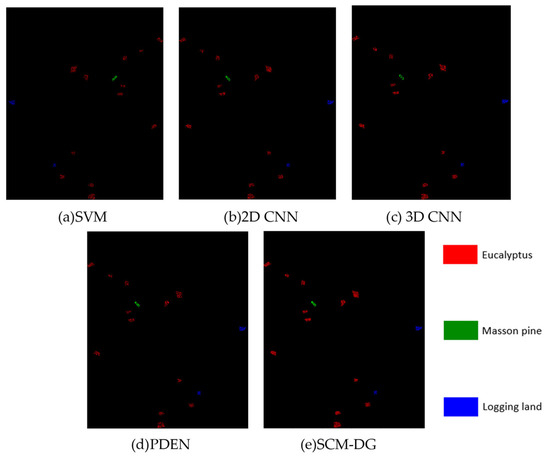
Figure 4.
Comparison of truth maps for different classification methods.
Three commonly used HSI classification algorithms, SVM, 2D CNN, and 3D CNN, and a domain generalization classification method, PDEN, in machine vision direction, were selected for comparative analysis in the experiment. The three commonly used HSI classification algorithms mentioned above increase in attention to data dimensions. The PDEN method, as a classic domain generalization method, can be compared with the method proposed in this paper to verify its effectiveness. The results demonstrate that the approach proposed in this paper outperforms several traditional methods directly applied to the target domain classification in terms of classification performance for each class. Notably, the precision for Masson Pine and Logging land exceeds 95%. However, the slightly inferior performance of Eucalyptus may be attributed to its relatively fewer samples for training in Region 1. Consequently, the network model may not have fully exploited the feature learning process, resulting in insufficient diversity in the generated samples to support detailed training. Nonetheless, the overall performance remains superior to other methods.
Compared to the domain generalization method PDEN, our proposed approach shows slightly lower classification accuracy specifically for the Logging Land class. This difference may stem from PDEN’s focus on radiometric and geometric transformations during data generation, which are less suited for capturing the morphological information predominant in Logging Land. In contrast, our method emphasizes capturing the unique spatial and spectral characteristics inherent in hyperspectral images (HSIs). Additionally, integrating a contrastive adversarial learning strategy enhances the model’s ability to learn robust and meaningful representations, leading to improved training outcomes and more favorable classification results in HSI applications.
5. Conclusions
This paper introduces a method for classifying subtropical tree species using domain generalization and hyperspectral imaging (HSI). The model is exclusively trained in the source domain and tested in the target domain. The method incorporates generator elements empowered with adversarial learning to enhance the utilization of spectral and spatial information in hyperspectral images. By ensuring diversity and reliability in generated samples, the approach enhances the network model’s learning process, resulting in improved classification accuracy. Compared to several traditional methods trained directly on the target domain, our proposed approach demonstrates superior classification performance. However, due to the fact that the training and testing samples belong to different regions of the same area, and the sample categories are highly correlated, there may be some deviation in the accuracy of the evaluation.
Author Contributions
Conceptualization, W.L. and X.W.; methodology, W.L.; software, X.W.; validation, X.W., W.L., and L.Z.; formal analysis, X.W.; investigation, W.L.; resources, L.Z.; data curation, L.Z.; writing—original draft preparation, X.W.; writing—review and editing, W.L. and Y.H.; visualization, Y.H.; supervision, W.L. and Y.H.; project administration, L.Z.; funding acquisition, W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 42071414, the Key Laboratory of Land Satellite Remote Sensing Application, Ministry of Natural Resources of the People’s Republic of China (Grant No. KLSMNR-K202201), the Open Fund of State Key Laboratory of Remote Sensing Science (Grant No. OFSLRSS202202), the Key Laboratory of Land Satellite Remote Sensing Application, the Ministry of Natural Resources of the People’s Republic of China (Grant No. 202305), and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (Grant No. KYCX24_1219).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are contained in this paper.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, Y.-L.; Zhou, G.-Y.; Zhang, D.-Q.; Wenigmann, K.O.; Otieno, D.; Tenhunen, J.; Zhang, Q.-M. Quantification of ecosystem carbon exchange characteristics in a dominant subtropical evergreen forest ecosystem. Asia-Pac. J. Atmos. Sci. 2012, 48, 1–10. [Google Scholar] [CrossRef]
- Li, S.; Song, W.; Fang, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Deep Learning for Hyperspectral Image Classification: An Overview. IEEE Trans. Geosci. Remote Sens. 2019, 57, 6690–6709. [Google Scholar] [CrossRef]
- Modzelewska, A.; Fassnacht, F.E.; Stereńczak, K. Tree Species Identification within an Extensive Forest Area with Diverse Management Regimes Using Airborne Hyperspectral Data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101960. [Google Scholar] [CrossRef]
- Trier Ø, D.; Salberg, A.B.; Kermit, M.; Rudjord, Ø.; Gobakken, T.; Næsset, E.; Aarsten, D. Tree Species Classification in Norway from Airborne Hyperspectral and Airborne Laser Scanning Data. Eur. J. Remote Sens. 2018, 51, 336–351. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- La Rosa, L.E.C.; Sothe, C.; Feitosa, R.Q.; de Almeida, C.M.; Schimalski, M.B.; Oliveira, D.A.B. Multi-Task Fully Convolutional Network for Tree Species Mapping in Dense Forests Using Small Training Hyperspectral Data. ISPRS J. Photogramm. Remote Sens. 2021, 179, 35–49. [Google Scholar] [CrossRef]
- Wang, J.; Lan, C.; Liu, C.; Ouyang, Y.; Qin, T. Generalizing to unseen domains: A survey on domain generalization. arXiv 2021, arXiv:2103.03097. [Google Scholar]
- Zhang, Y.; Li, W.; Sun, W.; Tao, R.; Du, Q. Single-source domain expansion network for cross-scene hyperspectral image classification. IEEE Trans. Image Process. 2023, 32, 1498–1512. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-encoding variational Bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Proc. Adv. Neural Inf. Process. Syst. 2014, 27, 53–65. [Google Scholar]
- Li, L.; Gao, K.; Cao, J.; Huang, Z.; Weng, Y.; Mi, X.; Yu, Z.; Li, X.; Xia, B. Progressive domain expansion network for single domain generalization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 224–233. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).