Abstract
Conventional cancer drugs are small molecules that target specific pathways. We introduced PCMS, a 26 kDa supramolecule combining sensors (S), molecular motors (M), and switching molecules (C), integrated within a fourth-generation PAMAM structure (P). PCMS identifies and deactivates cancer cell nucleus dynamics. A decade ago, we demonstrated programmable, clock-like interactions among the S-C-M components. In this study, we captured images of fractal patterns formed by chromosomal compartments and developed a theoretical model of their fractal dynamics. We showed that the nucleus behaves like a cavity, producing resonance effects similar to Chladni patterns. When the external agent, PCMS, interacts with this cavity, it generates a fractal pattern. We identified and mapped five key phase transitions that ultimately lead to the breakdown of cancer cell nuclei.
1. Introduction
Fractal analysis has emerged as a valuable tool for quantifying cancer cell morphology and nuclear characteristics. Studies have shown that the fractal dimension (FD) of chromatin increases during carcinogenesis and tumor progression, serving as a potential diagnostic and prognostic marker [1,2]. Fractals are widely studied in cancer research, particularly in medical physiology, pathophysiology, and pathology [3,4,5].
Asymptotic fractal analysis of nuclear profiles reveals significant differences between normal and malignant cells, with the parameters c, L, and Bm serving as effective discriminators. These parameters are derived from the formula Br = Bm {1 + (r/L) c} −1, where Br is the boundary length measured at ruler size r, Bm is the maximum boundary length, L is a scaling constant, and c is the asymptotic fractal dimension minus the topological dimension (D − Dt) [6,7]. This approach has shown promise in various cancers, including oral squamous cell carcinomas and multiple myelomas [1]. Additionally, lacunarity measurements, which characterize the distribution of gaps in fractals, have been extended to analyze cancer cell lines with irregular shapes, providing successful discrimination based on vacuole morphology [8]. Fractal analysis can be used to efficiently estimate the geometrical complexity and irregularity of lung cancer cell structures and tumor growth [9]. These fractal-based techniques offer potential molecular diagnostic applications for cancer prognosis and quantitative analysis of nuclear pleomorphism.
Fractal analysis is based on chromosomes that are not randomly distributed within the nucleus but occupy discrete sub-nuclear domains [10], with active genes preferentially located in the periphery of these territories [11]. Thus, when territories interact, translocations and transcription-dependent associations are developed [12]. Furthermore, the distinct volumes and shapes of chromosome territories (CTs), such as the X chromosome territory, indicate functional roles for these territories [13,14]. These vibroelastic studies (Online Text S1) carried out on various nuclear elements and the nucleus for over half a century have focused solely on the natural progression of cancer cells within living systems. To our knowledge, no prior research has tracked drug effects on CTs. This study marks the first in a series where we aim to report on this novel approach. It uniquely measures Chladni patterns within CTs, revealing how cavity effects create basic fractal patterns, followed by fractal pattern analysis to separate drug effects from the cell’s natural self-similarity. Interchromosomal domain (ICD) spaces, DNA-free channels between CTs, are key to nuclear organization [15]. These channels respond to cellular resonance, forming Chladni patterns and mappings that help us filter cooperative, logical interactions between ICDs and preferential genes.
2. Materials and Methods
2.1. Cell Lines
The human lung adenocarcinoma cell line A549 was obtained from Riken cell bank, Japan. A549 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Waltham, MA, USA) and supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Nichirei, Tokyo, Japan) and 1% Pen-Strep (Gibco, Waltham, MA, USA). The cells were maintained in a CO2 incubator at 37 °C, 5% CO2, and 95% humidity.
2.2. Confocal Microscopy
A549 cells were seeded at a density of 2000 cells/500 μL of media in the four chambers of 35 mm quadrant plates and maintained under optimum conditions for 24 h.
For staining the nucleus, live cells were incubated with DAPI (Invitrogen, Waltham, MA, USA), a fluorescent dye that selectively binds to the minor grooves of the adenine-thymine (A-T) regions of double stranded DNA. A working stock of 1 μg/mL DAPI was prepared in milli-Q water (Merck Millipore, Burlington, MA, USA). The cells were rinsed properly with 1 × PBS, and then 200 μL of 1 μg/mL DAPI was added to the cells and incubated in the dark at room temperature for 10 min. Next, the DAPI solution was discarded, and the cells were rinsed with 1 × PBS to remove excess DAPI. Subsequently, 200 μL of Live Cell Imaging Solution (Invitrogen, Waltham, MA, USA) was added to the cells. The cells were treated with different concentrations of PCMS and observed under TCS SP5 confocal microscope (Leica Microsystems, Leica, Germany).
For staining the RNA, SYTO RNASelect (Invitrogen, Waltham, MA, USA), a green fluorescent, cell-permeant nucleic acid staining dye that selectively stains RNA, was used. This dye exhibits bright green fluorescence when bound to RNA (absorption/emission maxima ∼490/530 nm), but only a weak fluorescent signal when bound to DNA. To prepare 1 mL of the labeling solution, a 5 μM intermediate stock was prepared by adding 1 μL of the 5 mM stock solution to 1 mL of the medium, mixing and then adding 100 μL of the 5 μM intermediate stock to 900 μL of the medium. This 500 nM labeling solution was pre-warmed at 37 °C prior to application to the cells. Cells were rinsed with 1 × PBS and then 200 μL of pre-warmed 500 nM of the dye is added to the cells and incubated for 10 min at 37 °C. Subsequently, 200 μL of Live Cell Imaging Solution was added to the cells. The cells were treated with different concentrations of PCMS and observed under TCS SP5 confocal microscope.
2.3. Chladni and Fractal Analysis Simulator Development
We have provided flowchart of the algorithm used to build two simulators and their differences in the Online Text S2: Chladni and fractal simulator: Differences.
First, we developed a Python-based software v1 that enables users to upload fluorescence images over time and perform two core analyses: fractal analysis and Chladni pattern analysis. The fractal analysis tool [16] applies multiple scientific protocols to examine the complex, self-similar structures of CTs and ICD within cancer cell nucleus. Key methods include the Box-Counting method [17], Hausdorff Dimension, and Lacunarity Analysis, each providing unique metrics, such as fractal dimensions [2] and spatial heterogeneity. Advanced techniques like the Wavelet Transform and Multifractal Analysis allow detailed examination of structural variations in loops and strings, offering insights into texture and roughness as variables. Mosaic analysis reveals intracellular dynamics [18]. Loop geometry and curved lines of CTs and ICDs were treated as separate objects, and image processing protocols were fine-tuned for them separately [19].
The Chladni pattern analysis module identifies resonant patterns within cellular structures by detecting nodal lines formed in response to vibrational forces as nuclear membrane acts as a cavity. Key steps involve preprocessing images to grayscale, smoothing noise, and using edge detection algorithms to enhance pattern visibility. Wavelet-based analyses were intensively used [20]. Tools like Radon and Fourier Transforms aid in detecting symmetry and periodic structures characteristic of Chladni patterns. Additionally, machine learning techniques, such as template matching and feature extraction, enhance recognition of known Chladni shapes. Combining fractal and Chladni analyses allows for nuanced tracking of compartmental-to-cellular interactions and resonance effects among CTs and ICDs. The local changes in symmetry and the collective symmetry of a cluster of cells were analyzed using single shot image processing.
3. Protocol of the Study
Cancer cells display altered chromatin structures that impact gene expression. To investigate the morphological effects of these changes, we used a well-known molecular nanobot, PCMS, engineered to monitor pH and molecular motor activity [21,22] within cancer cell nuclei (Figure 1a) [23,24]. As detailed in the Section 2, 1 pM of PCMS was introduced into lung cancer cells, cultured in vitro and tracked under fluorescence confocal microscope. The fluorescent image displays loops as distinct compartments, and our image processing algorithm, as described in the Section 2, systematically detects and identifies the changes in the loop structures, enabling precise characterization of their dynamics and alterations. In Figure 1b, we present two images, where three CTs are highlighted with arrows [25]. In the three CTs shown in Figure 1b, we observe that the loops condense progressively over time [26,27]. These images represent the penultimate phases leading up to the final phase what we have identified as the ’point of no return’. The interphase state, however, remains enigmatic and requires in depth studies to fully understand its dynamics [28].
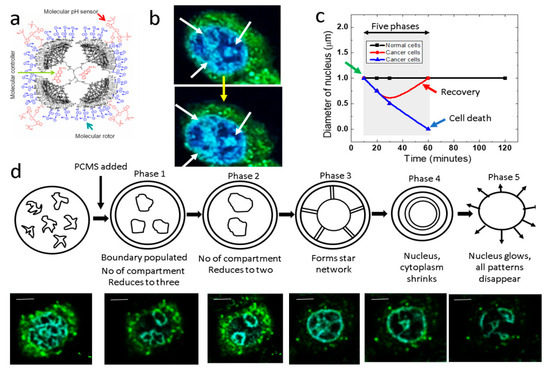
Figure 1.
PCMS and five phases in lung cancer cells: (a) The PCMS molecular structure, with components labeled as P = PAMAM, C = Controller, M = Molecular Motor, and S = Sensor. (b) White and yellow arrows highlight chromosomal compartments within a lung cancer cell and modifications in compartments, respectively. (c) Temporal variation of the cancer cell nucleus is shown, with the green arrow denoting the administration of PCMS and the red plot indicating instances where the nucleus reverts to its original shape, suggesting potential resistance to destruction. (d) Five phases of chromosome compartmentalization within the cancer cell nucleus are presented, with the top row showing schematics and the bottom row displaying corresponding fluorescence images, scale bar is 2 μm. Phases 4 and 5 represent points of no return.
After adding PCMS, the nuclei of cancer cells undergo five major phase transitions ultimately leading to nucleus disintegration. Interestingly, after the formation of three CTs, we observed a point where cell nucleus reverts to its initial state (Figure 1c). Notably, all cells respond differently to PCMS and progress through different phases at varying time intervals. One interesting aspect of this plot is that those cells that undergo a destruction under PCMS treatment, exhibit an approximately 200% increase in nuclear size, this particular enlarged nuclear phase is what we define as the ‘point of no return’.
CTs are a fundamental aspect of nuclear architecture, representing distinct regions within the nucleus where specific chromosomes are located [29]. These territories are crucial for organizing genetic material within the cell. Research indicates that CTs have a spatially constrained structure, with heterochromatic foci limiting long-range transformations of CTs, while euchromatic foci undergo positional oscillations, influencing local changes in chromosome shape [30].
4. Results
4.1. Five Phases of a Cancer Cell Nucleus
We have identified five phase transitions where we see the interplay of ICDs and CTs as shown in Figure 1d.
In the first phase, filamentary structures, form hairlike threads spread all over the nucleus. If we do not add PCMS in the culture plate, the random filamentary distributions would remain the same for upto 18 h, as observed in our experiments. However, when 50 μL of 1 pM PCMS is added, we see the formation of three large CTs, which we refer to as phase 1 (image number 7/22) (Figure S1). The shapes of the compartments vary across different cell culture plates; however, the covered areas remain consistent within a ±10% range, as demonstrated by our analysis of eight plates. Therefore, the energetics involved in the formation of these compartments are consistent over different culture plates. After a period of time, the cells spontaneously undergo a phase transition, during which the number of CTs is reduced from three to two. We were interested to determine whether one among the three CTs formed in phase 1 disappear resulting in two CTs in phase 2 (image number 10/22) (Figure S1). Contrastingly, our observations indicate that all the three CTs formed in phase 1 disappears and two new CTs are formed in phase 2. Thus, in Figure 1b, it is shown that the three CTs condense and disintegrates into filaments. To ensure consistency, we analyzed the surface area coverage of the two newly formed CTs across eight culture plates from different experiments. The results indicate that the total surface area coverage of the CT pairs remains consistent, varying by no more than ±10%. Thus, our observations align with the finding that chromosomal topology has a significant role in determining nuclear architecture [31].
The CT pairs disappear and a large nearly circular CT forms with ICDs branching out like a radiating lines or corona from this nearly circular CT, in phase 3. This higher order chromatin structure carries an interesting physics as well as biology [32]. Two experimental observations we would like to point out. First, accurately select the center of nucleus to build a pair of nearly circular cavities, and second, formation of corona connecting inner cavity to the external cavity. Therefore, the nucleus appears nearly two times bigger than the phase 2. Once the nucleus attains phase 3, it enters the ‘point of no return’ as described in Figure 1c. We have observed several cells where phase 3 formation fails, and the nucleus jumps from phase 2 to the normal state where filamentary structures are randomly distributed all over the nucleus. Therefore, phase 4 is a step towards disintegration of nucleus. Deformation in nucleus has been studied extensively as it reflects vital cellular functionality [33].
In phase 4, the corona like structure of ICDs that link inner circular CT to the external CT starts disappearing. Initially, the connecting lines fall part, primarily they delink from the external ring, and then, we see two isolated CT rings. In the final phase 5, gaps or discontinuities develop in the perimeter of outer ring and it shrinks to regain continuity. Similarly, both outer and inner rings acquire irreparable damage and the ICDs come out from the nucleus to the cell cytoplasm as nucleus gets disintegrated.
For analyzing fractal dynamics and Chladni pattern, we have divided the whole 50 min experiments into 22 parts and these 22 images were analyzed using a series of already established protocols (Figure S1). In this study, the difference in approach from conventional studies is that we are adding an external agent and that agent is triggering low entropy, highly symmetric structures step by step in the cell. Therefore, it is utmost essential that we identify the Chladni pattern that naturally generates in a cavity at different resonances. Undoubtedly, our repeated observations have shown that external agents and unique cellular functions trigger unique distributions of CT and ICDs. Non-spontaneous transitions serve as a marker for the presence of external agents and their functional activity within the cell.
4.2. Chladni Pattern Analysis
Figure 2 illustrates a detailed Chladni pattern analysis of seventeen lung cancer cells examined prior to the addition of PCMS. The analysis provides insights into the structural organization and complexity of the cellular architecture. We have used Sobel or Canny edge detection to identify the filaments and it helps in finding correct loops. Chladni pattern algorithm relies on the detection of a correct boundary. Figure 2a quantifies the density of loops or strings per unit area, using a color scale to represent values, with a maximum of 1, indicating the extent of spatial filament arrangement. Here we find that ~50% of the structures are homogeneously distributed in the nucleus before injecting PCMS.
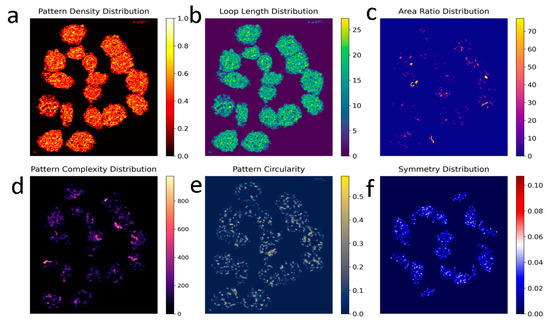
Figure 2.
Snapshot of Chladni pattern analysis live feed: Chladni pattern analysis of seventeen lung cancer cells observed before the addition of PCMS. Panel (a) shows the number of loops or strings per unit area relative to the total area, with a maximum value of 1, represented by a color scale. In panel (b), the distribution of filament length is depicted by the number of pixels along their length, color-coded to show average filament length or inter-compartmental distances (ICDs). Panel (c) highlights large loop structures in comparison to smaller loop structures, while panel (d) displays regions with intertwined loops and filaments, with a color scale indicating high and low intertwinement. Panel (e) assesses the completion percentage of filamentary loops, with bright colors marking more circular structures. Finally, panel (f) presents the degree of symmetry, with red regions indicating asymmetry and blue regions representing symmetry.
Figure 2b maps out the distribution of filament lengths, measuring the number of pixels used along the filament length and color-coding these values to show variations in average filament length or ICDs. Figure 2c represents the area ratio distribution where the program detects pattern contours, calculates their areas, and computes the average area across all patterns. It generates an area ratio distribution map by assigning each pattern a value based on the ratio of its area to the average, visualized with a ‘plasma’ color map (warmer colors for higher ratios, cooler for lower). This analysis reveals variations in pattern sizes, aiding in identifying size anomalies, uniformity, and regional size evolution. Applied wavelet transform for multiscale analysis to capture features of the Chladni patterns across different scales, it accounts for complexity of the pattern. This method is useful for detecting both large and small-scale features in the image. Figure 2d focuses on regions where loops and filaments are intertwined, using a color scale to signify areas of high and low intertwinement, reflecting the complexity of the cellular network.
Chladni patterns have distinct lines and areas where filaments or loops accumulate, so contour detection trace the outlines of nodal regions. Figure 2e evaluates the degree to which filamentary structures form completed loops, with brighter colors indicating more circular, completed formations. Loops are homogeneously distributed all over the nucleus in majority of the cells.
Chladni patterns are often symmetric. We have applied symmetry detection methods like Hough Transforms and Radon Transforms to detect radial symmetry and circular nodal lines in the image. Finally, Figure 2f examines the symmetry of these structures, identifying areas of order and disorder, where red regions denote asymmetry and blue regions signify symmetry, providing a visual representation of structural uniformity and complexity in the cancer cells. Furthermore, symmetry-associated entropy emerges as a key regulator of mammalian chromosome organization [34].
Figure 3 shows a detailed Chladni pattern analysis on seventeen lung cancer cells, focusing on structural changes over time in response to PCMS. The analysis involved capturing high-resolution images of each cell just before the introduction of PCMS, followed by monitoring the cells over a 50 min duration. This time window was divided into 22 equal intervals, yielding 22 sequential snapshots per cell. From these, six key snapshots were analyzed to illustrate different Chladni patterns, and four representative snapshots were selected to evaluate variations in density, complexity, number of patterns, and average area of the structures.
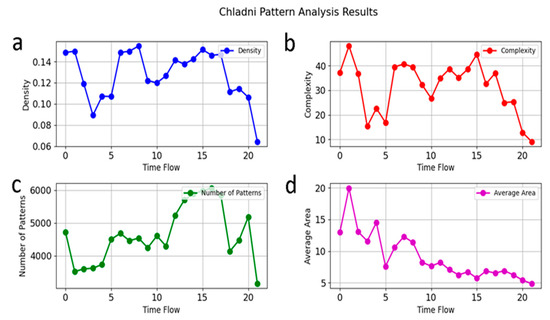
Figure 3.
Chladni pattern analysis on seventeen lung cancer cells: Seventeen lung cancer cells are zoomed in just before PCMS addition, followed by Chladni pattern analysis on each cell. The 50 min duration is divided into 22 intervals, capturing 22 snapshots at equal time points. As illustrated in Figure 2, six snapshots show different Chladni patterns, with four selected here to plot (a) variations in density; (b) complexity; (c) number of patterns; (d) average area.
Figure 3a demonstrates the nearly periodic oscillation of loop and filamentary structures’ density. This periodic behavior could be attributed to the regular communication between the lung cancer cells and the PCMS agent, suggesting a resonance-like interaction of CTs [20]. Interestingly, Figure 3b, which represents structural complexity, closely mirrors the oscillatory trends seen in Figure 3a. This similarity is expected, as the complexity of the patterns is directly influenced by the density of the filamentary structures, highlighting the interconnected nature of these two parameters.
In contrast, Figure 3c, depicting the number of Chladni patterns, reveals a distinct behavior. Unlike density and complexity, the number of patterns does not follow a periodic trend. Instead, there is a marked increase in the number of patterns just before the complete disintegration of the nucleus. This surge suggests a sudden destabilization or fragmentation phase, indicating that the cellular structure is nearing collapse.
Figure 3d represents the average area of the Chladni patterns, which shows a continuous decline over time. This trend indicates a progressive contraction of the structural features within the nucleus. However, a noteworthy observation occurs during phase 4, where a large structure briefly forms before the rapid disintegration of the nucleus. This phase could represent a critical transition, where the nucleus temporarily reorganizes before breaking down entirely under the influence of PCMS. Together, these observations provide valuable insights into how Chladni pattern dynamics and resonance interactions relate to the structural integrity and disintegration process in lung cancer cells, shedding light on the underlying physical mechanisms at play.
4.3. Fractal Analysis
Fractal analysis is a routine study to effectively assess the geometric complexity and irregularity of shapes and patterns in cancer cells [9]. However, the detailed analysis presented in Figure 4 offers critical insights into the self-organization and dynamic structural behavior of lung cancer cells in response to PCMS. By segmenting and labeling self-similar objects within the nucleus, we identified one hundred and sixty distinct compartments across a cluster of seventeen lung cancer cells. The initial segmentation process involved converting images to grayscale, which facilitated accurate compartment detection. Colors were then assigned based on compartment areas, revealing a diverse and complex distribution of compartmental symmetries within the cell cluster. This initial distribution provides a baseline for understanding how structural features evolve over time. The shapes of CTs evolve as a function of time and change in shape is associated with gene density [35].
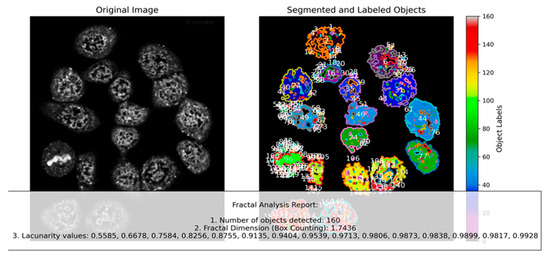
Figure 4.
Segmenting and labeling self-similar objects: Seventeen lung cancer cells are zoomed in just before PCMS addition, and Chladni pattern analysis is performed on each cell. The image is converted to grayscale for analysis (left). Each “compartment” represents a closed area; a total of one hundred and sixty objects are detected (right). Based on compartment areas, colors are assigned, revealing an initial distribution of compartmental symmetries within the cancer cell cluster.
Our observations revealed a fascinating trend: as PCMS is introduced, the assigned colors for segmented compartments display a coordinated and progressive change across a subset of cells. This phenomenon of evolving symmetry highlights a level of collective emergence, where localized structural transformations appear synchronized across multiple cells. Figure 2f illustrates this collective behavior, suggesting that structural changes are not isolated but rather interconnected, potentially mediated by cellular communication or shared biophysical constraints [36].
Furthermore, the phase transitions induced by PCMS, as depicted in Figure 1d, highlight the complexity of these interactions. During early transitions, we see that the nuclei of these cells undergo coordinated reorganization, influencing both the compartmental distribution and the filamentary structures within. This collective reconfiguration implies a potential phase-driven mechanism where neighboring cells respond in unison, adjusting their structural components and symmetry patterns in a way that could have significant implications for understanding cancer cell dynamics and the impact of therapeutic agents like PCMS. Such findings open avenues for exploring how phase transitions and compartmental symmetries contribute to cancer progression and treatment responses.
Figure 5 represents a comprehensive analysis of the temporal evolution of fractal patterns in lung cancer cells following the introduction of PCMS. The study focuses on understanding how compartmental and filamentary structures within the cells dynamically change over a 50 min period, divided into 22 equal time intervals (Figure S1). Figure 5a illustrates the significant transformation in the symmetry distribution and the number of compartments over time. Since complex and larger assemblies form as a function of time, the algorithm identifies the self-similar features across various scales. Initially, upon PCMS addition, there is a substantial increase in the number of compartments and filamentary structures. However, as PCMS interacts with the cell nucleus, these initially fragmented components undergo an organized restructuring, leading to a dramatic reduction in the number of compartments. However, at the same time, the nucleus size increases, larger assemblies form in the device, thus, number of filaments remain conserved. This observation underscores a critical transition from a chaotic to a more ordered and unified architecture. Translocations in oncogenic human cancer has been a point of interest and here we have seen abundant onsets of such translocations of CTs [37].
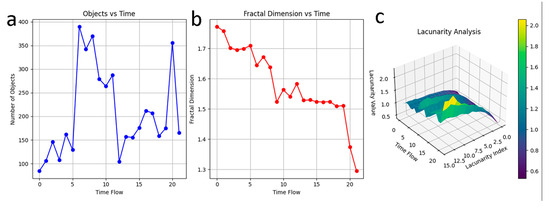
Figure 5.
Temporal evolution of fractal pattern: Seventeen lung cancer cells are zoomed in just before PCMS addition, and Fractal pattern analysis is performed on each cell. (a) After PCMS is introduced, the compartmental structure of each cell changes over time to estimate the scale free features. The total duration of 50 min is divided into 22 equal intervals, producing 22 snapshots. Upon PCMS addition, the symmetry distribution of each cell transforms. The changes in compartmental shapes and symmetry across cells are significant, with the number of compartments varying non-linearly. (b) Fractal dimension varies as a function of time, indicating structural complexity changes. (c) Lacunarity peaks just before nuclear disintegration, displayed by ripples in the graph.
Figure 5b captures the variation in fractal dimension as a function of time or spatial scale [38,39], reflecting the evolving structural complexity of the cancer cells and its scale free features. The fractal dimension significantly decreases as PCMS acts, signifying a shift from intricate, self-similar patterns to a more simplified and unified structure. This reduction highlights the PCMS-induced formation of a unique, singular architecture within the cells, emphasizing a loss of complexity and a movement towards structural uniformity.
Figure 5c provides insights into the lacunarity of the cellular patterns, which serves as a measure of texture and gap distribution. Lacunarity is a measure of how texture or fractal structures fill space and describes the gaps or holes within the fractal [8]. A low lacunarity indicates homogeneity, while a high lacunarity indicates heterogeneity. The data reveal periodic fluctuations in lacunarity, with pronounced peaks just before the nuclear disintegration. This spike in lacunarity indicates increased heterogeneity and instability in the structural arrangement, acting as a precursor to the cell’s ultimate breakdown. The rippling effect seen in the lacunarity graph emphasizes the unstable and transitional state of the cells, culminating in a dramatic disintegration phase. Collectively, these findings suggest that PCMS induces significant reorganization and destabilization of cellular architecture, with distinct fractal and spatial patterns emerging as key indicators of the cells’ response and eventual collapse.
Chromosomal filaments, though forming distinct large-scale structural oscillations (Chladni-like cavity effect) at particular scale, also exhibit fractal behavior of CTs and ICDs across scales due to their inherent molecular properties. Therefore, both the features coexist and slowly disappears together as PCMS induced effect that leads to disintegration of cancer cell nucleus.
5. Conclusions
Our study demonstrates the power of fractal analysis in understanding the morphological changes and structural dynamics of lung cancer cells under the influence of a molecular nanobot, PCMS. By employing advanced imaging and segmentation techniques, we captured detailed temporal evolutions of compartmental and filamentary structures over a 50 min period. The introduction of PCMS induced significant phase transitions within the nucleus, marked by a shift from highly fragmented, chaotic configurations to more organized and unified architectures, as evidenced by variations in fractal dimension, lacunarity, and Chladni pattern dynamics.
We observed that the number of compartments initially spiked before reorganizing into larger, cohesive structures, highlighting the cellular reorganization process. The fractal dimension decreased over time, indicating a reduction in structural complexity and a transition toward singular, simplified architectures. Lacunarity measurements revealed critical instability moments, with peaks preceding nuclear disintegration, underscoring the progressive breakdown of the cellular network. These findings suggest that PCMS not only disrupts cancer cell nuclei but also orchestrates a series of organized phase transitions that ultimately lead to cell collapse. The synchronization of compartmental transformations across multiple cells indicates potential collective behavior mediated by shared biophysical constraints or cellular communication.
Overall, this work introduces a novel approach to tracking drug effects on CTs and emphasizes the significance of fractal and Chladni pattern analysis in cancer research [40]. The insights gained here open new avenues for developing targeted cancer therapies that exploit the structural vulnerabilities of cancer cells, offering a promising diagnostic and prognostic toolset for future applications.
Supplementary Materials
The following supplementary materials can be downloaded at: https://www.mdpi.com/article/10.3390/fractalfract9010008/s1, Online Text S1: Vibroelastic studies of nuclear chromosomes and chromatins. Online Text S2: Chladni and fractal simulator: Differences. Figure S1: Experimental images showing nuclear transition. References [41,42,43,44] are cited in Supplementary Materials.
Author Contributions
A.B. and A.B.K. planned the research; A.S. and S.G. synthesized the PCMS molecule; P.D. cultured the cancer cell, imaged intake of PCMS; A.B. and P.D. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Acknowledgments
Parama Dey acknowledges National Institute for Materials Science (NIMS) for awarding the International Cooperative Graduate Program (ICGP) Fellowship under the “Indian Institute of Technology Guwahati—NIMS Cooperative Graduate Program”. Ajaikumar B. Kunnumakkara acknowledges professional development fund (BSBE/ABK/PDF) for supporting the present work.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Metze, K. Fractal dimension of chromatin: Potential molecular diagnostic applications for cancer prognosis. Expert Rev. Mol. Diagn. 2013, 13, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Metze, K.; Adam, R.; Florindo, J.B. The fractal dimension of chromatin—A potential molecular marker for carcinogenesis, tumor progression, and prognosis. Expert Rev. Mol. Diagn. 2019, 19, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, M.; Giuliani, A.; Cucina, A.; D’Anselmi, F.; Soto, A.M.; Sonnenschein, C. Fractal analysis in a systems biology approach to cancer. Semin Cancer Biol. 2011, 21, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Pantić, I.; Paunović-Pantić, J.; Radojevic-Skodric, S. Application of fractal and textural analysis in medical physiology, pathophysiology, and pathology. Med. Istraživanja 2022, 55, 43–51. [Google Scholar] [CrossRef]
- Baish, J.W.; Jain, R.K. Fractals and cancer. Cancer Res. 2000, 60, 3683–3688. [Google Scholar]
- Landini, G.; Rippin, J.W. An “asymptotic fractal” approach to the morphology of malignant cell nuclei. Fractals 1993, 1, 326–335. [Google Scholar] [CrossRef]
- Landini, G.; Rippin, J.W. Quantification of nuclear pleomorphism using an asymptotic fractal model. Anal. Quant. Cytol. Histol. 1996, 18, 167–176. [Google Scholar]
- Borys, P.; Krasowska, M.; Grzywna, Z.J.; Djamgoz, M.B.; Mycielska, M.E. Lacunarity as a novel measure of cancer cell behavior. BioSystems 2008, 94, 276–281. [Google Scholar] [CrossRef]
- Lennon, F.E.; Cianci, G.C.; Cipriani, N.A.; Hensing, T.A.; Zhang, H.J.; Chen, C.T.; Murgu, S.D.; Vokes, E.E.; Vannier, M.W.; Salgia, R. Lung cancer—A fractal viewpoint. Nat. Rev. Clin. Oncol. 2015, 12, 664–675. [Google Scholar] [CrossRef]
- Iwasaki, O.; Corcoran, C.; Noma, K. Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle. Nucleic Acids Res. 2015, 44, 3618–3628. [Google Scholar] [CrossRef]
- Kurz, A.; Lampel, S.; Nickolenko, J.; Bradl, J.; Benner, A.; Zirbel, R.; Lichter, P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 1996, 135, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Branco, M.; Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006, 4, e138. [Google Scholar] [CrossRef] [PubMed]
- Eils, R.; Dietzel, S.; Bertin, E.; Schröck, E.; Speicher, M.; Ried, T.; Cremer, T. Three-dimensional reconstruction of painted human interphase chromosomes: Active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 1996, 135, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- George, P.; Kinney, N.; Liang, J.; Onufriev, A.; Sharakhov, I. Three-dimensional organization of polytene chromosomes in somatic and germline tissues of malaria mosquitoes. Cells 2020, 9, 339. [Google Scholar] [CrossRef]
- Verschure, P.; Kraan, I.; Manders, E.; Driel, R. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 1999, 147, 13–24. [Google Scholar] [CrossRef]
- Bubnov, R.V.; Melnyk, I.M. The methods of fractal analysis of diagnostic images. Initial clinical experience. Dalʹnevostočnyj Med. Žurnal 2011, 3, 108–113. [Google Scholar]
- Karperien, A.L.; Jelinek, H.F. Box-counting fractal analysis: A primer for the clinician. In The Fractal Geometry of the Brain; Di Ieva, A., Ed.; Springer: New York, NY, USA, 2016; pp. 13–44. [Google Scholar]
- Landini, G.; Iannaccone, P.M. Modeling of mosaic patterns in chimeric liver and adrenal cortex: Algorithmic organogenesis? FASEB J. 2000, 14, 823–827. [Google Scholar] [CrossRef][Green Version]
- Dey, P. Basic principles and applications of fractal geometry in pathology: A review. Anal. Quant. Cytol. Histol. 2005, 27, 284–290. [Google Scholar]
- Ramirez-Cobo, P.; Vidakovic, B. A 2D wavelet-based multiscale approach with applications to the analysis of digital mammograms. Comput. Stat. Data Anal. 2013, 58, 71–81. [Google Scholar] [CrossRef]
- Singhania, A.; Ghosh, I.; Sahoo, P.; Fujita, D.; Ghosh, S.; Bandyopadhyay, A. Radio Waveguide–Double Ratchet Rotors Work in Unison on a Surface to Convert Heat into Power. Nano Lett. 2020, 20, 6891–6898. [Google Scholar] [CrossRef]
- Singhania, A.; Chatterjee, S.; Kalita, S.; Saha, S.; Chettri, P.; Gayen, F.R.; Saha, B.; Sahoo, P.; Bandyopadhyay, A.; Ghosh, S. An Inbuilt Electronic Pawl Gates Orbital Information Processing and Controls the Rotation of a Double Ratchet Rotary Motor. ACS Appl. Mater. Interfaces 2023, 15, 15595–15604. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chatterjee, S.; Roy, A.; Ray, K.; Swarnakar, S.; Fujita, D.; Bandyopadhyay, A. Resonant oscillation language of a futuristic nano-machine-module: Eliminating cancer cells & Alzheimer Aβ plaques. Curr. Top. Med. Chem. 2015, 15, 534–541. [Google Scholar] [PubMed]
- Ghosh, S.; Roy, A.; Singhania, A.; Chatterjee, S.; Swarnakar, S.; Fujita, D.; Bandyopadhyay, A. In-vivo and in-vitro toxicity test of molecularly engineered PCMS: A potential drug for wireless remote-controlled treatment. Toxicol. Rep. 2018, 5, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Szczepińska, T.; Rusek, A.; Plewczynski, D. Intermingling of chromosome territories. Genes Chromosomes Cancer 2019, 58, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Alonso, L.; Holland, P.; Le Gras, S.; Zhao, X.; Jost, B.; Bjørås, M.; Barral, Y.; Enserink, J.M.; Chymkowitch, P. Mitotic chromosome condensation resets chromatin to safeguard transcriptional homeostasis during interphase. Proc. Natl. Acad. Sci. USA 2023, 120, e2210593120. [Google Scholar] [CrossRef]
- Toné, S.; Sugimoto, K.; Tanda, K.; Suda, T.; Uehira, K.; Kanouchi, H.; Samejima, K.; Minatogawa, Y.; Earnshaw, W.C. Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp. Cell Res. 2007, 313, 3635–3644. [Google Scholar] [CrossRef]
- Rosa, A.; Everaers, R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008, 4, e1000153. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Maya-Mendoza, A.; Jackson, D. Labeling DNA replication foci to visualize chromosome territories in vivo. Curr. Protoc. Cell Biol. 2017, 75, e19. [Google Scholar] [CrossRef]
- Berríos, S. Nuclear architecture of mouse spermatocytes: Chromosome topology, heterochromatin, and nucleolus. Cytogenet. Genome Res. 2017, 151, 61–71. [Google Scholar] [CrossRef]
- Fudenberg, G.; Mirny, L. Higher-order chromatin structure: Bridging physics and biology. Curr. Opin. Genet. Dev. 2012, 22, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Awazu, A. Nuclear dynamical deformation induced hetero- and euchromatin positioning. Phys. Rev. E 2015, 92, 032709. [Google Scholar] [CrossRef] [PubMed]
- Finan, K.; Cook, P.; Marenduzzo, D. Non-specific (entropic) forces as major determinants of the structure of mammalian chromosomes. Chromosome Res. 2010, 19, 53–61. [Google Scholar] [CrossRef]
- Sehgal, N.; Fritz, A.; Morris, K.; Torres, I.; Chen, Z.; Xu, J.; Berezney, R. Gene density and chromosome territory shape. Chromosoma 2014, 123, 499–513. [Google Scholar] [CrossRef][Green Version]
- Heymans, O.; Fissette, J.; Vico, P.; Blacher, S.; Masset, D.; Brouers, F. Is fractal geometry useful in medicine and biomedical sciences? Med. Hypotheses 2000, 54, 360–366. [Google Scholar] [CrossRef]
- Zheng, J. Oncogenic chromosomal translocations and human cancer (review). Oncol. Rep. 2013, 30, 2011–2019. [Google Scholar] [CrossRef]
- Etehadtavakol, M.; Lucas, C.; Sadri, S.; Ng, E.Y.K. Analysis of breast thermography using fractal dimension to establish possible difference between malignant and benign patterns. J. Healthc. Eng. 2010, 1, 27–43. [Google Scholar] [CrossRef]
- Dumansky, Y.V.; Lyakh, Y.E.; Gorshkov, O.G.; Gurianov, V.G.; Prihodchenko, V.V. Fractal dimensionality analysis of normal and cancerous mammary gland thermograms. Chaos Solitons Fractals 2012, 45, 1494–1500. [Google Scholar] [CrossRef]
- John, A.M.; Elfanagely, O.; Ayala, C.A.; Cohen, M.; Prestigiacomo, C.J. The utility of fractal analysis in clinical neuroscience. Rev. Neurosci. 2015, 26, 633–645. [Google Scholar] [CrossRef]
- Marko, J.F. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008, 16, 469–497. [Google Scholar] [CrossRef]
- Houchmandzadeh, B.; Marko, J.F.; Chatenay, D.; Libchaber, A. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol. 1997, 139, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Almagro, S.; Dimitrov, S.; Hirano, T.; Vallade, M.; Riveline, D. Individual chromosomes as viscoelastic copolymers. EPL 2003, 63, 908–914. [Google Scholar] [CrossRef]
- Bertsch, G.F. Elasticity in the response of nuclei. Ann. Phys. 1974, 86, 138–146. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).