An Overview of AI-Guided Thyroid Ultrasound Image Segmentation and Classification for Nodule Assessment
Abstract
1. Introduction
- A comprehensive taxonomy of AI-guided methods, systematically organizing approaches from traditional image processing to state-of-the-art DL models like ViTs. Figure 1 presents a visual roadmap of the topics discussed, organizing the landscape of AI-guided thyroid US image analysis into its core components.
- A critical and comparative evaluation through consolidated strengths and weaknesses tables, summarizing over 100 distinct methods to illuminate research gaps and guide future work.
- Practical guidance designed to bridge the gap between academic research and clinical implementation. This includes a detailed discussion of public datasets, checklists for evaluating model alignment with clinical standards like TIRADS, and actionable “playbook” boxes for operationalizing key concepts such as explainability and clinical workflow integration.
2. Thyroid US Image Segmentation for Nodule Boundary Extraction
- 1.
- Clinical benefitsAssessment of nodule vascularity: The primary improvement is the ability to visualize and quantify blood flow within a nodule after the injection of a microbubble contrast agent. Certain vascular patterns, such as irregular, chaotic, or peripheral vascularity, as well as rapid contrast uptake and wash-out, may increase the suspicion of malignancy.Quantitative perfusion analysis: Models can be trained to extract quantitative features from the video, such as wash-in/wash-out curves and time-to-peak enhancement, providing objective and reproducible perfusion metrics that are difficult to quantify reliably through visual inspection alone.
- 2.
- Practical limitations and challengesHardware and workflow disruption: CEUS requires specialized US equipment and the intravenous administration of a contrast agent, which is not part of routine, first-line thyroid nodule screening in most clinical settings.Data scarcity and annotation complexity: The acquisition, storage, and, most importantly, annotation of video data—often requiring frame-level or temporal labeling of perfusion phases–are far more complex and labor-intensive than for static images. This has led to a profound lack of large, public datasets, hindering robust model development.Model-specific challenges: While models like DpRAN can capture multiscale temporal features, the inherent variability in perfusion can be challenging to model. These systems can lack the flexibility to robustly distinguish truly salient perfusion differences from background noise in highly heterogeneous nodules.
- 3.
- When to useTemporal and CEUS-based CAD methods are generally not intended for routine screening. Their ideal application is as a second-line diagnostic tool for characterizing equivocal or indeterminate nodules that are difficult to classify using B-mode features alone, or in specialized academic research settings focused on tumor vascularity.
| Reference | Method | Strengths | Weaknesses | Description |
|---|---|---|---|---|
| Maroulis et al. (2007) [7] | Active contour | Addresses smooth/blurred boundaries, robust to intensity inhomogeneity, inherent denoising | Cannot be applied on isoechoic nodules | Introduces the variable background active contour (VBAC), a Chan–Vese model [22] variant, which mitigates the effects of inhomogeneous tissue in US images by selectively excluding outlier regions from the contour evolution equations. |
| Savelonas et al. (2009) [23] | Active contour | Copes with isoechoic nodules, integrates textural features | Parameter adjustment (see Section 4.1) | Introduces the joint echogenicity-texture (JET) active contour, which incorporates statistical texture information into a modified energy functional, enabling it to cope with challenging isoechoic thyroid nodules. |
| Du et al. (2015) [24] | Active contour | Noise robustness | Region-based information is not considered, parameter adjustment (see Section 4.1) | Presents a pipeline centered on a distance-regularized level set guided by a local phase symmetry feature. This approach is designed to suppress speckle noise and prevent boundary leakage. |
| Koundal et al. (2016) [25,26] | Active contour/neutrosophic clustering | Integration of neutrosophic and clustering information, automated parameter adjustment | Dependency on rough region-of-interest (ROI) estimation, computational complexity/cost (see Section 4.1) | Integrates spatial information with neutrosophic L-means clustering to derive a robust region of interest (ROI), which then guides a distance-regularized level set [27] for precise delineation. Later, Koundal et al. [28] proposed a similar pipeline employing intuitionistic fuzzy c-means clustering. |
| Mylona et al. (2014) [19] | Active contour | Automated parameter adjustment | Potential robustness issues for gradient orientation estimation based on structure tensor (see Section 4.1) | Employs orientation entropy (OE) to automatically adjust the regularization and data fidelity parameters of region-based active contours, adapting the model’s behavior to local image structure. |
| Tsantis et al. (2006) [31] | Wavelets, local maxima chaining, Hough transform | Noise robustness, copes with iso-echoic nodules | Prior knowledge is simply expressed as circular shapes | A three-stage traditional method that combines wavelet edge detection for speckle reduction, a multiscale structure model for contour representation, and a Hough transform to distinguish nodule boundaries. |
| Chiu et al. (2014) [32] | Radial gradient | Noise robustness | User-defined ROI/boundary points (see Section 4.1) | A semi-automatic method that uses a radial gradient algorithm [33] and variance-reduction statistics to select cut points on the nodule boundary from a user-defined ROI, with additional filtering for outliers. |
| Nugroho et al. (2015) [21] | Bilateral filtering, active contour | Addresses smooth/blurred boundaries, inherent denoising, and topological adaptability | Assumes nearly homogeneous foreground and background | A traditional pipeline that applies bilateral filtering for image denoising and uses the Chan–Vese model [22] for nodule delineation, leveraging its inherent denoising and topological adaptability. |
| Le et al. (2015) [29] | Gradient and directional vector flow active contour | Increased contour flexibility, reduced computational time and complexity | Multiple parameters, no end-to-end optimization (see Section 4.1) | A variant of the dynamic directional gradient vector flow active contour [30] that redefines the energy functional by altering straight lines to fold lines in order to increase contour flexibility. It also introduces a vector field to reduce computational complexity. |
| Ma et al. (2017) [35] | CNN | Fully automatic | Ignores global context, lacks interpretability (see Section 4.2) | An early CNN-based approach that formulates segmentation as a patch-level classification task. It uses a multi-view strategy on patches from normal and nodular glands to generate a probability map. |
| Wang et al. (2023) [36] | CNN (DPAM-PSPNet) | Integrates multiscale context, brightness, contrast, and structural similarity | Model complexity (see Section 4.3) | Introduces a dual path attention mechanism (DPAM) into pyramid scene parsing network (PSPNet) [37]. This mechanism is designed to capture both global contextual information and fine-grained nodule edge structures simultaneously. |
| Zhou et al. (2018) [38] | CNN (U-Net) | Requires only limited human effort | Not fully automatic | U-Net is accompanied by an interactive segmentation stage. The model is guided during training and inference by four manually determined endpoints of major and minor nodule axes. |
| Nandamuri et al. (2019) [39] | FCN | Relatively efficient inference | Tested on a moderately sized dataset (see Section 4.4) | Introduces SUMNet, an FCN that learns the spatial relationship between classes. It uses feature concatenation and index-passing-based unpooling to enhance semantic segmentation. |
| Abdolali et al. (2020) [40] | Mask R-CNN | No complex postprocessing required | Moderately sized dataset, concerns about overfitting (see Section 4.4) | Applies a mask R-CNN model, extending the faster R-CNN object detector with a specialized loss function to perform instance segmentation of thyroid US images. |
| Koumar et al. (2020) [41] | VGG16 variant | Efficient inference, fully automatic | Moderate detection rate for cystic components, moderate accuracy | A VGG16 [42] variant that uses dilated convolutions to expand the receptive field. It features two separate outputs to simultaneously delineate both the normal thyroid gland and the nodule. |
| Wu et al. (2020) [43] | DenseNet-121 combined with ASPP | Captures multiscale information | Limited exploitation of boundary information | Combines atrous spatial pyramid pooling (ASPP) with depth-wise separable convolutions. This approach is designed to better capture contextual information while managing the size of the feature maps. |
| Webb et al. (2021) [44] | Deeplab v3+ with DenseNet-101 variant | Preserves spatial resolution, improved low-level feature extraction | Computational and memory cost (see Section 4.3) | Adapts the DeepLabv3+ [45] architecture with a ResNet101 backbone for thyroid US. It features a dual-output design to delineate the thyroid gland and nodules/cysts separately, improving handling of overlapping classes. |
| Nugroho et al. (2021) [46] | Res-UNet | Combines strengths of U-Net (preserves spatial resolution) and ResNet (robustness and depth) | Linear interpolation may degrade details | Employs a Res-UNet model, which integrates the residual connections of ResNet into the U-Net architecture, in order to combine the strengths of both (spatial preservation and deep feature extraction). |
| Xiangyu et al. (2022) [47] | DPCNN | Effective localization | Hyperparameter adjustment (see Section 4.1), no deep feature representation | A method based on a pulse-coupled neural network that first performs a rough localization, which is then refined using variance and covariance criteria to identify and segment the final lesion area. |
| Nguyen et al. (2022) [48] | U-Net | Refined segmentation obtained with the successive application of SN and EN | Computational cost (see Section 4.3), struggles in cases of small thyroid glands (see Section 4.1) | Introduces a dual network based on information fusion. It uses a “suggestion network” (SN) to generate an initial rough mask, which is then refined by an “enhancement network” (EN). |
| Yang et al. (2022) [49] | U-Net variant | Contextual understanding via PAM, captures boundary details via MRM | Computational complexity/cost (see Section 4.3), small-sized dataset (see Section 4.4) | Introduces DMU-Net, a dual-subnet architecture with a U-shaped and an inverse U-shaped path. It incorporates a pyramid attention module (PAM) and a margin refinement module (MRM) to capture both context and fine details. |
| Song et al. (2022) [50] | Faster R-CNN | Enhanced localization by adding a segmentation branch, fine-grained annotations are not required | Computational complexity/cost (see Section 4.3), the quality of pseudo-labels affects performance | Introduces FDnet, a feature-enhanced dual-branch network based on faster R-CNN. It adds a semantic segmentation branch and introduces a method for generating pseudo-labels (computationally generated masks) for training. |
| Chen et al. (2022) [51] | Trident network | Captures multiscale information, suppresses false positives, handles complex textures | Computational complexity/cost (see Section 4.3), generalization concerns (see Section 4.4) | Introduces MTN-Net, a multi-task network based on the trident network. It uses trident blocks with different receptive fields to detect nodules of varying sizes and includes a specialized non-maximum suppression (TN-NMS). |
| Gan et al. (2022) [52] | U-Net variant | Enhanced spatial attention via polarized self-attention | Computational complexity/cost (see Section 4.3), hyperparameter adjustment (see Section 4.1), generalization concerns (see Section 4.4) | A U-Net variant that uses an enhanced residual module with soft pooling in the encoder. It also incorporates a full-channel, attention-assisted skip connection based on polarized self-attention. |
| Jin et al. (2022) [53] | Boundary field regression branch integrated with existing networks | Can be integrated with various network architectures (U-Net, DeepLab v3+, etc.), enhances the accuracy of boundary extraction | Depends on the network backbone and on the quality of the utilized segmentation mask | Addresses the boundary imbalance issue by introducing a boundary field (BF) regression branch that is trained on a heatmap generated from existing masks to provide explicit boundary information to the network. |
| Shao et al. (2023) [54] | U-Net3+ variant | Reduced number of parameters, secondary feature maps derived by ghost modules may lack diversity | Inference time (see Section 4.3) | Introduces a U-Net3+ [55] variant that employs the “ghost” bottleneck module. The latter uses ghost modules (GMs) [56] and depth-wise convolutions [57] to expand and then reduce channel numbers efficiently. |
| Dai et al. (2024) [58] | U-Net++ variant | SK-based attention addresses diversity in nodules’ size and shape | Moderately sized dataset (see Section 4.4), oversegmentation in some cases | Introduces SK-Unet++, which starts from U-Net++ [59] and adds adaptive receptive fields based on the selective kernel (SK) attention mechanism to better handle nodules of varying sizes. |
| Zheng et al. (2023) [60] | U-Net variant | ResNeSt copes with the presence of small nodules, contextual awareness via ASPP | Inference time (see Section 4.3) | Presents DSRU-Net, which enhances U-Net with a ResNeSt backbone [61], atrous spatial pyramid pooling (ASPP) [43] for context, and deformable convolution (DC v3) [62] for adaptability to irregular gland and nodule shapes. |
| Wang et al. (2023) [63] | U-Net variant | MSAC captures both contextual and fine-grained information | Computational complexity/cost (see Section 4.3), concerns for generalization (see Section 4.4) | Introduces the multiscale attentional convolution (MSAC) module, which replaces standard convolutions in U-Net. This module uses cascaded convolutions and self-attention to concatenate features from various receptive fields. |
| Chen et al. (2023) [64] | CNN | Addresses the diversity in nodules’ size and shape | Cannot cope with multiple nodules | Introduces FDE-Net, which combines a CNN with a traditional image omics method. It employs a segmental frequency domain enhancement to reduce noise and strengthen contour details in the feature maps. |
| Ma et al. (2023) [65] | CycleGAN | Addresses domain shift due to multi-site US image collection | Computational complexity/cost (see Section 4.3) | A domain adaptation framework for multi-site datasets. It uses a CycleGAN-based [66] image translation module to unify image styles and a segmentation consistency loss to guide unsupervised learning on unlabeled target data. Their segmentation module is constructed into two symmetrical parts, which use EfficientNet [67]. |
| Bouhdiba et al. (2024) [68] | U-Net and RefineUNet | Captures both fine-grained and contextual information | Computational complexity/cost (see Section 4.3) | A deep architecture that combines U-Net with multi-resolution RefineUNet modules. This combination of residual blocks and chained residual pooling (CRP) is aimed at exploiting both fine-scale and contextual features. |
| Ma et al. (2024) [69] | CNN | Captures complex patterns via dense blocks, addresses small nodules and dataset imbalance | Generic backbone, not specialized for segmentation, struggles with heterogeneous nodules | Introduces MDenseNet, a densely connected CNN. The dense connectivity pattern is designed to capture complex patterns, address the small nodule problem, and mitigate dataset imbalance issues. |
| Liu et al. (2024) [70] | U-Net and ConvNeXt | Captures both edge detail and contextual information | Computational complexity/cost (see Section 4.3) | Builds on U-Net using a ConvNeXt [71] backbone and introduces four specialized modules: a boundary feature guidance module (BFGM), a multiscale perception fusion module (MSPFM), a depthwise separable ASPP, and a refinement module. |
| Xing et al. (2024) [72] | U-Net variant | Multiscale feature extraction, computational complexity/cost | Struggles in the presence of calcifications (see Section 4.1) | A U-Net variant that integrates three key modifications for efficiency and performance: dense connectivity for feature reuse, dilated convolutions for redesigning layers, and factorized convolutions to improve efficiency. |
| Yang et al. (2024) [73] | U-Net variant | Lightweight design reducing computational complexity/cost, dual attention aids capturing both local and global information | Has not been compared with some large models, such as SWIN U-Net | Introduces DACNet, a lightweight U-shaped network designed for efficiency. It uses depthwise convolutions with squeeze-and-excitation (DWSE) and split atrous with dual attention (ADA) to reduce parameters while capturing multi-scale features. |
| Xiao et al. (2025) [74] | DeepLab v3+, EfficientNet-B7 | Captures contextual information | Struggles with speckle noise | Replaces the standard backbone of the DeepLabv3+ architecture with the more efficient EfficientNet-B7. This combination leverages atrous convolution to capture context while handling morphological variability. |
| Ongole et al. (2025) [75] | FCM, ResNet101 variant | Addresses inhomogeneity, identifies calcium flecks | Computational complexity/cost (see Section 4.3) | Introduces Bi-MCS, a method combining fuzzy c-means and k-means clustering. This technique is incorporated into a ResNet101 variant (Bi-ResNet101) to enhance color sense-based segmentation by focusing on intensity variations. |
| Xiang et al. (2025) [76] | U-Net variant | Addresses datasets obtained from multiple sites by means of FL | Computational complexity/cost (see Section 4.1), limitations in adapting to multi-center scenarios of high variability (see Section 4.4) | A federated learning (FL) method based on a multi-attention guided U-Net (MAUNet). It uses a multiscale cross-attention (MSCA) module to handle variations in nodule shape and size across different institutions. |
| Yadav et al. (2022) [77] | SegNet, VGG16, U-Net | Addresses speckle noise | Computational complexity/cost (see Section 4.3) | Introduces Hybrid-UNet, a model that combines the architectural principles of SegNet, VGG16, and U-Net. The model is trained using transfer learning to delineate nodules and cystic components. |
| Ali et al. (2024) [78] | U-Net-inspired encoder–decoder | Captures contextual information | Computational complexity/cost (see Section 4.3) | Introduces CIL-Net, an encoder–decoder architecture that uses dense connectivity and a triplet attention block in the encoder, as well as a feature improvement block with dilated convolutions in the decoder to capture global context. |
| Sun et al. (2024) [79] | U-Net variant | Captures contextual information | Computational cost/complexity (see Section 4.3) | Introduces CRSANet, a U-Net-based network that uses class representations to describe the characteristics of thyroid nodules. A dual-branch self-attention module then refines the coarse segmentation results. |
| Wu et al. (2025) [80] | SWIN ViT | Captures global information, not high computational complexity/cost (for a ViT) | Global-local attention could be further enhanced | A pure ViT-based model that uses a SWIN transformer variant. It integrates depthwise convolutions into the transformer blocks to enhance global-local feature representations and uses a multi-level patch embedding. |
| Ma et al. (2023) [81] | SWIN ViT | Addresses blurred or uneven tissue regions, late fusion aids handling high-level features | Computational complexity/cost (see Section 4.3) | Introduces AMSeg, a SWIN transformer variant that exploits multiscale anatomical features for late-stage fusion. It uses an adversarial training scheme with separate segmentation and discrimination blocks to handle blurred tissue regions. |
| Bi et al. (2023) [82] | U-Net, ViT | Captures both fine-grained and contextual information, integrates low and high frequency | Computational complexity/cost (see Section 4.3) | A U-Net/ViT hybrid architecture named BPAT. It introduces a boundary point supervision module (BPSM) and an assembled transformer module (ATM) to explicitly enhance and constrain nodule boundaries. |
| Zheng et al. (2025) [83] | U-Net inspired, SWIN ViT | Addresses indistinct boundaries, captures both fine-grained and contextual information | Computational complexity/cost (see Section 4.3) | Introduces GWUNet, a U-shaped network combining a SWIN transformer with gated attention and a wavelet transform module. The latter is designed to aid in distinguishing indistinct nodule boundaries. |
| Li et al. (2023) [84] | CNN, ViT | Captures both fine-grained and contextual information, integrates low and high frequency | Computational complexity/cost (see Section 4.3), small-sized dataset with bias towards clear, well-defined boundaries (see Section 4.4) | A ViT/CNN hybrid network (TCNet) with two branches: a large kernel CNN branch extracts shape information, whereas an enhanced ViT branch models long-range dependencies, with both branches fused by a multiscale fusion module (MFM). |
| Ozcan et al. (2024) [85] | U-Net, ViT | Captures both fine-grained and contextual information | Computational complexity/cost (see Section 4.3) | Proposes Enhanced-TransUNet, which combines a ViT with U-Net. It adds an information bottleneck layer to the architecture, aiming to condense features and reduce overfitting. |
| Li et al. (2023) [86] | CNN (DLA-34), level-set | Reduces the annotation burden, requires only polygon contours, level-set considers fine-grained boundary information | Computational complexity/cost (see Section 4.3) | A weakly supervised deep active contour model. It uses a deep layer aggregation (DLA) network to deform an initial polygon contour by regressing vertex offsets, guided by a level-set-based loss function. |
| Sun et al. (2025) [87] | CLIP, ViT | Captures coarse-grained semantic features and fine-grained spatial details | Domain shift, since CLIP is trained on natural images (see Section 4.4) | Proposes CLIP-TNseg, which integrates a large, pre-trained CLIP model for coarse semantic features and a U-Net-style branch for fine-grained spatial features. |
| Lu et al. (2022) [88] | GAN, ResNet | Nodule localization via CAM | Limited GAN-guided deformations used for training | A GAN-based method guided by online class activation mapping (CAM). It uses ResNet for feature extraction and a deformable convolution module to capture discriminative features of nodular regions. |
| Kunapinun et al. (2023) [89] | GAN, DeepLab v3+, ResNet18 | Captures fine-grained information and maintains high-level consistency (via GAN), addresses GAN instability via PID | Computational complexity/cost (see Section 4.3) | Introduces StableSeg GAN, which combines supervised segmentation with unsupervised learning. It employs DeepLab v3+ as the generator, ResNet18 [90] as the discriminator, and a proportional-integral-derivative (PID) controller to stabilize GAN training and avoid mode collapse. |
| Wan et al. (2023) [91] | Aggregation network | Captures perfusion dynamics via CEUS, models multiscale temporal features | Lacks flexibility to capture salient perfusion differences | Introduces DpRAN for dynamic contrast-enhanced US (CEUS). It focuses on modeling perfusion dynamics by introducing a perfusion excitation (PE) gate and a cross-attention temporal aggregation (CTA) module. |
| Zhang et al. (2024) [92] | AKE, SSFA | Integrates domain knowledge, interpretability | Depends on the availability of pairs of radiologists’ reports and US images (see Section 4.4) | A hybrid method that exploits textual content from clinical reports. It uses an adversarial keyword extraction (AKE) module and a semantic-spatial feature aggregation (SSFA) module to integrate report information. |
3. Computational Assessment of Thyroid Nodules
4. Discussion
4.1. Methodological and Technical Challenges
4.2. Explainability, Interpretability, and Clinician Trust
- 1.
- Saliency/Heatmap Methods (achieving explainability)Principle: These post-hoc methods provide a spatial explanation via a heatmap.TIRADS Alignment: Indirect & inferred. The model itself remains a black box. The clinician must correlate the heatmap’s focus with suspicious TIRADS descriptors.Clinical Utility: Validate the model’s spatial plausibility by confirming it is focusing on relevant regions.
- 2.
- Concept-based & Prototypical Methods (achieving interpretability)Principle: These methods quantify predefined concepts to drive predictions, providing a semantic explanation.TIRADS Alignment: Direct & intrinsic. Concepts are explicitly mapped to TIRADS features. The output is a quantitative report, which directly mirrors the radiologist’s checklist.Clinical Utility: Enable a semantic audit of the model’s internal logic. This strengthens clinician trust.
- Example 1—Clinical Use of an Explainable (Heatmap-based) System
- Example 2—Clinical Use of an Interpretable (Concept-based) System
4.3. Computational Cost and Deployment Efficiency
4.4. Datasets, Generalization, and Annotation Challenges
4.5. Three-Dimensional Imaging, Doppler, Federated Learning, and Future Directions
- 1.
- Traditional ML (e.g., SVMs with handcrafted features):When to use: Best suited for smaller, well-curated datasets, where strong, interpretable features are already known or can be easily engineered (e.g., analyzing boundary irregularity as in [99]).Practical cost: Low computational requirements (no GPU needed for training), but high human effort is required for feature design and validation.Clinical constraint: Ideal for initial exploratory studies, resource-limited settings, or when a "white-box" model is required for regulatory or clinical trust reasons. Highly interpretable.
- 2.
- CNNs (e.g., U-Net, ResNet):When to use: The current workhorses for both segmentation and classification when a moderate-to-large annotated dataset (hundreds to thousands of images) is available. They excel at automatically learning hierarchical spatial features from images.Practical cost: Moderate-to-high computational cost (GPU is typically required for efficient training). Less interpretable than traditional ML.Clinical constraint: The standard choice for developing robust, high-performance CAD systems for deployment in clinical settings, provided sufficient data and hardware are available
- 3.
- ViTs (e.g., TransUNet, SWIN):When to use: Best for very large-scale datasets (many thousands of images), where capturing complex, long-range spatial dependencies across the entire image is critical.Practical cost: Very high computational and data cost. They are “data-hungry” and can overfit on smaller datasets unless sophisticated pretraining strategies are used.Clinical constraint: Currently best suited for well-funded academic research centers aiming for state-of-the-art performance and working with massive, multi-institutional datasets.
- 4.
- Hybrid methods (e.g., DL-extracted features + SVM classifier):When to use: A practical compromise when data is limited but you want to leverage the power of deep feature extraction without training a full end-to-end model.Practical cost: A balance between the two paradigms. Training is often faster and requires less data than a full end-to-end DL model.Clinical constraint: A pragmatic approach for many research groups or smaller clinical centers that have access to limited datasets but want to move beyond purely traditional methods.
5. Conclusions
5.1. Key Takeaways
- Paradigm shift: The field has decisively transitioned from traditional image analysis and ML methods—such as active contours, shape-based approaches, and handcrafted feature-based classification—to end-to-end DL models. While traditional methods remain interpretable and often effective in constrained settings, they are frequently limited by their dependency on manual input and handcrafted features, as well as by their sensitivity to parameter settings and initialization. In contrast, DL-based methods, with CNNs being particularly prominent, have demonstrated superior performance in both segmentation and classification tasks, largely due to their ability to learn hierarchical representations from raw US data, though this increased performance comes at the cost of generalization, interpretability, and data requirements. ViTs have recently emerged as a compelling alternative to CNNs by capturing long-range dependencies through self-attention mechanisms. Although still in early stages of exploration for thyroid US imaging, ViTs show potential to model complex spatial relationships, particularly in challenging scenarios involving heterogeneous nodule appearance.
- Hybrid methods as a pragmatic solution: A significant and growing line of research focuses on hybrid methods that combine the strengths of traditional ML and DL methods for enhanced performance and interpretability. One class of hybrid methods employs DL architectures for automatic feature extraction, followed by classical ML classifiers, such as SVMs or shallow feed-forward neural networks for the final classification. These combinations often improve robustness and reduce overfitting, especially on small datasets. Another class of hybrid methods integrates handcrafted features into DL pipelines, enriching the model with image-specific descriptors that are invariant to scale and rotation. A third class of hybrid methods incorporates expert-defined heuristics or rule-based logic to guide the learning process, for example, by embedding thyroid-specific knowledge or TIRADS-based thresholds, thereby enhancing both performance and interpretability.
- Data-centric bottleneck: While novel architectures continue to emerge, the most significant barrier to progress is no longer algorithmic innovation but the data ecosystem. The field’s primary bottleneck has shifted towards solving the challenges of data scarcity, quality, annotation efficiency, and cross-institutional generalization. Future breakthroughs are likely to come from better data strategies than from minor architectural tweaks.
- The definition of “state-of-the-art” is expanding beyond accuracy: A model’s success is no longer judged solely on its segmentation or classification accuracy on a benchmark dataset. The definition of state-of-the-art is expanding to include clinical utility and trustworthiness. Future CAD methods will be increasingly evaluated on their interpretability, their explicit alignment with clinical frameworks like TIRADS, and their ability to be safely operationalized within a human-in-the-loop workflow.
5.2. Key Challenges
- Dependency on human intervention (mostly in traditional methods): active contour and shape-based methods often require manual initialization and parameter tuning, complicating full automation and clinical deployment.
- Data diversity and availability: existing datasets are often small, lack diversity, and often miss clinically relevant classes [134].
- Generalization across institutions: variability in US equipment, acquisition protocols, and patient populations can lead to significant domain shifts. Solutions such as domain adaptation, transfer learning, and FL are promising.
- Small nodule detection: DL models often struggle to detect small nodules embedded in complex anatomical backgrounds, where echogenicity may resemble that of surrounding tissues.
- Explainability and trust: DL models are often perceived as “black-boxes”. Post hoc explainability tools like Grad-CAM and SHAP provide partial insight but need better alignment with clinical reasoning.
- 3D and Doppler imaging: the shift to 3D US and Doppler modalities offers more nuanced visual cues but also incre
- ases model complexity and data scarcity [14];
- Limitations in labeled data availability: annotating US images is time-consuming and costly. Semi-supervised, weakly supervised, and AL methods offer ways to reduce annotation burden.
- Privacy and collaboration: FL enables model training across institutions without sharing patient data but introduces technical challenges related to anonymization and communication overhead.
5.3. Clinical Integration
- Before adopting a CAD method, clinicians and departments should respond to the following questions on the depth, robustness, and safety of TIRADS alignment:
- 1.
- What is the depth of the TIRADS alignment?Risk-stratification alignment: Does the model only provide a final, high-level risk score (e.g., “TR4” or “high suspicion”)?Feature-level alignment: Does it provide granular, feature-level scores for each of the five TIRADS categories (composition, echogenicity, shape, margin, and echogenic foci)? This type of alignment is more transparent and clinically useful.
- 2.
- What is the validated performance granularity?Feature-Specific Accuracy: Has the system’s performance been validated for each individual TIRADS feature? A common failure mode is high overall accuracy that masks a critical weakness in detecting a specific high-risk feature (e.g., the model may excel at “composition” but fail on “echogenic foci”). Request a feature-by-feature performance breakdown.Handling of Ambiguity: How is the model’s performance on ambiguous or borderline cases quantified? Inquire about its accuracy in distinguishing between clinically similar but distinct categories, such as an "ill-defined" margin (lower risk) versus an "irregular" margin (higher risk).
- 3.
- How was the model validated against real-world variability?Was the model’s alignment validated on a diverse, multi-institutional dataset that includes different US machine vendors and patient populations? A model that performs well on clean, single-center data may fail when faced with noisy images acquired from diverse equipment. How does it perform on rare but clinically important nodule subtypes?
- 4.
- What was the quality and scale of the annotation data used for training?Was the data annotated by a single expert or by a consensus of multiple experienced radiologists? High-quality, consensus-based annotations are critical for building a robust model. How many examples of each specific feature (especially high-risk ones) were used in training?
- 5.
- Does the system explain its feature-level conclusions?If the model assigns a high-risk score for "margin," does it provide a heatmap or other type of representation to localize the suspicious feature along the nodule boundary?
- 1.
- Establishment of a shared responsibility policyBefore deployment, a formal policy should be established to clearly delineate liability. This policy should define the responsibilities of the AI vendor, the clinician (as the ultimate authority for the final diagnosis), and the healthcare institution (for providing adequate training and quality control).
- 2.
- Implementation of a mandatory human oversight protocolThe “assist, not replace” paradigm should be reinforced by formalizing the clinician-in-the-loop workflow. The protocol should mandate that any AI-generated finding, score, or segmentation be reviewed, verified, and explicitly accepted or rejected by a qualified radiologist before being entered into the patient’s official report.
- 3.
- Development of a continuous training and feedback programTo address clinician concerns and build trust, a transparent training program should be developed. This program should extend beyond basic use to include education on the limitations and common failure modes of AI. A clear feedback channel should also be established for clinicians to report and document cases of incorrect or ambiguous AI outputs.
- 4.
- Institution of regular quality control auditsThe performance of the CAD system should not be taken solely on the vendor’s claims. Periodic audits should be instituted to evaluate the system’s performance on a curated set of local cases with known pathological outcomes. This ensures the model remains robust on the department’s specific patient population and US equipment.
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burman, K.D.; Wartofsky, L. Thyroid Nodules. N. Engl. J. Med. 2015, 373, 2347–2356. [Google Scholar] [CrossRef]
- Cao, C.L.; Li, Q.L.; Tong, J.; Shi, L.N.; Li, W.X.; Xu, Y.; Cheng, J.; Du, T.T.; Li, J.; Cui, X.W. Artificial intelligence in thyroid ultrasound. Front. Oncol. 2023, 13, 1060702. [Google Scholar] [CrossRef]
- Wu, X.; Tan, G.; Luo, H.; Chen, Z.; Pu, B.; Li, S.; Li, K. A knowledge-interpretable multi-task learning framework for automated thyroid nodule diagnosis in ultrasound videos. Med. Image Anal. 2024, 91, 103039. [Google Scholar] [CrossRef]
- Shahroudnejad, A.; Vega, R.; Forouzandeh, A.; Balachandran, S.; Jaremko, J.; Noga, M.; Hareendranathan, A.R.; Kapur, J.; Punithakumar, K. Thyroid nodule segmentation and classification using deep convolutional neural network and rule-based classifiers. In Proceedings of the International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Online, 1–5 November 2021; pp. 3118–3121. [Google Scholar]
- Kang, S.; Lee, E.; Chung, C.W.; Jang, H.N.; Moon, J.H.; Shin, Y.; Kim, K.; Li, Y.; Shin, S.M.; Kim, Y.H.; et al. A beneficial role of computer-aided diagnosis system for less experienced physicians in the diagnosis of thyroid nodule on ultrasound. Sci. Rep. 2021, 11, 20448. [Google Scholar]
- Fresilli, D.; Grani, G.; De Pascali, M.L.; Alagna, G.; Tassone, E.; Ramundo, V.; Ascoli, V.; Bosco, V.; Biffoni, M.; Bononi, M.; et al. Computer-aided diagnostic system for thyroid nodule sonographic evaluation outperforms the specificity of less experienced examiners. J. Ultrasound 2020, 23, 169–174. [Google Scholar] [CrossRef]
- Maroulis, D.E.; Savelonas, M.A.; Iakovidis, D.K.; Karkanis, S.A.; Dimitropoulos, N. Variable background active contour model for computer-aided delineation of nodules in thyroid ultrasound images. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 537–543. [Google Scholar] [CrossRef]
- Li, L.R.; Du, B.; Liu, H.Q.; Chen, C. Artificial intelligence for personalized medicine in thyroid cancer: Current status and future perspectives. Front. Oncol. 2021, 10, 604051. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Kim, Y.J.; Choi, Y.; Hur, S.J.; Park, K.S.; Kim, H.J.; Seo, M.; Lee, M.K.; Jung, S.L.; Jung, C.K. Deep convolutional neural network for classification of thyroid nodules on ultrasound: Comparison of the diagnostic performance with that of radiologists. Eur. J. Radiol. 2022, 152, 110335. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Lee, E.; Han, K.; Kim, E.K.; Son, E.J.; Sohn, Y.M.; Seo, M.; Kwon, M.R.; Yoon, J.H.; Lee, J.H.; et al. Diagnosis of thyroid nodules on ultrasonography by a deep convolutional neural network. Sci. Rep. 2020, 10, 15245. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; You, H.; Li, K. A review of thyroid gland segmentation and thyroid nodule segmentation methods for medical ultrasound images. Comput. Methods Programs Biomed. 2020, 185, 105329. [Google Scholar] [CrossRef] [PubMed]
- Chambara, N.; Ying, M. The diagnostic efficiency of ultrasound computer–aided diagnosis in differentiating thyroid nodules: A systematic review and narrative synthesis. Cancers 2019, 11, 1759. [Google Scholar] [CrossRef] [PubMed]
- Anand, V.; Koundal, D. Computer-Assisted Diagnosis of Thyroid Cancer Using Medical Images: A Survey. In Recent Innovations in Computing; Singh, P.K., Kar, A.K., Singh, Y., Kolekar, M.H., Tanwar, S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 597, pp. 543–559. [Google Scholar]
- Gulame, M.B.; Dixit, V.V.; Suresh, M. Thyroid nodules segmentation methods in clinical ultrasound images: A review. Mater. Today Proc. 2021, 45, 2270–2276. [Google Scholar]
- Sorrenti, S.; Dolcetti, V.; Radzina, M.; Bellini, M.I.; Frezza, F.; Munir, K.; Grani, G.; Durante, C.; D’Andrea, V.; David, E.; et al. Artificial intelligence for thyroid nodule characterization: Where are we standing? Cancers 2022, 14, 3357. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Ludwig, B.; Mikuła, A.; Biernat, S.; Rudnicki, J.; Kaliszewski, K. The use of artificial intelligence in the diagnosis and classification of thyroid nodules: An update. Cancers 2023, 15, 708. [Google Scholar] [CrossRef]
- Yadav, N.; Dass, R.; Virmani, J. A systematic review of machine learning based thyroid tumor characterisation using ultrasonographic images. J. Ultrasound 2024, 27, 209–224. [Google Scholar] [CrossRef]
- Mylona, E.A.; Savelonas, M.A.; Maroulis, D. Automated adjustment of region-based active contour parameters using local image geometry. IEEE Trans. Cybern. 2014, 44, 2757–2770. [Google Scholar] [CrossRef]
- Mylona, E.A.; Savelonas, M.A.; Maroulis, D. Self-parameterized active contours based on regional edge structure for medical image segmentation. SpringerPlus 2014, 3, 424. [Google Scholar] [CrossRef]
- Nugroho, H.A.; Nugroho, A.; Choridah, L. Thyroid nodule segmentation using active contour bilateral filtering on ultrasound images. In Proceedings of the ΙΕΕΕ International Conference on Quality in Research (QiR), Lombok, Indonesia, 10–13 August 2015; pp. 43–46. [Google Scholar]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef]
- Savelonas, M.A.; Iakovidis, D.K.; Legakis, I.; Maroulis, D. Active Contours guided by Echogenicity and Texture for Delineation of Thyroid Nodules in Ultrasound Images. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 519–527. [Google Scholar] [CrossRef]
- Du, W.; Sang, N. An effective method for ultrasound thyroid nodules segmentation. In Proceedings of the International Symposium on Bioelectronics and Bioinformatics (ISBB), Beijing, China, 14–17 October 2015; pp. 207–210. [Google Scholar]
- Koundal, D.; Gupta, S.; Singh, S. Automated delineation of thyroid nodules in ultrasound images using spatial neutrosophic clustering and level set. Appl. Soft Comput. 2016, 40, 86–97. [Google Scholar] [CrossRef]
- Koundal, D.; Gupta, S.; Singh, S. Computer aided thyroid nodule detection system using medical ultrasound images. Biomed. Signal Process. Control 2018, 40, 117–130. [Google Scholar]
- Li, C.; Xu, C.; Gui, C.; Fox, M.D. Distance regularized level set evolution and its application to image segmentation. IEEE Trans. Image Process. 2010, 19, 3243–3254. [Google Scholar] [CrossRef] [PubMed]
- Koundal, D.; Sharma, B.; Guo, Y. Intuitionistic based segmentation of thyroid nodules in ultrasound images. Comput. Biol. Med. 2020, 121, 103776. [Google Scholar] [CrossRef] [PubMed]
- Le, Y.; Xu, X.; Zha, L.; Zhao, W.; Zhu, Y. Tumour localisation in ultrasound-guided high-intensity focused ultrasound ablation using improved gradient and direction vector flow. IET Image Process. 2015, 9, 857–865. [Google Scholar]
- Cheng, J.; Foo, S.W. Dynamic directional gradient vector flow for snakes. IEEE Trans. Image Process. 2006, 15, 1563–1571. [Google Scholar] [PubMed]
- Tsantis, S.; Dimitropoulos, N.; Cavouras, D.; Nikiforidis, G. A hybrid multi-scale model for thyroid nodule boundary detection on ultrasound images. Comput. Methods Programs Biomed. 2006, 84, 86–98. [Google Scholar] [CrossRef]
- Chiu, L.Y.; Chen, A. A variance-reduction method for thyroid nodule boundary detection on ultrasound images. In Proceedings of the IEEE International Conference on Automation Science and Engineering (CASE), Taipei, Taiwan, 18–22 August 2014; pp. 681–685. [Google Scholar]
- Madabhushi, A.; Metaxas, D.N. Combining low-, high-level and empirical domain knowledge for automated segmentation of ultrasonic breast lesions. IEEE Trans. Med. Imaging 2003, 22, 155–169. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. Med. Image Comput. Comput. Assist. Interv. 2015, 9351, 234–241. [Google Scholar]
- Ma, J.; Wu, F.; Jiang, T.; Zhao, Q.; Kong, D. Ultrasound image-based thyroid nodule automatic segmentation using convolutional neural networks. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1895–1910. [Google Scholar] [CrossRef]
- Wang, S.; Li, Z.; Liao, L.; Zhang, C.; Zhao, J.; Sang, L.; Qian, W.; Pan, G.; Huang, L.; Ma, H. DPAM-PSPNet: Ultrasonic image segmentation of thyroid nodule based on dual-path attention mechanism. Phys. Med. Biol. 2023, 68, 165002. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Zhou, S.; Wu, H.; Gong, J.; Le, T.; Wu, H.; Chen, Q.; Xu, Z. Mark-guided segmentation of ultrasonic thyroid nodules using deep learning. In Proceedings of the ACM International Symposium on Image Computing and Digital Medicine, Chengdu, China, 13–14 October 2018; pp. 21–26. [Google Scholar]
- Nandamuri, S.; China, D.; Mitra, P.; Sheet, D. SUMNet: Fully convolutional model for fast segmentation of anatomical structures in ultrasound volumes. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019; pp. 1729–1732. [Google Scholar]
- Abdolali, F.; Kapur, J.; Jaremko, J.L.; Noga, M.; Hareendranathan, A.R.; Punithakumar, K. Automated thyroid nodule detection from ultrasound imaging using deep convolutional neural networks. Comput. Biol. Med. 2020, 122, 103871. [Google Scholar] [CrossRef]
- Kumar, V.; Webb, J.; Gregory, A.; Meixner, D.D.; Knudsen, J.M.; Callstrom, M.; Fatemi, M.; Alizad, A. Automated segmentation of thyroid nodule, gland, and cystic components from ultrasound images using deep learning. IEEE Access 2020, 8, 63482–63496. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. very deep convolutional networks for large-scale image recognition. Int. J. Comput. Assist. Radiol. Surg. 2015, 9351, 234–241. [Google Scholar]
- Wu, Y.; Shen, X.; Bu, F.; Tian, J. Ultrasound image segmentation method for thyroid nodules using ASPP fusion features. IEEE Access 2020, 8, 172457–172466. [Google Scholar] [CrossRef]
- Webb, J.M.; Meixner, D.D.; Adusei, S.A.; Polley, E.C.; Fatemi, M.; Alizad, A. Automatic deep learning semantic segmentation of ultrasound thyroid cineclips using recurrent fully convolutional networks. IEEE Access 2021, 9, 5119–5127. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Nugroho, H.A.; Frannita, E.L.; Nurfauzi, R. An automated detection and segmentation of thyroid nodules using Res-UNet. In Proceedings of the IEEE International Conference on Electrical Engineering, Computer Science and Informatics (EECSI), Semarang, Indonesia, 20–21 October 2021; pp. 181–185. [Google Scholar]
- Xiangyu, D.; Huan, Z.; Yahan, Y. Ultrasonic image segmentation algorithm of thyroid nodules based on DPCNN. In Lecture Notes in Electrical Engineering, Proceedings of the International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD), Online, 25–26 March 2021; Su, R., Zhang, Y.D., Liu, H., Eds.; Springer: Singapore, 2021; Volume 784, pp. 163–174. [Google Scholar]
- Nguyen, D.T.; Choi, J.; Park, K.R. Thyroid nodule segmentation in ultrasound image based on information fusion of suggestion and enhancement networks. Mathematics 2022, 10, 3484. [Google Scholar] [CrossRef]
- Yang, Q.; Geng, C.; Chen, R.; Pang, C.; Han, R.; Lyu, L.; Zhang, Y. DMU-Net: Dual-route mirroring U-Net with mutual learning for malignant thyroid nodule segmentation. Biomed. Signal Process. Control 2022, 77, 103805. [Google Scholar] [CrossRef]
- Song, R.; Zhu, C.; Zhang, L.; Zhang, T.; Luo, Y.; Liu, J.; Yang, J. Dual-branch network via pseudo-label training for thyroid nodule detection in ultrasound image. Appl. Intell. 2022, 52, 11738–11754. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, W.; Hu, W. MTN-Net: A multi-task network for detection and segmentation of thyroid nodules in ultrasound images. In Knowledge Science, Engineering and Management KSEM 2022 Lecture Notes in Computer Science; Memmi, G., Yang, B., Kong, L., Zhang, T., Qiu, M., Eds.; Springer: Cham, Switzerland, 2022; Volume 13370. [Google Scholar]
- Gan, J.; Zhang, R. Ultrasound image segmentation algorithm of thyroid nodules based on improved U-Net network. In Proceedings of the ACM International Conference on Control, Robotics and Intelligent System, Online, 26–28 August 2022; pp. 61–66. [Google Scholar]
- Jin, Z.; Li, X.; Zhang, Y.; Shen, L.; Lai, Z.; Kong, H. Boundary regression-based deep neural network for thyroid nodule segmentation in ultrasound images. Neural Comput. Appl. 2022, 34, 22357–22366. [Google Scholar] [CrossRef]
- Shao, J.; Pan, T.; Fan, L.; Li, Z.; Yang, J.; Zhang, S.; Zhang, J.; Chen, D.; Zhu, X.; Chen, H.; et al. FCG-Net: An innovative full-scale connected network for thyroid nodule segmentation in ultrasound images. Biomed. Signal Process. Control 2023, 86, 105048. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Chen, Y.; Wu, J. UNet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Online, 4–8 May 2020. [Google Scholar]
- Han, K.; Wang, Y.; Tian, Q.; Guo, J.; Xu, C.; Xu, C. GhostNet: More features from cheap operations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020; pp. 1580–1589. [Google Scholar]
- Sifre, L.; Mallat, S. Rigid-motion scattering for texture classification. arXiv 2014, arXiv:1403.1687. [Google Scholar] [CrossRef]
- Dai, H.; Xie, W.; Xia, E. SK-Unet++: An improved Unet++ network with adaptive receptive fields for automatic segmentation of ultrasound thyroid nodule images. Med. Phys. 2024, 51, 1798–1811. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A nested U-Net architecture for medical image segmentation. In Proceedings of the International Workshop in Deep Learning in Medical Image Analysis (DLMIA) and Multimodal Learning for Clinical Decision (ML-CDS), Held in Conjunction with Medical Image Computing and Computer Assisted Intervention (MICCAI), Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Zheng, T.; Qin, H.; Cui, Y.; Wang, R.; Zhao, W.; Zhang, S.; Geng, S.; Zhao, L. Segmentation of thyroid glands and nodules in ultrasound images using the improved U-Net architecture. BMC Med. Imaging 2023, 23, 56. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Zhang, Z.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; et al. ResNeSt: Split-attention networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 19–20 June 2022; pp. 2735–2745. [Google Scholar]
- Wang, W.; Dai, J.; Chen, Z.; Huang, Z.; Li, Z.; Zhu, X.; Hu, X.; Lu, T.; Lu, L.; Li, H.; et al. InternImage: Exploring large-scale vision foundation models with deformable convolutions. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 17–24 June 2023; pp. 2735–2745e. [Google Scholar]
- Wang, R.; Zhou, H.; Fu, P.; Shen, H.; Bai, Y. A multiscale attentional Unet model for automatic segmentation in medical ultrasound images. Ultrason. Imaging 2023, 45, 159–174. [Google Scholar] [CrossRef]
- Chen, H.; Yu, M.; Chen, C.; Zhou, K.; Qi, S.; Chen, Y.; Xiao, R. FDE-net: Frequency-domain enhancement network using dynamic-scale dilated convolution for thyroid nodule segmentation. Comput. Biol. Med. 2023, 153, 106514. [Google Scholar] [CrossRef]
- Ma, W.; Li, X.; Zou, L.; Fan, C.; Wu, M. Symmetrical awareness network for cross-site ultrasound thyroid nodule segmentation. Front. Public Health 2023, 11, 1055815. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Bouhdiba, K.; Meddeber, L.; Meddeber, M.; Zouagui, T. A hybrid deep neural network architecture RefineUnet for ultrasound thyroid nodules segmentation. In Proceedings of the International Conference on Advanced Electrical Engineering (ICAEE), Sidi-Bel-Abbes, Algeria, 5–7 November 2024; pp. 1–6. [Google Scholar]
- Ma, J.; Kong, D.; Wu, F.; Bao, L.; Yuan, J.; Liu, Y. Densely connected convolutional networks for ultrasound image based lesion segmentation. Comput. Biol. Med. 2024, 168, 107725. [Google Scholar] [CrossRef]
- Liu, J.; Mu, J.; Sun, H.; Dai, C.; Ji, Z.; Ganchev, I. BFG&MSF-Net: Boundary feature guidance and multi-scale fusion network for thyroid nodule segmentation. IEEE Access 2024, 12, 78701–78713. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Xing, G.; Wang, S.; Gao, J.; Li, X. Real-time reliable semantic segmentation of thyroid nodules in ultrasound images. Phys. Med. Biol. 2024, 69, 025016. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, H.; Shao, Y.; Chen, B. DAC-Net: A light-weight U-shaped network based efficient convolution and attention for thyroid nodule segmentation. Comput. Biol. Med. 2024, 180, 108972. [Google Scholar] [CrossRef]
- Xiao, N.; Kong, D.; Wang, J. Ultrasound thyroid nodule segmentation algorithm based on DeepLabV3+ with EfficientNet. J. Digit. Imaging 2025. [Google Scholar] [CrossRef]
- Ongole, D.; Saravanan, S. Advanced thyroid nodule detection using ultrasonography image analysis and bilateral mean clustering strategies. Comput. Biol. Med. 2025, 186, 109647. [Google Scholar] [CrossRef]
- Xiang, Z.; Tian, X.; Liu, Y.; Chen, M.; Zhao, C.; Tang, L.N.; Xue, E.S.; Zhou, Q.; Shen, B.; Li, F.; et al. Federated learning via multi-attention guided UNet for thyroid nodule segmentation of ultrasound images. Neural Netw. 2025, 181, 106754. [Google Scholar] [CrossRef]
- Yadav, N.; Dass, R.; Virmani, J. Objective assessment of segmentation models for thyroid ultrasound images. J. Ultrasound 2022, 26, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Wang, M.; Xie, J. CIL-Net: Densely connected context information learning network for boosting thyroid nodule segmentation using ultrasound images. Con. Comput. 2024, 16, 1176–1197. [Google Scholar] [CrossRef]
- Sun, S.; Fu, C.; Xu, S.; Wen, Y.; Ma, T. CRSANet: Class representations self-attention network for the segmentation of thyroid nodules. Biomed. Signal Process. Control 2024, 91, 105917. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, L.; Yang, T. Thyroid nodule ultrasound image segmentation based on improved SWIN transformer. IEEE Access 2025, 13, 19788–19795. [Google Scholar] [CrossRef]
- Ma, X.; Sun, B.; Liu, W.; Sui, D.; Chen, J.; Tian, Z. AMSeg: A novel adversarial architecture based multi-scale fusion framework for thyroid nodule segmentation. IEEE Access 2023, 11, 72911–72924. [Google Scholar] [CrossRef]
- Bi, H.; Cai, C.; Sun, J.; Jiang, Y.; Lu, G.; Shu, H.; Ni, X. BPAT-UNet: Boundary preserving assembled transformer UNet for ultrasound thyroid nodule segmentation. Comput. Methods Programs Biomed. 2023, 238, 107614. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Yu, S.; Wang, Y.; Wen, J. GWUNet: A UNet with gated attention and improved wavelet transform for thyroid nodules segmentation. In MultiMedia Modeling MMM 2025 Lecture Notes in Computer Science; Ide, I., Kompatsiaris, I., Xu, C., Yanai, K., Chu, W.T., Nitta, N., Riegler, M., Yamasaki, T., Eds.; Springer: Singapore, 2025; Volume 15521. [Google Scholar]
- Li, G.; Chen, R.; Zhang, J.; Liu, K.; Geng, C.; Lyu, L. Fusing enhanced transformer and large kernel CNN for malignant thyroid nodule segmentation. Biomed. Signal Process. Control 2023, 83, 104636. [Google Scholar] [CrossRef]
- Ozcan, A.; Tosun, Ö.; Donmez, E.; Sanwal, M. Enhanced-TransUNet for ultrasound segmentation of thyroid nodules. Biomed. Signal Process. Control 2024, 95, 106472. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, S.; Chang, C.; Wang, Y.; Guo, Y. A weakly supervised deep active contour model for nodule segmentation in thyroid ultrasound images. Pattern Recognit. Lett. 2023, 165, 128–137. [Google Scholar]
- Sun, X.; Wei, B.; Jiang, Y.; Mao, L.; Zhao, Q. CLIP-TNseg: A multi-modal hybrid framework for thyroid nodule segmentation in ultrasound images. IEEE Signal Process. Lett. 2025, 32, 1625–1629. [Google Scholar]
- Lu, J.; Ouyang, X.; Shen, X.; Liu, T.; Cui, Z.; Wang, Q.; Shen, D. GAN-guided deformable attention network for identifying thyroid nodules in ultrasound images. IEEE J. Biomed. Health Inform. 2022, 26, 1582–1590. [Google Scholar]
- Kunapinun, A.; Dailey, M.N.; Songsaeng, D.; Parnichkun, M.; Keatmanee, C.; Ekpanyapong, M. Improving GAN learning dynamics for thyroid nodule segmentation. Ultrasound Med. Biol. 2023, 49, 416–430. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Wan, P.; Xue, H.; Liu, C.; Chen, F.; Kong, W.; Zhang, D. Dynamic perfusion representation and aggregation network for nodule segmentation using contrast-enhanced US. IEEE J. Biomed. Health Inform. 2023, 27, 3431–3442. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Li, X.; Shen, L.; Lai, Z.; Kong, H. Adversarial keyword extraction and semantic-spatial feature aggregation for clinical report guided thyroid nodule segmentation. In Pattern Recognition and Computer Vision (PRCV), Lecture Notes in Computer Science; Liu, Q., Wang, H., Ma, Z., Zheng, W., Zha, H., Chen, X., Wang, L., Ji, R., Eds.; Springer: Singapore, 2024; Volume 14437. [Google Scholar]
- Chambara, N.; Liu, S.Y.W.; Lo, X.; Ying, M. Diagnostic performance evaluation of different TI-RADS using ultrasound computer-aided diagnosis of thyroid nodules: An experience with adjusted settings. PLoS ONE 2021, 16, e0245617. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Ha, E.J.; Park, J.H. Computer-aided diagnostic system for thyroid nodules on ultrasonography: Diagnostic performance based on the thyroid imaging reporting and data system classification and dichotomous outcomes. AJNR Am. J. Neuroradiol. 2021, 42, 559–565. [Google Scholar]
- Xie, F.; Luo, Y.K.; Lan, Y.; Tian, X.Q.; Zhu, Y.Q.; Jin, Z.; Zhang, Y.; Zhang, M.B.; Song, Q.; Zhang, Y. Differential diagnosis and feature visualization for thyroid nodules using computer-aided ultrasonic diagnosis system: Initial clinical assessment. BMC Med. Imaging 2022, 22, 153. [Google Scholar] [CrossRef]
- Gomes Ataide, E.J.; Jabaraj, M.S.; Schenke, S.; Petersen, M.; Haghghi, S.; Wuestemann, J.; Illanes, A.; Friebe, M.; Kreissl, M.C. Thyroid nodule detection and region estimation in ultrasound images: A comparison between physicians and an automated decision support system approach. Diagnostics 2023, 13, 2873. [Google Scholar] [CrossRef]
- Wang, Y.; Yue, W.; Li, X.; Liu, S.; Guo, L.; Xu, H.; Zhang, H.; Yang, G. Comparison study of radiomics and deep learning-based methods for thyroid nodules classification using ultrasound images. IEEE Access 2020, 8, 52010–52017. [Google Scholar] [CrossRef]
- Zhao, W.J.; Fu, L.R.; Huang, Z.M.; Zhu, J.Q.; Ma, B.Y. Effectiveness evaluation of computer-aided diagnosis system for the diagnosis of thyroid nodules on ultrasound: A systematic review and meta-analysis. Medicine 2019, 98, e16379. [Google Scholar] [CrossRef]
- Savelonas, M.A.; Iakovidis, D.K.; Dimitropoulos, N.; Maroulis, D. Computational characterization of thyroid tissue in the Radon domain. In Proceedings of the IEEE International Symposium on Computer-Based Medical Systems (CBMS), Maribor, Slovenia, 20–22 June 2007; pp. 189–192. [Google Scholar]
- Seabra, J.C.R.; Fred, A.L.N. A Biometric identification system based on thyroid tissue echo-morphology. In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing, Porto, Portugal, 14–17 January 2009; pp. 186–193. [Google Scholar]
- Savelonas, M.; Maroulis, D.; Sangriotis, M. A Computer-aided system for malignancy risk assessment of nodules in thyroid US images based on boundary features. Comput. Methods Programs Biomed. 2009, 96, 25–32. [Google Scholar] [CrossRef]
- Legakis, I.; Savelonas, M.A.; Maroulis, D.; Iakovidis, D.K. Computer-based nodule malignancy risk assessment in thyroid ultrasound images. Int. J. Comput. Appl. 2011, 33, 29–35. [Google Scholar] [CrossRef]
- Iakovidis, D.K.; Keramidas, E.G.; Maroulis, D. Fusion of fuzzy statistical distributions for classification of thyroid ultrasound patterns. Artif. Intell. Med. 2010, 50, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Katsigiannis, S.; Keramidas, E.; Maroulis, D. A Contourlet transform feature extraction scheme for ultrasound thyroid texture classification. Eng. Intell. Syst. 2010, 18, 171. [Google Scholar]
- Keramidas, E.G.; Maroulis, D.; Iakovidis, D.K. ΤND: A thyroid nodule detection system for analysis of ultrasound images and videos. J. Med. Syst. 2012, 36, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.; Dudhe, A. Texture analysis of thyroid ultrasonography images for diagnosis of benign & malignant nodule using feed forward neural network. Int. J. Comput. Engin. Manag. 2018, 5, 205–211. [Google Scholar]
- Kale, S.; Punwatkar, K. Texture analysis of ultrasound medical images for diagnosis of thyroid nodule using support vector machine. Int. J. Comput. Sc. Mob. Comput. 2013, 2, 71–77. [Google Scholar]
- Vadhiraj, V.V.; Simpkin, A.; O’Connell, J.; Singh Ospina, N.; Maraka, S.; O’Keeffe, D.T. Ultrasound image classification of thyroid nodules using machine learning techniques. Medicina 2021, 57, 527. [Google Scholar] [CrossRef]
- Nanda, S.; Sukumar, M. Thyroid nodule classification using steerable pyramid–based features from ultrasound images. J. Clin. Eng. 2018, 43, 149–158. [Google Scholar] [CrossRef]
- Kononenko, I. Estimating attributes: Analysis and extensions of RELIEF. In Proceedings of the European Conference on Machine Learning (ECML), Catania, Italy, 6–8 April 1994; pp. 171–182. [Google Scholar]
- Acharya, U.R.; Chowriappa, P.; Fujita, H.; Bhat, S.; Dua, S.; Koh, J.E.W.; Eugene, L.W.J.; Kongmebhol, P.; Ng, K.H. Thyroid lesion classification in 242 patient population using Gabor transform features from high resolution ultrasound images. Knowl. -Based Syst. 2016, 107, 235–245. [Google Scholar]
- Raghavendra, U.; Acharya, U.R.; Gudigar, A.; Tan, J.H.; Fujita, H.; Hagiwara, Y.; Molinari, F.; Kongmebhol, P.; Ng, K.H. Fusion of spatial gray level dependency and fractal texture features for the characterization of thyroid lesions. Ultrasonics 2017, 77, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Chen, H.; Li, Q.; Zhou, M.; Chen, L.; Cai, Z.; Fang, Y.; Zhou, H. Ultrasound-based differentiation of malignant and benign thyroid Nodules: An extreme learning machine approach. Comput. Methods Programs Biomed. 2017, 147, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Prochazka, A.; Gulati, S.; Holinka, S.; Smutek, D. Classification of thyroid nodules in ultrasound images using direction-independent features extracted by two-threshold binary decomposition. Technol. Cancer Res. Treat. 2019, 18, 1533033819830748. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, U.; Gudigar, A.; Maithri, M.; Gertych, A.; Meiburger, K.M.; Yeong, C.H.; Madla, C.; Kongmebhol, P.; Molinari, F.; Ng, K.H.; et al. Optimized multi-level elongated quinary patterns for the assessment of thyroid nodules in ultrasound images. Comput. Biol. Med. 2018, 95, 55–62. [Google Scholar] [CrossRef]
- Gomes Ataide, E.J.; Ponugoti, N.; Illanes, A.; Schenke, S.; Kreissl, M.; Friebe, M. Thyroid nodule classification for physician decision support using machine learning-evaluated geometric and morphological features. Sensors 2020, 20, 6110. [Google Scholar] [CrossRef]
- Barzegar-Golmoghani, E.; Mohebi, M.; Gohari, Z.; Aram, S.; Mohammadzadeh, A.; Firouznia, S.; Shakiba, M.; Naghibi, H.; Moradian, S.; Ahmadi, M.; et al. ELTIRADS framework for thyroid nodule classification integrating elastography, TIRADS, and radiomics with interpretable machine learning. Sci. Rep. 2025, 15, 8763. [Google Scholar] [CrossRef]
- Chi, J.; Walia, E.; Babyn, P.; Wang, J.; Groot, G.; Eramian, M. Thyroid nodule classification in ultrasound images by fine-tuning deep convolutional neural network. J. Digit. Imaging 2017, 30, 477–486. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Varhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Liang, X.; Yu, J.; Liao, J.; Chen, Z. Convolutional neural network for breast and thyroid nodules diagnosis in ultrasound imaging. BioMed Res. Int. 2020, 2020, 1763803. [Google Scholar] [CrossRef]
- Zhu, Y.; Fu, Z.; Fei, J. An image augmentation method using convolutional network for thyroid nodule classification by transfer learning. In Proceedings of the IEEE International Conference on Computer and Communications (ICCC), Chengdu, China, 13–16 December 2017. [Google Scholar]
- Song, R.; Zhang, L.; Zhu, C.; Liu, J.; Yang, J.; Zhang, T. Thyroid nodule ultrasound image classification through hybrid feature cropping network. IEEE Access 2020, 8, 64064–64074. [Google Scholar] [CrossRef]
- Zhang, S.; Du, H.; Jin, Z.; Zhu, Y.; Zhang, Y.; Xie, F.; Zhang, M.; Tian, X.; Zhang, J.; Luo, Y. A novel interpretable computer-aided diagnosis system of thyroid nodules on ultrasound based on clinical experience. IEEE Access 2020, 8, 53223–53231. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Wang, L.; Zhou, X.; Nie, X.; Lin, X.; Li, J.; Zheng, H.; Xue, E.; Chen, S.; Chen, C.; Du, M.; et al. A multi-scale densely connected convolutional neural network for automated thyroid nodule classification. Front. Neurosci. 2022, 16, 878718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Q.; Zhao, J.; Yu, H.; Wang, F.; Zhang, J. Automatic ultrasound diagnosis of thyroid nodules: A combination of deep learning and KWAK TI-RADS. Phys. Med. Biol. 2023, 68, 205021. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.W.; Choi, I.J.; Kang, J.Y.; Jang, W.I.; Lee, G.H.; Lee, M.C. Ultrasonographic thyroid nodule classification using a deep convolutional neural network with surgical pathology. J. Digit. Imaging 2020, 33, 1202–1208. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, C.; Wang, Q.; Zhang, H.; Shi, L.; Zhang, Z. A deep learning-based method for detecting and classifying the ultrasound images of suspicious thyroid nodules. Med. Phys. 2021, 48, 7959–7970. [Google Scholar] [CrossRef]
- Han, B.; Zhang, M.; Gao, X.; Wang, Z.; You, F.; Li, H. Automatic classification method of thyroid pathological images using multiple magnification factors. Neurocomputing 2021, 460, 231–242. [Google Scholar] [CrossRef]
- Chatfield, K.; Simonyan, K.; Vedaldi, A.; Zisserman, A. Return of the devil in the details: Delving deep into convolutional nets. arXiv 2014, arXiv:1405.3531. [Google Scholar] [CrossRef]
- Liu, Z.; Zhong, S.; Liu, Q.; Xie, C.; Dai, Y.; Peng, C.; Chen, X.; Zou, R. Thyroid nodule recognition using a joint convolutional neural network with information fusion of ultrasound images and radiofrequency data. Eur. Radiol. 2021, 31, 5001–5011. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, C.; Wu, D.; Zeng, Y.; Zhong, S.; Qiu, W. Automatic segmentation and classification of thyroid nodules in ultrasound images with convolutional neural networks. In Segmentation, Classification, and Registration of Multi-Modality Medical Imaging Data MICCAI 2020 Lecture Notes in Computer Science; Shusharina, N., Heinrich, M.P., Huang, R., Eds.; Springer: Cham, Switzerland, 2021; Volume 12587. [Google Scholar]
- Kang, Q.; Lao, Q.; Li, Y.; Jiang, Z.; Qiu, Y.; Zhang, S.; Li, K. Thyroid nodule segmentation and classification in ultrasound images through intra- and inter-task consistent learning. Med. Image Anal. 2022, 79, 102443. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, R.; Zhou, M.; Fu, P.; Bai, Y. A deep learning-based cascade automatic classification system for malignant thyroid nodule recognition in ultrasound image. J. Phys. Conf. Ser. 2022, 2363, 012029. [Google Scholar] [CrossRef]
- Wang, Y.; Gan, J. Benign and malignant classification of thyroid nodules based on ConvNeXt. In Proceedings of the ACM International Conference on Control, Robotics and Intelligent System, Online, 26–28 August 2022; pp. 56–60. [Google Scholar]
- Yang, J.; Shi, X.; Wang, B.; Qiu, W.; Tian, G.; Wang, X.; Wang, P.; Yang, J. Ultrasound image classification of thyroid nodules based on deep learning. Front. Oncol. 2022, 12, 905955. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2019, 128, 336–359. [Google Scholar] [CrossRef]
- Wang, B.; Yuan, F.; Lv, Z.; He, Y.; Chen, Z.; Hu, J.; Yu, J.; Zheng, S.; Liu, H. Hierarchical deep learning networks for classification of ultrasonic thyroid nodules. J. Imaging Sci. Technol. 2022, 66, 040409. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. arXiv 2017, arXiv:1703.06870. [Google Scholar]
- Xing, G.; Miao, Z.; Zheng, Y.; Zhao, M. A multi-task model for reliable classification of thyroid nodules in ultrasound images. Biomed. Eng. Lett. 2024, 14, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.; Dass, R.; Virmani, J. Deep learning-based CAD system design for thyroid tumor characterization using ultrasound images. Multimed. Tools Appl. 2024, 83, 43071–43113. [Google Scholar] [CrossRef]
- Al-Jebrni, A.H.; Ali, S.G.; Li, H.; Lin, X.; Li, P.; Jung, Y.; Kim, J.; Feng, D.D.; Sheng, B.; Jiang, L.; et al. SThy-Net: A feature fusion-enhanced dense-branched modules network for small thyroid nodule classification from ultrasound images. Vis. Comput. 2023, 39, 3675–3689. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Cao, J.; Zhu, Y.; Tian, X.; Wang, J. Tnc-Net: Automatic classification for thyroid nodules lesions using convolutional neural network. IEEE Access 2024, 12, 84567–84578. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Bay, H.; Tuytelaars, T.; Van Gool, L. SURF: Speeded up robust features. In Proceedings of the European Conference on Computer Vision (ECCV), Graz, Austria, 7–13 May 2006; pp. 404–417. [Google Scholar]
- Aboudi, N.; Khachnaoui, H.; Moussa, O.; Khlifa, N. Bilinear pooling for thyroid nodule classification in ultrasound imaging. Arab. J. Sci. Eng. 2023, 48, 10563–10573. [Google Scholar] [CrossRef]
- Göreke, V. A novel deep-learning-based CADx architecture for classification of thyroid nodules using ultrasound images. Interdiscip. Sci. Comput. Life Sci. 2023, 15, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, C.; Xu, Z.; Xu, L.; Zhan, W.; Xiao, J.; Hou, Y.; Huang, B.; Huang, L.; Li, S. An interpretable two-branch bi-coordinate network based on multi-grained domain knowledge for classification of thyroid nodules in ultrasound images. Med. Image Anal. 2024, 97, 103255. [Google Scholar] [CrossRef]
- Xie, J.; Guo, L.; Zhao, C.; Li, X.; Luo, Y.; Jianwei, L. A hybrid deep learning and handcrafted features based approach for thyroid nodule classification in ultrasound images. J. Phys. Conf. Ser. 2020, 1693, 012160. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Mäenpää, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Hang, Y. Thyroid nodule classification in ultrasound images by fusion of conventional features and Res-GAN deep features. J. Healthc. Eng. 2021, 2021, 9917538. [Google Scholar] [CrossRef] [PubMed]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition (CVPR), San Diego, CA, USA, 20–26 June 2005; pp. 886–893. [Google Scholar]
- Shankarlal, B.; Sathya, P.D.; Sakthivel, V.P. Computer-aided detection and diagnosis of thyroid nodules using machine and deep learning classification algorithms. IETE J. Res. 2023, 69, 995–1006. [Google Scholar] [CrossRef]
- Swathi, G.; Altalbe, A.; Kumar, R.P. QuCNet: Quantum-inspired convolutional neural networks for optimized thyroid nodule classification. IEEE Access 2024, 12, 27829–27842. [Google Scholar] [CrossRef]
- Duan, X.; Duan, S.; Jiang, P.; Li, R.; Zhang, Y.; Ma, J.; Zhao, H.; Dai, H. An ensemble deep learning architecture for multilabel classification on TI-RADS. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 576–582. [Google Scholar]
- Liu, W.; Lin, C.; Chen, D.; Niu, L.; Zhang, R.; Pi, Z. Shape-margin knowledge augmented network for thyroid nodule segmentation and diagnosis. Comput. Methods Programs Biomed. 2024, 244, 107999. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.X.; Chen, Y.; Yang, K.F.; Luo, Y.; Ma, B.Y.; Li, Y.J. A local and global feature disentangled network: Toward classification of benign-malignant thyroid nodules from ultrasound image. IEEE Trans. Med. Imaging 2022, 41, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, X.; Shi, G.; Xiao, N.; Song, K.; Zhao, J.; Hao, R.; Li, K. Semantic consistency generative adversarial network for cross-modality domain adaptation in ultrasound thyroid nodule classification. Appl. Intell. 2022, 52, 10369–10383. [Google Scholar] [CrossRef] [PubMed]
- Avola, D.; Cinque, L.; Fagioli, A.; Filetti, S.; Grani, G.; Rodola, E. Multimodal feature fusion and knowledge-driven learning via experts consult for thyroid nodule classification. IEEE Trans. Circuits Syst. Video Technol. 2022, 32, 2527–2534. [Google Scholar] [CrossRef]
- Deng, P.; Han, X.; Wei, X.; Chang, L. Automatic classification of thyroid nodules in ultrasound images using a multi-task attention network guided by clinical knowledge. Comput. Biol. Med. 2022, 150, 106172. [Google Scholar] [CrossRef]
- Bai, Z.; Chang, L.; Yu, R.; Li, X.; Wei, X.; Yu, M.; Liu, Z.; Gao, J.; Zhu, J.; Zhang, Y.; et al. Thyroid nodules risk stratification through deep learning based on ultrasound images. Med. Phys. 2020, 47, 6355–6365. [Google Scholar] [CrossRef]
- Yang, W.; Dong, Y.; Du, Q.; Qiang, Y.; Wu, K.; Zhao, J.; Yang, X.; Zia, M.B. Integrate domain knowledge in training multi-task cascade deep learning model for benign–malignant thyroid nodule classification on ultrasound images. Eng. Appl. Artif. Intell. 2021, 98, 104064. [Google Scholar] [CrossRef]
- Shi, G.; Wang, J.; Qiang, Y.; Yang, X.; Zhao, J.; Hao, R.; Yang, W.; Du, Q.; Kazihise, N.G.F. Knowledge-guided synthetic medical image adversarial augmentation for ultrasonography thyroid nodule classification. Comput. Methods Programs Biomed. 2020, 196, 105611. [Google Scholar] [CrossRef]
- Iakovidis, D.K.; Savelonas, M.A.; Karkanis, S.A.; Maroulis, D.E. A genetically optimized level set approach to segmentation of thyroid ultrasound images. Appl. Intell. 2007, 27, 193–203. [Google Scholar] [CrossRef]
- Ma, J.; Wu, F.; Jiang, T.; Zhu, J.; Kong, D. Cascade convolutional neural networks for automatic detection of thyroid nodules in ultrasound images. Med. Phys. 2017, 44, 1678–1691. [Google Scholar] [CrossRef]
- Molnár, K.; Kálmán, E.; Hári, Z.; Giyab, O.; Gáspár, T.; Rucz, K.; Bogner, P.; Tóth, A. false-positive malignant diagnosis of nodule mimicking lesions by computer-aided thyroid nodule analysis in clinical ultrasonography practice. Diagnostics 2020, 10, 378. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. In Proceedings of the International Conference on Learning Representations (ICLR), Virtual , 3–7 May 2021. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. SWIN Transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Online, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Pedraza, L.; Vargas, C.; Narváez, F.; Durán, O.; Muñoz, E.A.; Romero, E. An open access thyroid ultrasound image database. In Proceedings of the International Symposium on Medical Information Processing and Analysis, Cartagena de Indias, Colombia, 14–16 October 2015; pp. 1–6. [Google Scholar]
- Gong, H.; Chen, J.; Chen, G.; Li, H.; Li, G.; Chen, F. Thyroid region prior guided attention for ultrasound segmentation of thyroid nodules. Comput. Biol. Med. 2022, 155, 106389. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Sahu, A.K.; Zaheer, M.; Sanjabi, M.; Talwalkar, A.; Smith, V. Federated optimization in heterogeneous networks. In Proceedings of the Machine Learning Systems (MLSys) Conference, Austin, TX, USA, 2–4 March 2020. [Google Scholar]
- Karimireddy, S.P.; Kale, S.; Mohri, M.; Reddi, S.; Stich, S.; Suresh, A.T. SCAFFOLD: Stochastic Controlled Averaging for Federated Learning. In Proceedings of the International Conference on Machine Learning (ICML), Cambridge, MA, USA, 13–18 July 2020; pp. 5132–5143. [Google Scholar]
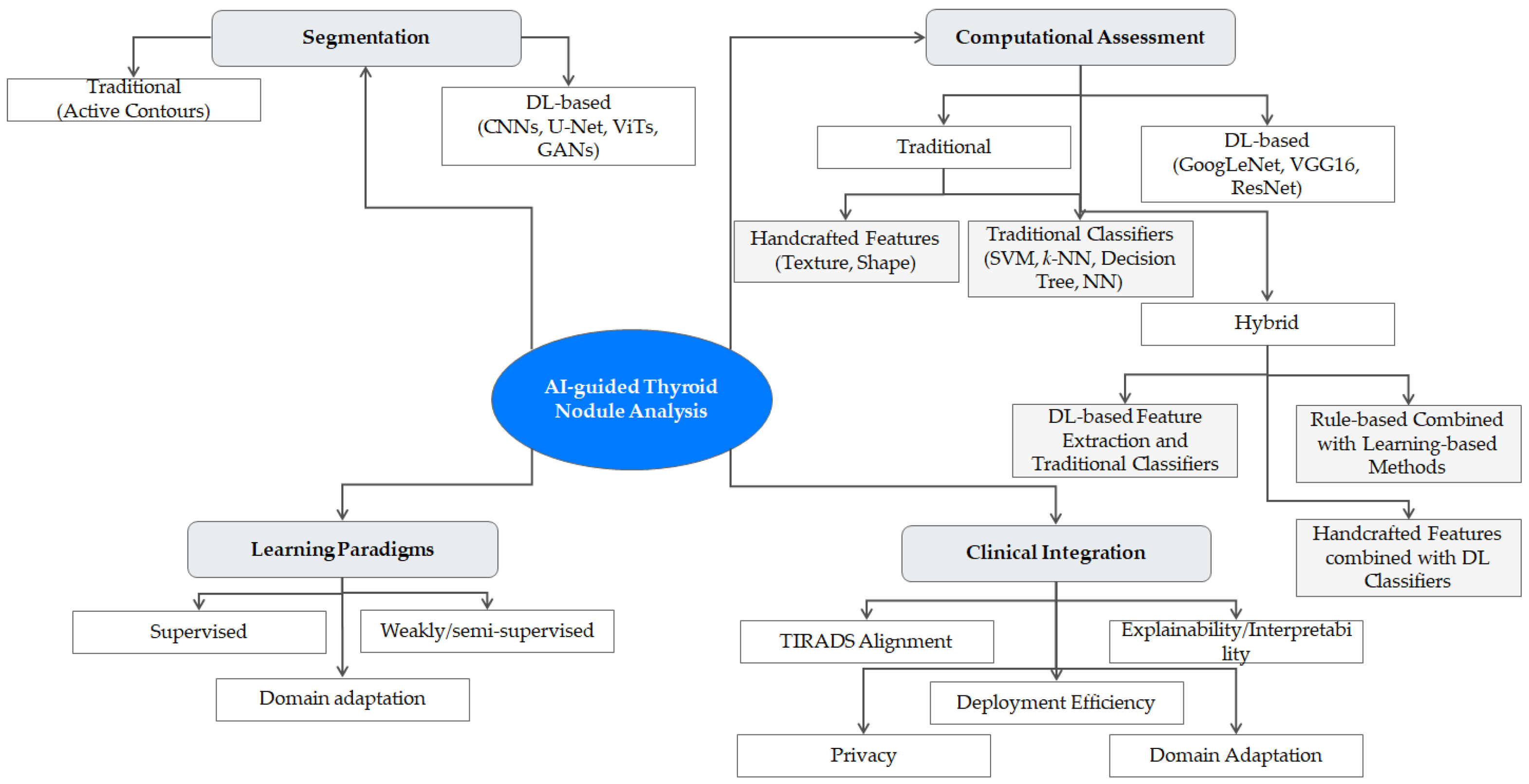
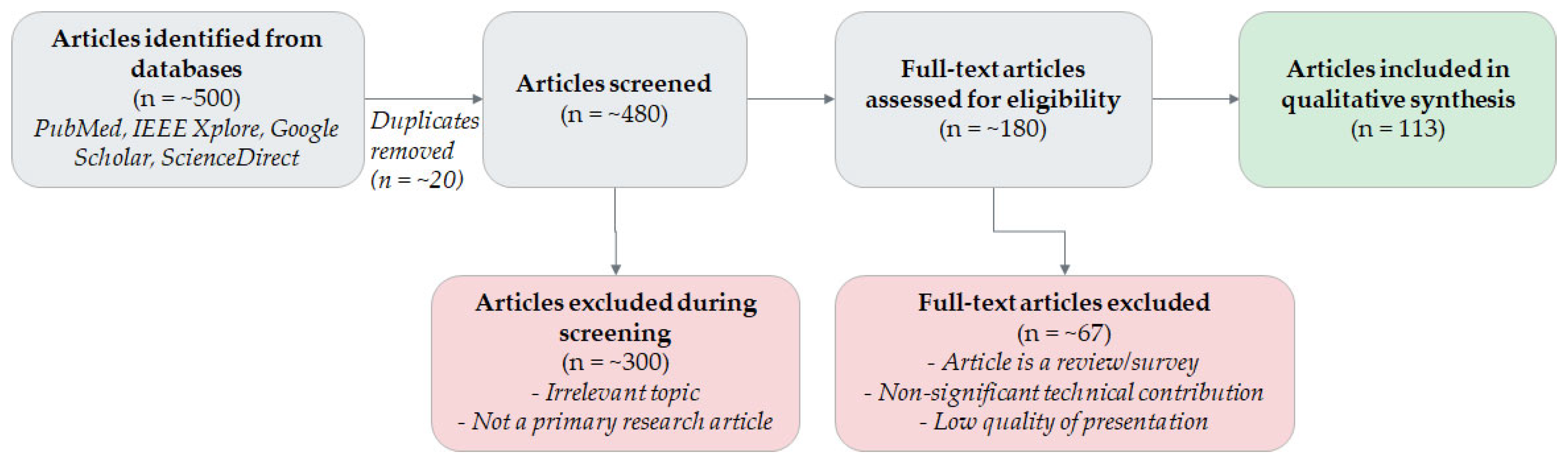
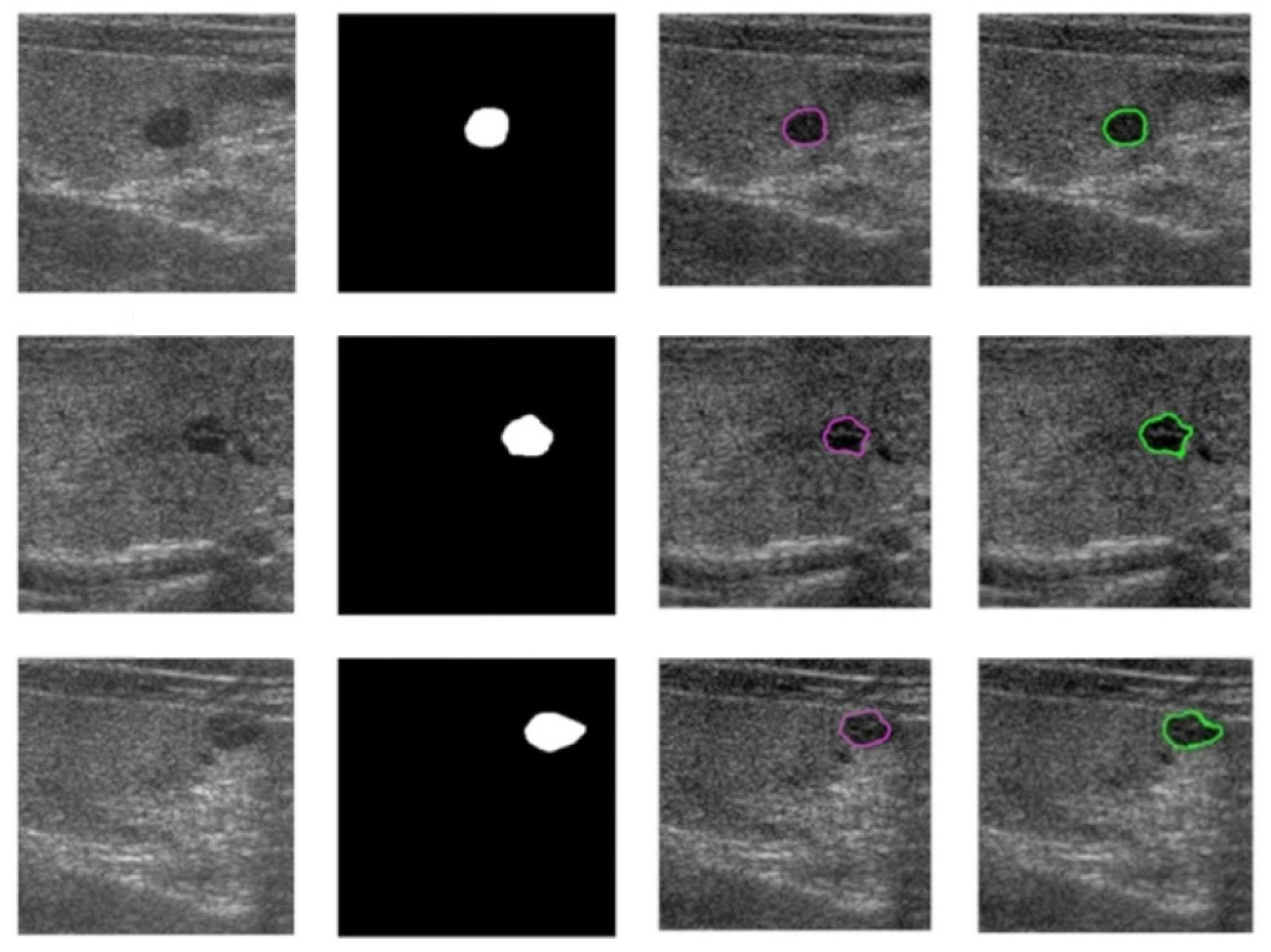
| Reference | Method | Strengths | Weaknesses | Description |
|---|---|---|---|---|
| Savelonas et al. (2007) [99] | Radon transform, SVM, k-NN | Physically interpretable hypothesis: different tissue types exhibit patterns of distinct directionality, limited computational complexity/cost | Standalone Radon features provide only partial texture representation, no end-to-end optimization (see Section 4.1), empirical parameter adjustment (see Section 4.1) | Differentiates thyroid tissue based on directionality patterns, encoded using Radon transform features and classified with k-NN and SVM. |
| Seabra et al. (2009) [100] | Radon transform, wavelet transform, Rayleigh-based, PCA, k-NN, MAP | Leverages complementary descriptors, limited computational complexity/cost | No end-to-end optimization (see Section 4.1), empirical parameter adjustment (see Section 4.1) | A multi-descriptor scheme that combines Radon transform features with Rayleigh parameters, mid-distance term, and wavelet coefficients, using PCA for dimensionality reduction before classification. |
| Savelonas et al. (2009) [101] | Boundary features, SVM, k-NN | Aligned with TIRADS malignancy risk stratification, distinguishes medium-risk and high-risk nodules, limited computational complexity/cost | Standalone boundary features cannot capture tissue texture (see Section 4.1) | A malignancy risk assessment method based on nodule boundary features. It uses features that correlate with TIRADS descriptors, like boundary irregularity and local echogenicity variance. |
| Legakis et al. (2011) [102] | Active contour, boundary and textural features, SVM | Unified for full workflow (detection, segmentation, assessment), combines boundary and textural features | No end-to-end optimization (see Section 4.1), empirical parameter adjustment (see Section 4.1) | A multi-stage pipeline that integrates processes for thyroid gland boundary detection, nodule detection and delineation (using active contours), and malignancy risk assessment. |
| Iakovidis et al. (2010) [103] | Fuzzy local binary patterns, fuzzy intensity histograms, SVM | Noise robustness, limited computational complexity/cost | No end-to-end optimization (see Section 4.1), empirical parameter adjustment (see Section 4.1) | A texture-based method that represents sonographic features, using a noise-resistant combination of fuzzy local binary patterns (LBP) for texture and fuzzy intensity histograms for echogenicity. |
| Katsigianis et al. (2010) [104] | Contourlet transform, SFFS | Contourlet transform captures multidirectional and multiscale texture patterns, noise robustness | No end-to-end optimization (see Section 4.1), empirical parameter adjustment (see Section 4.1) | A texture representation scheme based on the Contourlet transform. It uses thresholding to select the most significant transform coefficients, followed by sequential forward feature selection (SFFS). |
| Keramidas et al. (2012) [105] | Fuzzy local binary patterns, fuzzy intensity histograms, SVM | Addresses both ROI detection and assessment, noise robustness, limited computational complexity/cost | No end-to-end optimization (see Section 4.1), empirical parameter adjustment (see Section 4.1) | Introduces the thyroid nodule detection (TND) system, which combines automatic rough region-of-interest (ROI) extraction via unsupervised gland boundary detection with a fuzzy approach that represents US patterns by means of fuzzy LBP and fuzzy gray-level histograms. |
| Kale et al. (2018, 2013) [106,107], Vadhiraj et al. (2021) [108] | GLCM, two-layer feed-forward neural network | Limited computational complexity/cost, SVM variants ([107,110]) are less prone to overfitting | No end-to-end optimization (see Section 4.1), GLCM is sensitive to noise (see Section 4.1) | This series of works uses gray-level co-occurrence matrix (GLCM) features for nodule assessment, exploring classification with both feed-forward neural networks and SVMs. |
| Nanda et al. (2018) [109] | GLCM, ReliefF, SVM | Captures directional and multiscale information, addresses overfitting (via ReliefF and SVM) | No end-to-end optimization (see Section 4.1), GLCM is sensitive to noise (see Section 4.1) | Combines GLCM with textural features derived from a steerable pyramid decomposition, which captures information at different scales and orientations. It uses ReliefF [110] for feature selection. |
| Acharya et al. (2016) [111] | Gabor transform, ReliefF, various classifiers | Captures directional and frequency-based information (via Gabor), addresses dataset imbalance, addresses overfitting (via ReliefF and SVM) | No end-to-end optimization (see Section 4.1), computational complexity/cost of Gabor transform/entropy calculations (for a non-DL-based method) | An automated method that extracts entropy features from a Gabor transform of high-resolution US images. It uses locality sensitive discriminant analysis (LSDA) for feature reduction and over-sampling strategies to handle data imbalance. |
| Raghvendra et al. (2017) [112] | SGLDF/fractal features | Captures spatial dependencies, self-similar patterns, and irregularities, can be integrated with various classifiers | No end-to-end optimization (see Section 4.1), boundary features may be overlooked | Fuses spatial gray-level dependence features (SGLDF) with fractal texture features to represent the intrinsic structure of thyroid lesions, using graph-based marginal Fisher analysis (MFA) for feature reduction. |
| Xia et al. (2017) [113] | ELM, ReliefF | Low computational complexity/cost | Sensitivity to initialization/hyperparameter adjustment (see Section 4.1), lower capacity than DL-based methods (see Section 4.1) | Employs an extreme learning machine (ELM) [114], an efficient algorithm for single-hidden-layer feedforward networks, to discriminate between malignant and benign nodules. |
| Prochazka et al. (2019) [115] | Fractal features, SVM, random forests | Captures self-similar patterns and irregularities, robust to probe orientation and inclination | Requires manual segmentation (see Section 4.1), small dataset size (see Section 4.4) | A fractal-based method that uses histogram analysis and segmentation-based fractal texture analysis to extract direction-independent features, aiming for invariance to the US probe’s orientation. |
| Raghavendra et al. (2018) [116] | Elongated quinary patterns, PSO, SVM | Captures local directional textural information, addresses overfitting (with PSO/SVM) | Depends on preprocessing, difficult to interpret (see Section 4.2) | Uses optimized multi-level elongated quinary patterns to extract higher-order spectral (HOS) entropy features for classification. |
| Gomes Ataide et al. (2020) [117] | Random forest, geometrical/morphological features | Clinically interpretable features aligned with TIRADS, low computational complexity/cost | Textural/echogenicity features are not considered | Extracts a comprehensive set of 27 geometric and morphological features and selects a subset of 11 features that are evaluated as significant according to the TIRADS framework. |
| Barzegar-Golmoghani et al. (2025) [118] | SVM, random forest, SHAP, PDP | Clinically interpretable features aligned with TIRADS | Requires US elastography | Introduces ELTIRADS, a multimodal framework that integrates US elastography data with standard TIRADS scores and radiomic features. It employs Shapley additive explanations (SHAP) and partial dependence plots (PDP) for enhanced interpretability. |
| Chi et al. (2017) [119] | GoogLeNet, random forest | Addresses dataset imbalance | No end-to-end optimization (see Section 4.1), limited interpretability (see Section 4.2) | An early hybrid, CNN-based method that fine-tunes a pre-trained GoogLeNet [120] model and uses the extracted deep features in a cost-sensitive random forest classifier. |
| Liang et al. (2020) [121] | CNN | Data-driven feature extraction | No end-to-end optimization (see Section 4.1), limited interpretability (see Section 4.2) | A similar early hybrid, CNN-based method that also uses a pre-trained model to extract features, which are then used to train a separate classifier for the final assessment. |
| Zhu et al. (2017) [122] | ResNet18 | Data-driven feature extraction | Limited interpretability (see Section 4.2) | Fine-tunes a ResNet18 architecture using transfer learning and a CNN-based data augmentation strategy to generate new training samples. |
| Song et al. (2020) [123] | CNN | Multi-branch network, captures both local and global information | Computational complexity/cost (see Section 4.3), limited interpretability (see Section 4.2) | Introduces a hybrid multi-branch CNN. It combines a global branch for overall classification with a feature cropping branch that performs multi-cropping on feature maps to focus on local discriminative details. |
| Duan et al. (2020) [157] | EfficientNet, feature pyramid network | Clinically interpretable features aligned with TIRADS, captures multiscale information | Computational complexity/cost (see Section 4.3) | An ensemble architecture that integrates three modules for multi-class TIRADS feature assessment: an EfficientNet, a feature engineering module for geometric/textural features, and a feature pyramid network. |
| Zhang et al. (2020) [124] | ResNet50 | Clinically interpretable features aligned with TIRADS | Computational complexity/cost (see Section 4.3) | Develops an interpretable CAD system (iCADs) comprising ResNet50 [90] trained on ImageNet [125] for benign/malignant classification and an ensemble of six specialized sub-networks, each trained to assess a specific TIRADS feature. |
| Wang et al. (2022) [126] | CNN, HAC | Captures multiscale information, enhanced contextual awareness | Computational complexity/cost (see Section 4.3) | Introduces n-ClsNet, a CNN model featuring a multiscale classification layer, multiple skip blocks, and a hybrid atrous convolution (HAC) block to enhance the learning of spatial information at different scales. |
| Zhang et al. (2023) [127] | CNN | Multilabel output, aligned with TIRADS | Computational complexity/cost (see Section 4.3) | Multi-label, two-stage pipeline. It first uses a U-Net++ variant for segmentation, followed by a multi-task CNN that classifies benign/malignant status and four specific malignant features aligned with TIRADS. |
| Wang et al. (2020) [97] | VGG16 | Combines low-level textural information with high-level semantic information | VGG16 lacks residual connections, no end-to-end optimization (see Section 4.1) | A method that combines high-throughput statistical feature extraction with a fine-tuned, pre-trained VGG16 model, using linear discriminant analysis (LDA) for dimensionality reduction. |
| Kwon et al. (2020) [128] | VGG16 | Addresses overfitting | VGG16 lacks residual connections, limited interpretability (see Section 4.2) | Employs a pre-trained VGG16 model with transfer learning, using data augmentation and global average pooling to mitigate overfitting. |
| Zhao et al. (2021) [129] | ResNet50, CBAM | End-to-end optimization, captures multiscale information | Small-sized dataset used, sensitive to dataset imbalance, annotations were performed by radiologists of various levels of experience (see Section 4.4) | A two-stage framework. The first stage uses a single-shot detection (SSD) network for nodule detection, and the second stage employs a convolution-based attention module (CBAM) to aid a ResNet50 in focusing on relevant features for classification. |
| Han et al. (2021) [130] | VGG-f | Inspired by radiologists’ cognitive process, captures multiscale information, copes with annotation effort by means of AL | Complex pipeline, VGG-f is outdated when compared to more recent backbones (e.g., ResNet, EfficientNet) | An attention-driven framework inspired by the radiologist’s cognitive process (“zoom-in”). It uses VGG-f [131] for low-magnification suspicious region detection, as well as for high-magnification patch classification. |
| Liu et al. (2021) [132] | Two-branched CNN | Uses complementary, bimodal information, end-to-end optimization | Requires RF signal data, which are often not accessible | An information fusion-based joint CNN (IF-JCNN) that evaluates both US images and radiofrequency (RF) signals. It uses a two-branched CNN to extract features from each modality, which are fused at the backend. |
| Wang et al. (2021) [133] | Dual-attention ResNet variant | End-to-end optimization | Computational complexity/cost (see Section 4.3) | Introduces a cascaded dual-attention ResNet-based network that integrates segmentation and classification in a single pipeline, aiming for efficient and automatic end-to-end analysis. |
| Kang et al. (2022) [134] | U-Net | End-to-end optimization, consistency constraints address task conflict | Computational complexity/cost (see Section 4.3), requires annotations for multiple tasks | A multi-task learning framework that jointly addresses segmentation and classification. It introduces intra- and inter-task consistent learning to enforce consistent predictions among different but related tasks. |
| Zhou et al. (2022) [135] | Res-U-Net, CVAE | Addresses small dataset size and categorical imbalance, aligned with TIRADS | No end-to-end optimization (see Section 4.1), computational complexity/cost (see Section 4.3) | Introduces a cascaded automatic classification system (CACS), which uses a Res-U-Net for ROI extraction and a conditional variational autoencoder (CVAE) to generate synthetic US images for data augmentation before final classification. |
| Wang et al. (2022) [136] | ConvNeXt | End-to-end optimization | Interpretability (see Section 4.2) | Benign/malignant classification based on transfer learning using a pre-trained ConvNeXt [71]. |
| Yang et al. (2022) [137] | ResNet18, Grad-CAM | Computational complexity/cost, interpretability | ResNet18 may underperform when compared to deeper networks | Employs ResNet18 with transfer learning and uses gradient-weighted class activation mapping (Grad-CAM) [138] to visualize the model’s attention. |
| Wang et al. (2022) [139] | Mask-RCNN | End-to-end optimization | Computational complexity/cost (see Section 4.3) | A mask-guided hierarchical DL (MHDL) framework. It first uses a mask R-CNN [140] to locate the nodule ROI, then a residual attention network to extract depth features, and finally an attention drop-based CNN for classification. |
| Xing et al. (2024) [141] | CNN, ASPP | End-to-end optimization, captures contextual information | Biased dataset (see Section 4.4), complex architecture | A multi-task framework that combines dense connectivity, squeeze-and-excitation (SE) connectivity, and an atrous spatial pyramid pooling (ASPP) layer to enhance feature extraction and optimize feature reuse. |
| Yadav et al. (2024) [142] | CNN, PCA-SVM | Direct, comparative benchmark of various CNN backbones for feature extraction | No end-to-end optimization (see Section 4.1) | A wide range of pre-trained CNNs (e.g., AlexNet, VGG, GoogLeNet, DenseNet, ResNet) are compared as feature extractors. The extracted deep features are classified by a PCA-SVM pipeline. |
| Al-Jebrni et al. (2023) [143] | Inception v3/ResNet-inspired | End-to-end optimization, addresses small nodules, captures multiscale information | Computational complexity/cost (see Section 4.3), dataset is small/biased (see Section 4.4) | Introduces SThy-Net, a network explicitly designed for small thyroid nodules. It is inspired by Inception-v3 [144] and features a dense-branched module and a Gaussian-enhanced feature fusion module. |
| Li et al. (2023) [84] | CNN/ViT | Captures multiscale information and nodule shape features | Small-sized dataset (see Section 4.4) | A DL-based CAD method (TCNet) that fuses a large kernel CNN branch (for shape features) with an enhanced ViT branch (for remote connections) via a multiscale fusion module (MFM). |
| Cao et al. (2024) [145] | SP-LeNet, ConvNeXt | Addresses small data volume and categorical imbalance, captures both contextual and fine-grained information | Computational complexity/cost (see Section 4.3), not aligned with TIRADS (see Section 4.2) | Introduces Tnc-Net, which consists of three parts: an SP-LeNet backbone for global features on small datasets, a ConvNeXt branch for local features, and a feature aggregation module. |
| Wu et al. (2024) [3] | YOLO, BiLSTM | Uses US video, exploits spatio-temporal information, inspired by radiologists’ cognitive processes | Computational complexity/cost (see Section 4.3), depends on US video availability | A framework for thyroid nodule assessment in US video. It is based on a “you-only-look-once” (YOLO) architecture and an attention-driven bidirectional LSTM (BiLSTM) to collect spatio-temporal information between video frames. |
| Aboudi et al. (2023) [148] | CNN, SVM | Rich feature representation | Computational complexity/cost (see Section 4.3), small-sized dataset (see Section 4.4), interpretability (see Section 4.4) | Investigates the use of bilinear models, which consist of a fusion of the outputs of two different CNNs (e.g., VGG19 and ResNet50) using outer products, which are then classified by a linear SVM. |
| Göreke (2023) [149] | VGG16, SVM | Integrates domain knowledge, captures both textural and morphological features | No end-to-end optimization (see Section 4.1) | A hybrid method, where the final feature vector used by an SVM is formed by combining textural and morphological features that have been extracted by a pre-trained VGG16 network. |
| Wang et al. (2024) [150] | CNN, FPN | Integrates domain knowledge, aligned with TIRADS, interpretability | Computational complexity/cost (see Section 4.3), depends on a large amount of expert annotations (see Section 4.4) | Introduces multi-grained domain knowledge representation (MG-DKR) to convert image features into classification labels and masks. A two-branch bi-coordinate network (TB2C-Net) learns from this representation. |
| Xie et al. (2020) [151] | Two-branched ResNet, LBP | LBP induces inductive bias, aiding in cases of small-sized datasets | Computational complexity/cost (see Section 4.3), risk of feature redundancy | A hybrid method that combines DL with handcrafted features. It uses a two-stream deep neural network that processes both the source US image and its corresponding LBP feature map. |
| Hang (2021) [153] | Res-GAN, LBP, SIFT, SURF, random forest | Handcrafted features induce inductive bias, aiding in cases of small-sized datasets | Computational complexity/cost (see Section 4.3), risk of feature redundancy | A hybrid method that fuses traditional handcrafted features (histogram-of-gradients (HoG) [154], LBP, SIFT, SURF) with deep features extracted from a residual GAN (Res-GAN). |
| Shankarlal et al. (2023) [155] | Kirch’s edge detector, DTCT, CANFES, CNN | DTCT captures multiscale features, CANFES aids interpretability | No end-to-end optimization (see Section 4.1), interpretability (see Section 4.2) | A hybrid CAD method that uses Kirsch’s edge detector and a dual-tree contourlet transform (DTCT) for feature extraction. The extracted features are used by a co-active adaptive neuro-fuzzy expert system (CANFES). |
| Swathi et al. (2024) [156] | Quanvolutional filter, CNN | Captures multiscale information | Interpretability (see Section 4.2) | Introduces QuCNet, a quantum-inspired CNN. It combines features extracted by a “quanvolutional” filter, which uses equations from random quantum circuits, with a typical CNN for classification. |
| Duan et al. (2020) [157] | EfficientNet, FPN | Granular, multilabel classification, aligned with TIRADS, interpretability | No end-to-end optimization (see Section 4.1), computational complexity/cost (see Section 4.3) | An ensemble architecture for multi-class TIRADS assessment that integrates three distinct feature extraction modules: EfficientNet, handcrafted geometric/textural features, and a multi-scale feature pyramid network (FPN). |
| Liu et al. (2024) [158] | CNN, ViT | Captures multiscale information, interpretability via CAM heatmaps | Computational complexity/cost (see Section 4.3), prior knowledge is limited to shape/margin | Introduces SKA-Net, a shape-margin knowledge augmented network. It is a dual-branch architecture for joint segmentation and diagnosis that uses an exponential mixture module to combine convolutional and self-attention feature maps. |
| Zhao et al. (2022) [159] | CNN | Disentangles tissue and anatomical information, aiding interpretability, aligned with radiologists’ cognitive process, end-to-end optimization | Computational complexity/cost (see Section 4.3) | Introduces LoGo-Net, a local and global feature disentangled network inspired by human vision. It uses a tissue-anatomy disentangled (TAD) block to decouple local tissue features from global anatomical features. |
| Shahroudnejad et al. (2021) [4] | U-Net variant | Aligned with TIRADS | Requires manually extracted ROI, struggles with heterogeneous nodules (see Section 4.1) | Analyzes videos and incorporates two rule-based classifiers (for composition and echogenicity) that are directly aligned with TIRADS classifications, in addition to the main DL model. |
| Zhao et al. (2022) [160] | SCGAN | Augments US image information with textual information, addresses domain shifts with semantic consistency loss | Computational complexity/cost (see Section 4.1), depends on the availability of paired text-US image data (see Section 4.4) | Introduces SCGAN, a deeply fused semantic consistency GAN. It organically incorporates domain knowledge from accompanying text reports, using self-attention and metric learning. |
| Avola et al. (2022) [161] | LBP, DWT, CNN variants (DenseNet, AlexNet, ResNet, VGG) ensemble | Handcrafted features and the use of a network ensemble aid training | Computational complexity/cost (see Section 4.3), interpretability (see Section 4.2) | A CAD method that leverages cues from an ensemble of expert networks (pre-trained on ImageNet) to guide the learning phase of a final DenseNet classifier, reducing the number of samples required for training. |
| Deng et al. (2022) [162] | ResNet50 variant | ACR TIRADS aligned, multitask learning aids generalization | Computational complexity/cost (see Section 4.3), interpretability (see Section 4.2), limited stratification of echogenic foci, requires ACR TIRADS annotations (see Section 4.2) | A multi-task branching attention network that incorporates domain knowledge by explicitly classifying each of the ACR TIRADS descriptors. It uses a variant of ResNet50 as its feature extraction backbone. |
| Bai et al. (2020) [163] | CNN | ACR TIRADS aligned, incorporates radiologists’ cognitive process | Requires ACR TIRADS annotations (see Section 4.2), focuses on boundary features, and ignores features in nodule areas | Introduces risk stratification network (RS-Net), an automatic hierarchical method that incorporates a CNN with domain knowledge aligned with ACR TIRADS, in order to perform risk stratification. |
| Yang et al. (2021) [164] | U-Net, DScGAN, S3VM | Addresses limitations in the availability of annotated data, integrates domain knowledge | Computational complexity/cost (see Section 4.3), depends on the availability of pairs of B-mode and elastography images | A multi-task cascade DL model (MCDLM) that integrates radiologists’ domain knowledge (DK) by using U-Net to transfer boundary information and assists a dual-path semi-supervised GAN (DScGAN) for classification. |
| Shi et al. (2020) [165] | CNN, LSTM, ACGAN | Integrates domain knowledge via term encoder, addresses data scarcity and dataset imbalance, aligned with TIRADS | Depends on the availability of pairs of radiologists’ reports and US images (see Section 4.4) | A knowledge-guided adversarial augmentation method. It employs term and image encoders in order to exploit radiologists’ domain knowledge in the form of conditions to constrain the auxiliary classifier GAN (ACGAN) framework for the synthesis of thyroid nodule images. |
| Architecture | Task | Parameters (M) | FPS ** | VRAM (GB) *** |
|---|---|---|---|---|
| VGG16 [42] | Classification | 138 | 60 | 3.0 |
| ResNet50 [90] | Classification | 25.6 | 120 | 2.5 |
| EfficientNet-B0 [67] | Classification | 5.3 | 150 | 1.5 |
| U-Net [34] **** | Segmentation | 31 | 90 | 3.0 |
| DeepLab-v3+ [45] | Segmentation | 41 | 80 | 3.5 |
| ViT Base [169] ***** | Classification | 86 | 75 | 4.0 |
| SWIN Tiny [170] ***** | Classification | 29 | 100 | 2.8 |
| Characteristic | DDTI (Digital Database for Thyroid Images) | TN3K (Thyroid Nodule 3K) |
|---|---|---|
| Reference | Pedraza et al. [171] | Gong et al. [172] |
| Size | 99 patients, 416 images | >3500 patients, >4500 images |
| Classes | Benign, malignant, normal tissue | Benign, malignant; TIRADS categories (2–5) |
| Imaging Modality | B-mode US | B-mode US |
| Image Resolution | 576 × 768 pixels | Varies |
| Annotation Type | Nodule boundary masks | Nodule boundary masks |
| Licensing | Open access for research | Open access for research |
| Typical Use | Foundational segmentation and classification tasks | Large-scale DL model training, TIRADS-aligned tasks |
| Reference | Method | Task | Dataset | IoU | Dice | Accuracy * | Precision * | Recall * |
|---|---|---|---|---|---|---|---|---|
| Wang et al. (2023) [36] | CNN (DPAM-PSPNet) | Segmentation | DDTI | 0.69 | 0.80 | 0.95 | - | - |
| Wang et al. (2023) [36] | CNN (DPAM-PSPNet) | Segmentation | TN3K | 0.75 | 0.85 | 0.97 | - | - |
| Shao et al. (2023) [54] | U-Net3+ variant | Segmentation | DDTI | 0.60 | 0.75 | |||
| Bouhdiba et al. (2024) [68] | U-Net and RefineUNet | Segmentation | TN3K | 0.96 | 0.81 | 0.94 | 0.88 | 0.82 |
| Liu et al. (2024) [70] | U-Net and ConvNeXt | Segmentation | DDTI | 0.69 | 0.81 | - | 0.81 | 0.84 |
| Liu et al. (2024) [70] | U-Net and ConvNeXt | Segmentation | TN3K | 0.79 | 0.87 | - | 0.89 | 0.87 |
| Yang et al. (2024) [73] | U-Net variant | Segmentation | DDTI | 0.74 | 0.84 | 0.96 | 0.86 | - |
| Yang et al. (2024) [73] | U-Net variant | Segmentation | TN3K | 0.78 | 0.87 | 0.98 | 0.85 | - |
| Xiao et al. (2025) [74] | DeepLab v3+, EfficientNet-B7 | Segmentation | DDTI | 0.75 | 0.85 | 0.96 | 0.85 | 0.88 |
| Ali et al. (2024) [78] | U-Net-inspired encoder–decoder | Segmentation | DDTI | 0.76 | 0.87 | 0.96 | 0.87 | 0.88 |
| Ali et al. (2024) [78] | U-Net-inspired encoder–decoder | Segmentation | TN3K | 0.69 | 0.82 | - | - | - |
| Sun et al. (2024) [79] | U-Net variant | Segmentation | DDTI | 0.79 | 0.74 | 0.97 | 0.76 | 0.75 |
| Wu et al. (2025) [80] | SWIN ViT | Segmentation | DDTI | 0.68 | 0.79 | - | - | - |
| Wu et al. (2025) [80] | SWIN ViT | Segmentation | TN3K | 0.73 | 0.82 | - | - | - |
| Ma et al. (2023) [81] | SWIN ViT | Segmentation | DDTI | 0.61 | 0.75 | - | 0.76 | - |
| Ma et al. (2023) [81] | SWIN ViT | Segmentation | TN3K | 0.72 | 0.84 | - | 0.88 | - |
| Ozcan et al. (2024) [85] | U-Net, ViT | Segmentation | DDTI | 0.91 | 0.95 | - | - | - |
| Ozcan et al. (2024) [85] | U-Net, ViT | Segmentation | TN3K | 0.71 | 0.83 | - | - | - |
| Li et al. (2023) [86] | CNN (DLA-34), level-set | Segmentation | DDTI | 0.69 | 0.82 | - | - | - |
| Song et al. (2020) [123] | CNN | Assessment | DDTI | - | - | 0.96 | 0.93 | 0.97 |
| Swathi et al. (2024) [156] | Quanvolutional filter, CNN | Assessment | DDTI | - | - | 0.94 | 0.95 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savelonas, M. An Overview of AI-Guided Thyroid Ultrasound Image Segmentation and Classification for Nodule Assessment. Big Data Cogn. Comput. 2025, 9, 255. https://doi.org/10.3390/bdcc9100255
Savelonas M. An Overview of AI-Guided Thyroid Ultrasound Image Segmentation and Classification for Nodule Assessment. Big Data and Cognitive Computing. 2025; 9(10):255. https://doi.org/10.3390/bdcc9100255
Chicago/Turabian StyleSavelonas, Michalis. 2025. "An Overview of AI-Guided Thyroid Ultrasound Image Segmentation and Classification for Nodule Assessment" Big Data and Cognitive Computing 9, no. 10: 255. https://doi.org/10.3390/bdcc9100255
APA StyleSavelonas, M. (2025). An Overview of AI-Guided Thyroid Ultrasound Image Segmentation and Classification for Nodule Assessment. Big Data and Cognitive Computing, 9(10), 255. https://doi.org/10.3390/bdcc9100255





