Growth Performance of a Newly Isolated and Culturable Thraustochytrid Strain from Sea Squirt Colonies
Abstract
1. Introduction
2. Materials and Methods
2.1. Thraustochytrid Culture Conditions
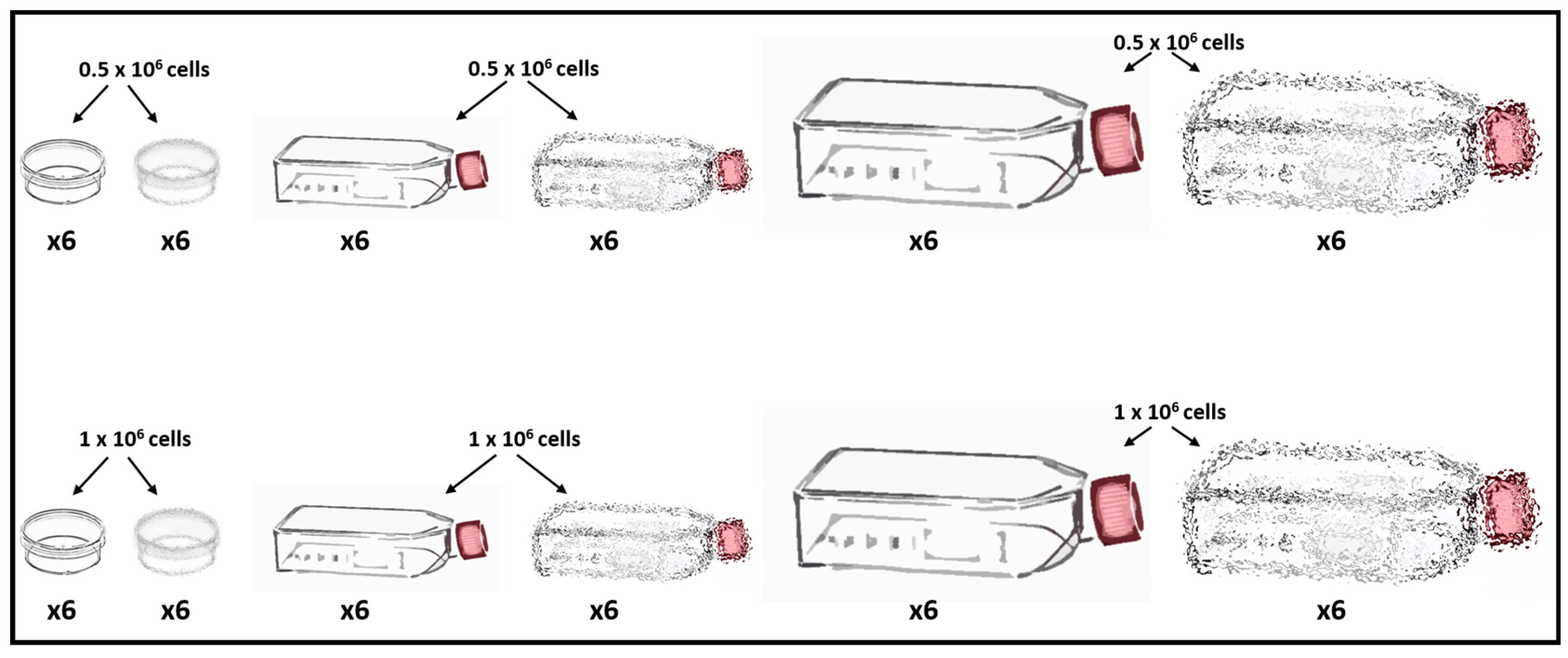
2.2. Experimental Design
2.3. Cell Counting
2.4. Thraustochytrid Cell Characterization
2.5. DNA Extraction, PCR Conditions and Phylogenetic Trees
3. Results
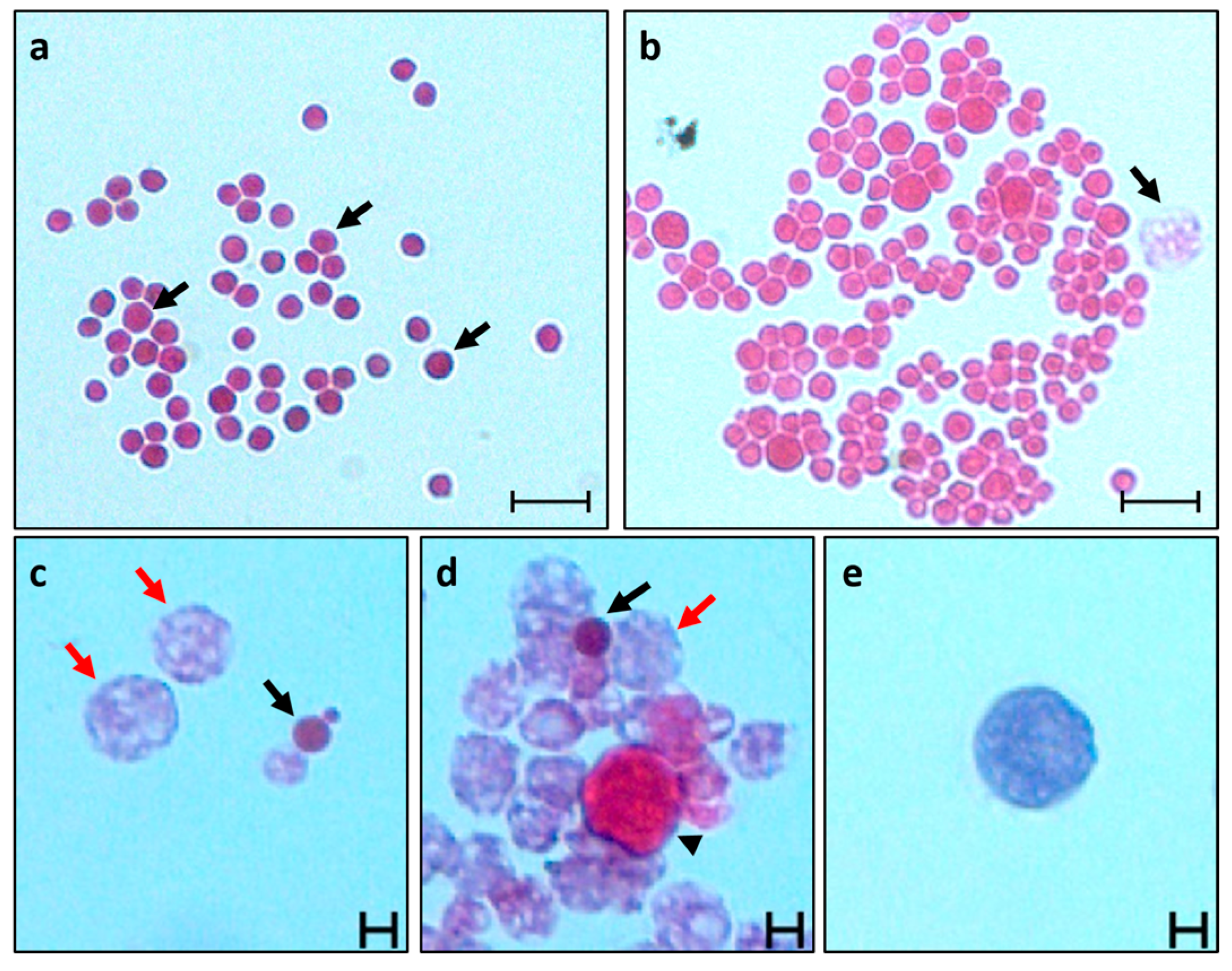
3.1. Emerged Thraustochytrid Cells in B. schlosseri Primary Cultures
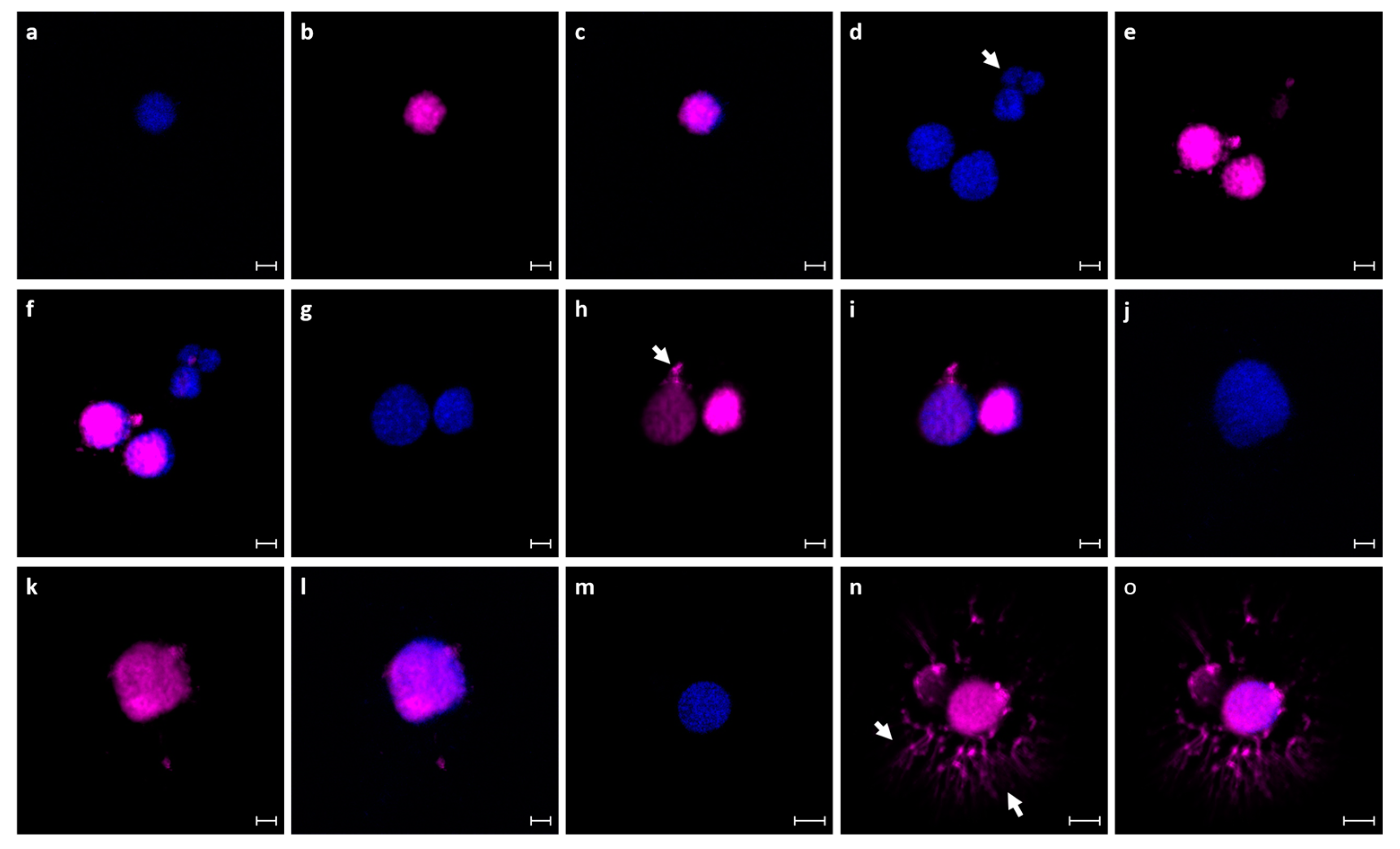
3.2. Thraustochytrid Primary Culture Characteristics
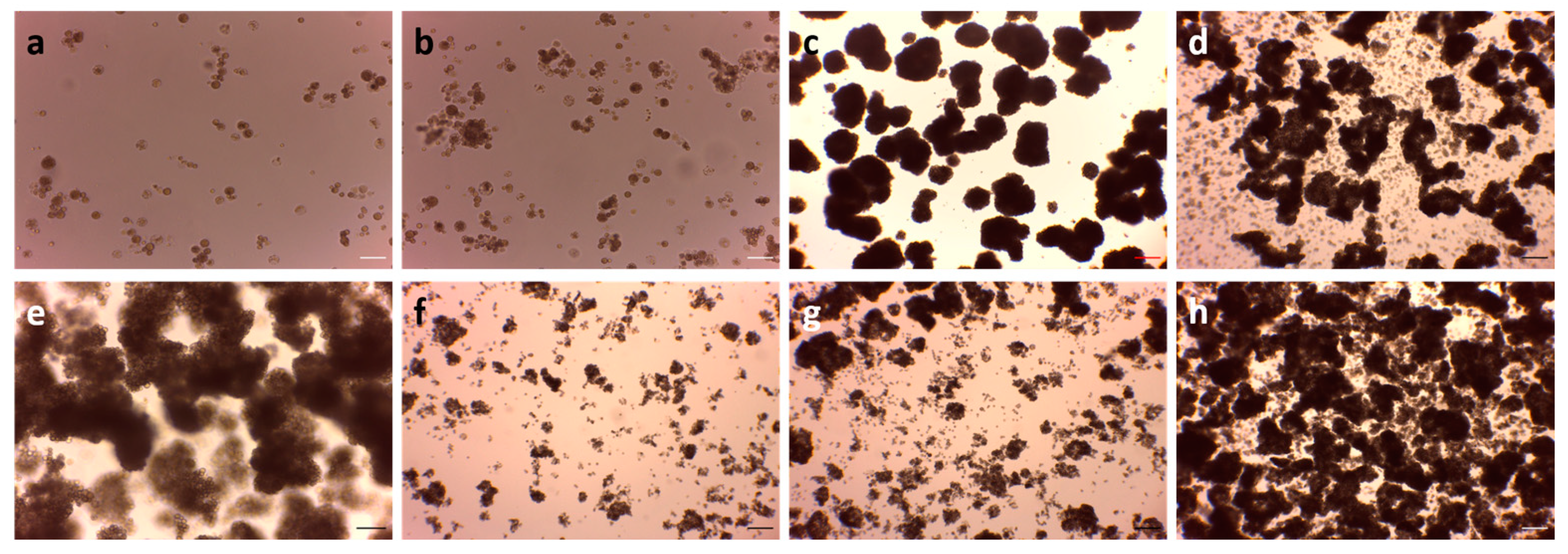
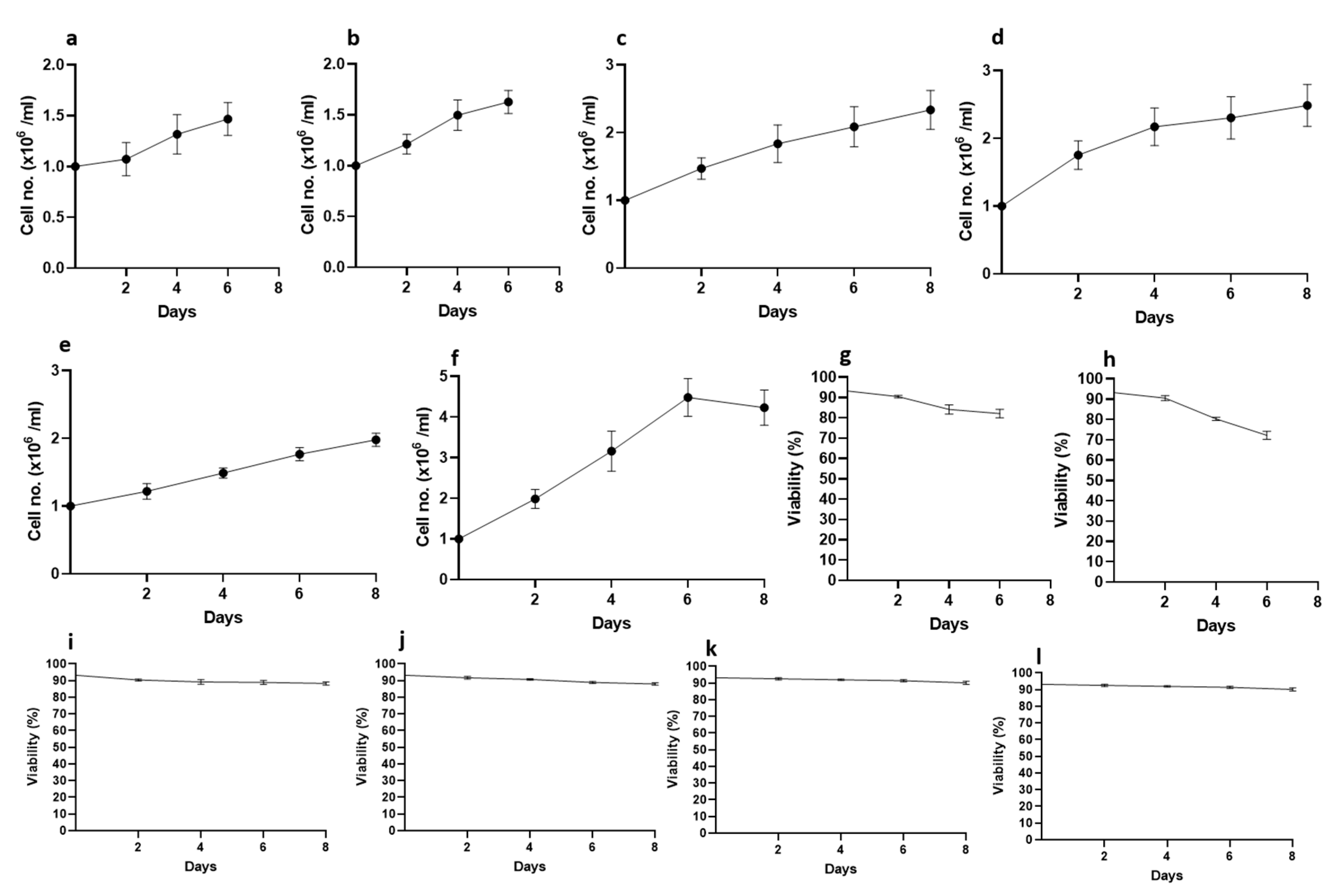
3.3. Thraustochytrid Culture Growth under Various Maintenance Conditions
3.4. Molecular Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ellenbogen, B.B.; Aaronson, S.; Goldstein, S.; Belsky, M. Polyunsaturated fatty acids of aquatic fungi: Possible phylogenetic significance. Comp. Biochem. Physiol. 1969, 29, 805–811. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Allsopp, M.T.E.P.; Chao, E.E. Thraustochytrids are chromists, not fungi: 18s rRNA signatures of Heterokonta. Philos. Trans. R. Soc. Lond. B 1994, 346, 387–397. [Google Scholar] [CrossRef]
- Honda, D.; Yokochi, T.; Nakahara, T.; Raghukumar, S.; Nakagiri, A.; Schaumann, K.; Takanori, H. Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18s ribosomal RNA gene. J. Eukaryot. Microbiol. 1999, 46, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Tsui, C.K.M.; Marshall, W.; Yokoyama, R.; Honda, D.; Lippmeier, J.C.; Craven, K.D.; Peterson, P.D.; Berbee, M.L. Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol. Phylogenetics Evol. 2009, 50, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Rinkevich, B. A simple, reliable and fast protocol for thraustochytrids DNA extraction. Mar. Biotechnol. 2001, 3, 100–102. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Douek, J.; Rinkevich, B. Development of a PCR strategy for thraustochytrids identification based on 18S-rDNA sequence. Mar. Biol. 2002, 140, 883–889. [Google Scholar] [CrossRef]
- Marchan, L.F.; Chang, K.J.L.; Nichols, P.D.; Mitchell, W.J.; Polglase, J.L.; Gutierrez, T. Taxonomy, ecology and biotechnological applications of thraustochytrids: A review. Biotechnol. Adv. 2018, 36, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Strassert, J.F.; Jamy, M.; Mylnikov, A.P.; Tikhonenkov, D.V.; Burki, F. New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote tree of life. Mol. Biol. Evol. 2019, 36, 757–765. [Google Scholar] [CrossRef]
- Lyu, L.; Wang, Q.; Wang, G. Cultivation and diversity analysis of novel marine thraustochytrids. Mar. Life Sci. Technol. 2021, 3, 263–275. [Google Scholar] [CrossRef]
- Chang, K.J.L.; Nichols, C.M.; Blackburn, S.I.; Dunstan, G.A.; Koutoulis, A.; Nichols, P.D. Comparison of thraustochytrids Aurantiochytrium sp., Schizochytrium sp., Thraustochytrium sp., and Ulkenia sp. for production of biodiesel, long-chain omega-3 oils, and exopolysaccharide. Mar. Biotechnol. 2014, 16, 396–411. [Google Scholar] [CrossRef]
- Liu, Y.; Singh, P.; Liang, Y.; Li, J.; Xie, N.; Song, Z.; Daroch, M.; Leng, K.; Johnson, Z.I.; Wang, G. Abundance and molecular diversity of thraustochytrids in coastal waters of southern China. FEMS Microbiol. Ecol. 2017, 93, fix070. [Google Scholar] [CrossRef] [PubMed]
- Qarri, A.; Rinkevich, Y.; Rinkevich, B. Employing marine invertebrate cell culture media for isolation and cultivation of thraustochytrids. Bot. Mar. 2021, 64, 447–454. [Google Scholar] [CrossRef]
- Bai, M.; Sen, B.; Wen, S.; Ye, H.; He, Y.; Zhang, X.; Wang, G. Culturable diversity of Thraustochytrids from coastal waters of Qingdao and their fatty acids. Mar. Drugs 2022, 20, 229. [Google Scholar] [CrossRef]
- Raghukumar, S. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur. J. Protistol. 2002, 38, 127–145. [Google Scholar] [CrossRef]
- Raghukumar, S.; Damare, V.S. Increasing evidence for the important role of labyrinthulomycetes in marine ecosystems. Bot. Mar. 2011, 54, 3–11. [Google Scholar] [CrossRef]
- Naganuma, T.; Takasugi, H.; Kimura, H. Abundance of thraustochytrids in coastal plankton. Mar. Ecol. Prog. Ser. 1998, 162, 105–110. [Google Scholar] [CrossRef]
- Porter, D. Phylum Labryinthulomycota. In Handbood of Protoctista, 3nd ed.; Margulis, L., Corliss, J.O., Melkonian, M., Chapman, D.J., Eds.; Jones and Bartlett: Boston, MA, USA, 1990; pp. 388–398. [Google Scholar]
- Mass, P.A.Y.; Kleinschuster, S.J.; Dykstra, M.J.; Smolowitz, R.; Parent, J. Molecular characterization of QPX (quahog parasite unknown), a pathogen of Mercenaria mercenaria. J. Shellfish. Res. 1999, 18, 561–567. [Google Scholar]
- Polglase, J.L. A preliminary report on the thraustochytrid(s) and labyrinthulid(s) associated with a pathological condition in the lesser octopus Eledone cirrhosa. Bot. Mar. 1980, 23, 699–706. [Google Scholar] [CrossRef]
- Grasela, J.J.; Pomponi, S.A.; Rinkevich, B.; Grima, J. Efforts to develop a cultured sponge cell line: Revisiting an intractable problem. Vitr. Cell. Dev. Biol. -Anim. 2012, 48, 12–20. [Google Scholar] [CrossRef]
- Rinkevich, B.; Frank, U.; Gateño, D.; Rabinowitz, C. The establishment of various cell lines from colonial marine invertebrates. In Use of Aquatic Invertebrates as Tools for Monitoring of Environmental Hazards; Mueller, W.E.G., Ed.; Gustav Fischer Verlag: Stuttgart, Germany, 1994; pp. 253–263. [Google Scholar]
- Rabinowitz, C.; Douek, J.; Weisz, R.; Shabtay, A.; Rinkevich, B. Isolation and characterization of four novel thraustochytrid strains from a colonial tunicate. IJMS 2006, 35, 341–350. [Google Scholar]
- Garcia-Vedrenne, A.E.; Groner, M.; Page-Karjian, A.; Siegmund, G.F.; Singhal, S.; Sziklay, J.; Roberts, S. Development of genomic resources for a thraustochytrid pathogen and investigation of temperature influences on gene expression. PLoS ONE 2013, 8, e74196. [Google Scholar] [CrossRef]
- Rinkevich, B. Cell cultures from marine invertebrates: Obstacles, new approaches and recent improvements. J. Biotechnol. 1999, 70, 133–153. [Google Scholar] [CrossRef]
- Lewis, T.E.; Nichols, P.D.; McMeekin, T.A. The biotechnological potential of thraustochytrids. Mar. Biotechnol. 1999, 1, 580–587. [Google Scholar] [CrossRef]
- Raghukumar, S. Thraustochytrid marine protists: Production of PUFAs and other emerging technologies. Mar. Biotechnol. 2008, 10, 631–640. [Google Scholar] [CrossRef]
- Gupta, A.; Barrow, C.J.; Puri, M. Omega-3 biotechnology: Thraustochytrids as a novel source of omega-3 oils. Biotechnol. Adv. 2012, 30, 1733–1745. [Google Scholar] [CrossRef]
- Rotter, A.; Barbier, M.; Bertoni, F.; Bones, A.M.; Cancela, M.L.; Carlsson, J.; Carvalho, M.F.; Cegłowska, M.; Chirivella-Martorell, J.; Conk Dalay, M.; et al. A new network for the advancement of marine biotechnology in Europe and beyond. Front. Mar. Sci. 2020, 7, 278. [Google Scholar] [CrossRef]
- de La Broise, D.; Ventura, M.; Chauchat, L.; Guerreiro, M.; Michez, T.; Vinet, T.; Gautron, N.; Le Grand, F.; Bideau, A.; Goïc, N.L.; et al. Scale-up to pilot of a non-axenic culture of thraustochytrids using digestate from methanization as nitrogen source. Mar. Drugs 2022, 20, 499. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Liu, Y.; Li, L.; Wang, G. Ecological dynamics and biotechnological implications of Thraustochytrids from marine habitats. Appl. Microbiol. Biotechnol. 2014, 98, 5789–5805. [Google Scholar] [CrossRef]
- Aasen, I.M.; Ertesvåg, H.; Heggeset, T.M.B.; Liu, B.; Brautaset, T.; Vadstein, O.; Ellingsen, T.E. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. App. Microbiol. Biotechnol. 2016, 100, 4309–4321. [Google Scholar] [CrossRef]
- Hong, W.K.; Heo, S.Y.; Park, H.M.; Kim, C.H.; Sohn, J.H.; Kondo, A.; Seo, J.W. Characterization of a squalene synthase from the thraustochytrid microalga Aurantiochytrium sp. krs101. J. Microbiol. Biotechnol. 2013, 23, 759–765. [Google Scholar] [CrossRef]
- Barta, D.G.; Coman, V.; Vodnar, D.C. Microalgae as sources of omega-3 polyunsaturated fatty acids: Biotechnological aspects. Algal Res. 2021, 58, 102410. [Google Scholar] [CrossRef]
- Pradeepkiran, J.A. Aquaculture role in global food security with nutritional value: A review. Transl. Anim. Sci. 2019, 3, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Fukada, H.; Renato, K.; Junpei, S.; Haruka, M.; Toshiro, M. Effects of complete replacement of fish oil with plant oil mixtures and algal meal on growth performance and fatty acid composition in juvenile yellowtail Seriola quinqueradiata. Fish. Sci. 2020, 86, 107–118. [Google Scholar] [CrossRef]
- Lee Chang, K.J.; Parrish, C.C.; Simon, C.J.; Revill, A.T.; Nichols, P.D. Feeding Whole Thraustochytrid Biomass to Cultured Atlantic Salmon (Salmo salar) Fingerlings: Culture Performance and Fatty Acid Incorporation. J. Mar. Sci. Eng. 2020, 8, 207. [Google Scholar] [CrossRef]
- Davies, S.J.; Roderick, E.; Brudenell-Bruce, T.; Bavington, C.D.; Hartnett, F.; Hyland, J.; de Souza Valente, C.; Wan, A.H.L. Delivering a nutritionally enhanced tilapia fillet using a pre-harvest phase omega-3 thraustochytrids protist enriched diet. Eur. J. Lipid Sci. Technol. 2022, 124, 2100153. [Google Scholar] [CrossRef]
- Turchini, G.M.; Bente, E.T.; Wing-Keong, N. Fish oil replacement in finfish nutrition. Rev. Aquac. 2009, 1, 10–57. [Google Scholar] [CrossRef]
- Miller, M.R.; Nichols, P.D.; Carter, C.G. Replacement of fish oil with thraustochytrid Schizochytrium sp. L. oil in Atlantic salmon parr (Salmo salar L.) diets. Comp. Biochem. Physiol. Part A 2007, 148, 382–392. [Google Scholar] [CrossRef]
- Qarri, A.; Kültz, D.; Gardell, A.M.; Rinkevich, B.; Rinkevich, Y. Improved Media Formulations for Primary Cell Cultures Derived from a Colonial Urochordate. Cells 2023, 12, 1709. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating the human-ape split by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3327. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Burja, A.M.; Radianingtyas, H.; Windust, A.; Barrow, C.J. Isolation and characterization of polyunsaturated fatty acid producing Thraustochytrium species: Screening of strains and optimization of omega-3 production. Appl. Microbiol. Biotechnol. 2006, 72, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Morabito, C.; Bournaud, C.; Maës, C.; Schuler, M.; Aiese Cigliano, R.; Dellero, Y.; Maréchal, E.; Amato, A.; Rébeillé, F. The lipid metabolism in thraustochytrids. Prog. Lipid Res. 2019, 76, 101007. [Google Scholar] [CrossRef]
- Nham Tran, T.L.; Miranda, A.F.; Gupta, A.; Puri, M.; Ball, A.S.; Adhikari, B.; Mouradov, A. The nutritional and pharmacological potential of new Australian thraustochytrids isolated from mangrove sediments. Mar. Drugs 2020, 18, 151. [Google Scholar] [CrossRef]
- FioRito, R.; Leander, C.; Leander, B. Characterization of three novel species of Labyrinthulomycota isolated from ochre sea stars (Pisaster ochraceus). Mar. Biol. 2016, 163, 170. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qarri, A.; Rinkevich, Y.; Douek, J.; Sardogan, A.; Rinkevich, B. Growth Performance of a Newly Isolated and Culturable Thraustochytrid Strain from Sea Squirt Colonies. Fishes 2024, 9, 22. https://doi.org/10.3390/fishes9010022
Qarri A, Rinkevich Y, Douek J, Sardogan A, Rinkevich B. Growth Performance of a Newly Isolated and Culturable Thraustochytrid Strain from Sea Squirt Colonies. Fishes. 2024; 9(1):22. https://doi.org/10.3390/fishes9010022
Chicago/Turabian StyleQarri, Andy, Yuval Rinkevich, Jacob Douek, Aydan Sardogan, and Baruch Rinkevich. 2024. "Growth Performance of a Newly Isolated and Culturable Thraustochytrid Strain from Sea Squirt Colonies" Fishes 9, no. 1: 22. https://doi.org/10.3390/fishes9010022
APA StyleQarri, A., Rinkevich, Y., Douek, J., Sardogan, A., & Rinkevich, B. (2024). Growth Performance of a Newly Isolated and Culturable Thraustochytrid Strain from Sea Squirt Colonies. Fishes, 9(1), 22. https://doi.org/10.3390/fishes9010022





