Abstract
Enterocytozoon hepatopenaei (EHP), a microsporidian responsible for hepatopancreatic microsporidiosis, is a major pathogen in commercial shrimp production. Among the affected species, Palaemon carinicauda (formerly Exopalaemon carinicauda) is commercially important in China and represents a potential research model for studying crustaceans. However, little information is available on its response to EHP infection. Hence, this study analyzed the transcriptome and metabolome of P. carinicauda’s hepatopancreas using high-throughput sequencing and liquid chromatograph-mass spectrometry (LC-MS) to determine its response during the early stage of infection. The transcriptomic analyses identified 730 differentially expressed genes, of which those associated with EHP infection were enriched in metabolic pathways as well as detoxification and antioxidant pathways. In addition, 144 differential metabolites were identified using a combination of positive and negative ion modes in LC-MS. The Kyoto encyclopedia of genes and genomes pathway analyses further indicated that the degradation of aromatic compounds, the AMP-activated protein kinase signaling pathway and C5-branched dibasic acid metabolism were significantly enriched after EHP infection. These results could provide useful insights into the effects of EHP on shrimps during the early stages of infection and help to understand the mechanisms underlying the stunted growth of shrimps after infection.
1. Introduction
Infectious diseases represent the main factor that limits shrimp cultivation around the world, and for Asian countries in particular, they have been reported to induce shrimp mortality or retarded growth during aquaculture [1]. In this context, hepatopancreatic microsporidiosis (HPM), caused by Enterocytozoon hepatopenaei (EHP), is currently among the main diseases which affect shrimp production [2]. EHP is a microsporidian parasite that was first reported in 2004 and officially characterized and named in 2009; this microsporidian has become a prominent pathogen which infects the hepatopancreas of hosts such as ridgetail white prawn (Palaemon carinicauda) (formerly Exopalaemon carinicauda), blue shrimp (Penaeus stylirostris), Pacific whiteleg shrimp (P. vannamei) and black tiger shrimp (P. monodon) to cause HPM [3,4,5,6,7]. EHP can spread rapidly via spores or by cannibalism of infected shrimp. Under laboratory conditions, shrimp can be successfully infected after being exposed to the infected tissue homogenate for 48 h [2]. However, unlike other infectious diseases of shrimp which have visible clinical signs, infection by EHP does not result in mass mortality. Instead, during the later stages of infection, it slows down growth and causes significant variations in shrimp size, thus leading to severe economic losses which threaten sustainable shrimp aquaculture [8].
The EHP-induced growth retardation of shrimps has been attracting the attention of researchers for several years; subsequently, efforts have been devoted to revealing the underlying mechanisms involved in the process. More recently, transcriptomic, proteomic and metabolic studies of EHP-infected shrimps showed that the infection process was associated with immune protein/gene expression, growth and energy synthesis pathways, and this could partly explain the slowed growth observed in infected shrimps [9,10,11]. A previous study has also shown that the survival of EHP-infected shrimps could be three months, with retarded growth generally visible from the second month after infection [8]. This finding is an indication that the host’s response might be different at different stages of infection. To the best of our knowledge, most studies have focused on the latter stages of the infection, where growth retardation is visible. However, little information is available on how hosts respond during the early infection stage, which could provide important information about the mechanisms of growth retardation and host response. In order to identify the significant genes and pathways involved in the response to EHP infection during the early stage, the application of omics-based approaches for the analysis of shrimp hepatopancreas may be useful. Such data would be valuable not only to identify the biological processes involved in EHP infection, but also to understand the mechanisms behind growth retardation.
The Palaemon carinicauda, also referred to as ridgetail white prawn, is assigned to the family of Palaemonidae within the suborder Pleocyemata, and it is one of the most commercially important shrimps in East China [12,13]. In addition, because of its transparent body, rapid growth, short reproductive cycle and strong adaptability, P. carinicauda is widely used as a model organism in crustacean research [14].
In this work, to investigate the molecular response of P. carinicauda after EHP infection, the infection status of shrimps (EHP-infected vs. uninfected) was first confirmed by PCR and through the examination of EHP spores. This was followed by transcriptomic and metabolomic profiling to identify differentially expressed genes and metabolites between the two groups of shrimps during the early stage of infection. Eventually, the expression patterns of genes in the hepatopancreas were verified by quantitative real-time PCR (qRT-PCR). It is expected that the results of the current study will provide useful information on the genes and signaling pathways that respond to EHP infection. Moreover, a deep understanding of the mechanism involved in the growth retardation of shrimps as a result of infection by intracellular pathogens.
2. Materials and Methods
2.1. Experiment Design and Sample Collection
Healthy P. carinicauda (body weight: 1.54 ± 0.22 g; body length: 4.95 ± 0.26 cm) were provided by commercial shrimp ponds in Lianyungang, Jiangsu Province, China. PCR tests were performed on animals to check for the presence of EHP prior to the start of the experiment, with only negative ones selected for subsequent steps. Shrimps (n = 180) in the treatment group (in triplicate), were initially given EHP-infected tissues to get infected and then fed with commercial feed during the whole experiment, while for the control group (n = 180) only commercial feed was provided. The experiment was carried out under controlled conditions (temperature: 26 °C, salinity: 27‰, pH: 7.8 ± 0.3), and 14 days after initiating infection, the shrimps were sampled and the seawater was wiped off their surfaces before collecting individual hepatopancreas. For each group, to reduce the sampling error caused by individual differences, five hepatopancreas were combined into a single sample, with three and six replicates used for transcriptomic and metabolomic analyses, respectively. After collection, each sample was immediately frozen in liquid nitrogen for storage at −80 °C until required for the extracting of RNA and metabolites.
2.2. PCR and Spore Examination for Detecting EHP
The EasyPure Genomic DNA Kit, EE101 (TransGen, Beijing, China) was used, as specified by the manufacturer, to extract total DNA from the collected hepatopancreas samples. This was followed by PCR amplification using the following primers to detect EHP: Forward: 5′-GCCTGAGAGATGGCTCCCACGT-3′; Reverse: 5′-GCGTACTATCCCCAGAGCCCGA-3′ [15]. In this case, amplification was carried out in 25 μL reaction volumes with a 2 × Taq PCR MasterMix II kit, KT201 (TransGen, Beijing, China); the PCR conditions were as follows: 95 °C for 5 min, followed by 35 cycles, each with an initial denaturation for 30 s at 95 °C, annealing for 30 s at 60 °C and extension for 30 s at 72 °C. The amplification process ended with a final extension at 72 °C for 10 min. The resulting PCR products were then examined by electrophoresis on 1.2% agarose gels to determine the infection status of shrimps.
To isolate EHP spores, the hepatopancreas of shrimps from the experimental group was mixed with a phosphate buffered solution and ground completely. After filtration, the spores were separated from the homogenate by performing differential centrifugation three times, including centrifugation at 2000 r/min for 10 min and centrifugation at 3000 r/min for 20 min followed by 8000 r/min for 10 min. Then the crude spore’s supernatant was obtained. After that, 30, 60 and 90% sucrose density gradient centrifugation was performed to isolate spores, and spores from different layers were collected. A small drop (about 10 μL) of the purified spores was placed on a microscope slide followed by the addition of 5 μL of Phloxin B. The slide was incubated at room temperature for 10 min, and observed under a light microscope [16].
2.3. Assembly of Transcriptomic Data and Analysis of Differentially Expressed Genes
For transcriptome analysis, total RNA extraction was carried out from EHP-infected (T-H) and control hepatopancreas samples (CK-H) using a Trizol reagent kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and checked by performing RNase free agarose gel electrophoresis. After that, samples were sent to the Gene denovo Company, Guangzhou, China, where a cDNA library was constructed prior to sequencing on an Illumina NovaSeq 6000 platform as previously described [17]. All RNA-Seq data were submitted to the NCBI SRA database (BioProject number: PRJNA903220). The transcriptome data were analyzed, including assembly and annotation. Briefly, the transcript was assembled using the StringTie software (version 1.3.4) program [18]. Functions were annotated with the assembled unigenes by comparing similarity levels using the BLAST algorithm against the following databases: COG/KOG, SwissProt and Nr (e-value < 0.00001). This was followed by the analysis of enriched Gene Ontology (GO) terms (http://bioinfo.cau.edu.cn/agriGO/, accessed on 15 March 2022) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (https://www.kegg.jp/kegg/pathway.html, accessed on 15 March 2022).
For each transcription region, an FPKM (fragment per kilobase of transcript per million fragments mapped) value was calculated to quantify its expression abundance and variations using StringTie software. RNAs differential expression analysis was performed using the DESeq2 software (version 1.22.2) with a comparison of two different groups [19]. Differentially expressed genes (DEGs) were then identified; the parameter of absolute Log2 FC (fold change) value of ≥ 1 and a False Discovery Rate (FDR) of < 0.05. Moreover, the expression patterns of eight vital genes were validated by qRT-PCR (Table S1).
2.4. Metabolite Extraction and Liquid Chromatograph-Mass Spectrometry Analysis
The samples were homogenized, and then suspended in 500 μL of prechilled 80% methanol. After that, the samples were kept on ice for 5 min prior to centrifugation for 20 min at 15,000× g and 4 °C. Part of the supernatant was then diluted in liquid chromatograph-mass spectrometry (LC-MS) grade water to a final concentration containing 53% methanol. After transferring the samples to a new centrifuge tube, centrifugation was again performed as before, with the resulting extracted metabolites eventually analyzed by LC-MS [20].
LC-MS was carried out using a Vanquish UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA), coupled with an Orbitrap Q ExactiveTM HF-X mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), at Gene Denovo Co., Ltd., Guangzhou, China.
2.5. Differential Metabolites Identification
A variable importance in projection (VIP) score of orthogonal projections to latent structures (OPLS) model was applied to rank the metabolites that best distinguished between two groups. The threshold of VIP was set to 1. In addition, the t-test was also used to provide a univariate analysis for screening differential metabolites. Those with a p value of t-test < 0.05 and VIP ≥ 1 were considered differential metabolites between two groups.
2.6. Quantitative Real-Time PCR
The total RNA isolated from the hepatopancreas (n =3 ) of the EHP-infected and control groups was used to synthesize cDNA using the PrimeScript® RT Reagent Kit, RR037 (TaKaRa, Shiga, Japan), according to the manufacturer’s protocols. The amplification was carried out using the StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and the SYBR Premix Ex Taq (TaKaRa, Shiga, Japan), with 18S rRNA selected as the reference gene [21]. The relative expression levels of target genes were eventually calculated with the 2–ΔΔCt method [22].
2.7. Statistical Analysis
All data were analyzed by performing one-way ANOVA, followed by Duncan’s test in SPSS software (version 21.0), with differences considered to be statistically significant at p < 0.05.
3. Results
3.1. Detection of EHP Infection in Shrimps
Fourteen days after initiating infection, PCR tests were carried out on shrimps from the EHP-infected and control groups. For the control group, the absence of electrophoretic bands was noted, while for the EPH-infected group, the target band was observed (Figure 1A). Spore examination was also carried out for the two groups, with a large number of EHP spores isolated only from the hepatopancreas of infected shrimps (Figure 1B).
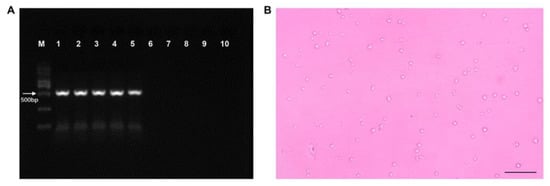
Figure 1.
Detection of EHP infection of Palaemon carinicauda. (A) PCR detection of EHP in P. carinicauda. M, marker; 1–5, positive samples; 6–10, negative samples. (B) EHP spores collected from EHP-infected shrimp, scale bar 20 μm.
3.2. Transcriptomic Alteration of P. carinicauda in Response to EHP Infection
RNA sequencing of hepatopancreas samples from CK-H and T-H was performed to identify the DEGs responsible for EHP infection in P. carinicauda. After the removal of low-quality sequences, an average of 40,934,471 and 44,962,924 reads was obtained for each sample from the CK-H and T-H groups, respectively. Overall, a total of 69,322 unigenes were identified for the two groups, with an N50 of 1645 bp as well as an average length of 862 bp for all unigenes, indicating that the sequencing and assembly were of a high quality (Table S2). Of all expressed genes, 730 DEGs (421 upregulated and 309 downregulated) in the hepatopancreas were found in the T-H group in comparison with the CK-H group (Table S3).
A functional annotation of the DEGs involved in EHP infection was performed by GO and KEGG analyses. In this case, all DEGs were significantly enriched in three GO terms (molecular function, cellular component and biological process) (Figure S1), and of those classified as biological processes, the main subcategories included single-organism, cellular and metabolic processes. More specifically, the DEGs were mainly involved in the “carboxylic acid metabolic process” (GO: 0019752), “oxoacid metabolic process” (GO: 0043436), “organic acid biosynthetic process” (GO: 0016053), “organic acid metabolic process” (GO: 006082) and “carboxylic acid biosynthetic process” (GO: 0046394) (Figure S2). Similarly, among the KEGG pathways which were significantly enriched as a result of EHP infection, the top 20 included the metabolic pathways, glycolysis/gluconeogenesis, starch and sucrose metabolism, carbon metabolism, biosynthesis of amino acids, pentose phosphate pathway and metabolism of xenobiotics by cytochrome P450 (Figure 2). Changes were also noted in other pathways, such as arginine and proline metabolism, in the fatty acid metabolism and the HIF-1 signaling pathway after EHP infection (Figure 2).
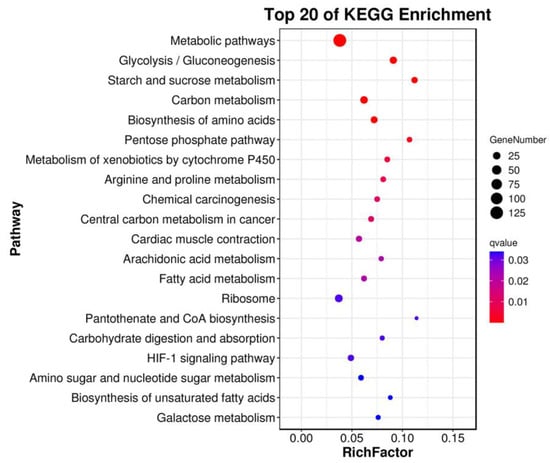
Figure 2.
The most enriched pathways of the differentially expressed genes in the hepatopancreas of Palaemon carinicauda after EHP infection.
In this study, some DEGs that were associated with metabolism were identified (Table 1). In particular, the heat map of the gene expression profiles for the T-H and CK-H groups revealed that genes related to enzymes involved in carbohydrate metabolism (e.g., GAPDH, PFKM, PKM, Aldoa, GPI and LDHA) were significantly down-regulated. In contrast, those involved in lipid metabolism, including ACAC, Scda2 and Fasn, were up-regulated (Figure 3).

Table 1.
Changes in metabolism-related differentially expressed genes in the hepatopancreas of Palaemon carinicauda in response to EHP infection.
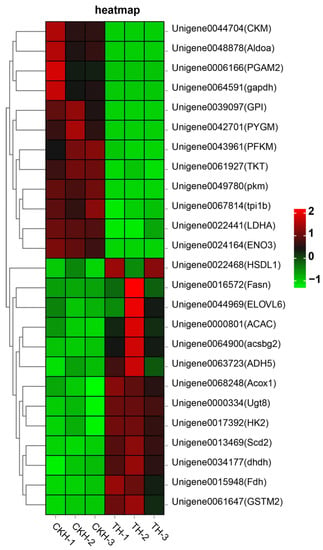
Figure 3.
Heatmap of significantly differentially expressed genes involved in metabolism pathways after EHP infection.
In addition, the RNA-Seq data were validated using the qRT-PCR method; for this purpose, eight metabolism-related DEGs were analyzed by qRT-PCR. The results were consistent with those obtained by RNA-seq, thereby confirming the reliability of the RNA-seq analysis (Figure 4).
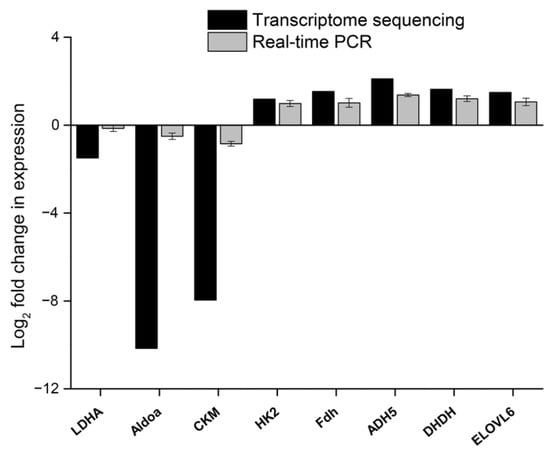
Figure 4.
Expression of eight differentially expressed genes from the transcriptome validated by qRT-PCR.
3.3. Identification of Differential Metabolites in P. carinicauda after EHP Infection
To further study the effects of EHP infection on metabolism, the differential metabolites in the hepatopancreas of sampled shrimps were analyzed with LC-MS. In this case, orthogonal projections to latent structures-discriminant analysis (OPLS-DA) was used to compare the control and EHP-infected groups (Figure 5); after 200 permutations, the R2 and Q2 were 0.996 and 0.789, respectively, in the positive ion mode (Figure 5A), while in the negative ion mode, they were 0.989 and 0.648, respectively, indicating that the established model was stable and reliable (Figure 5B).
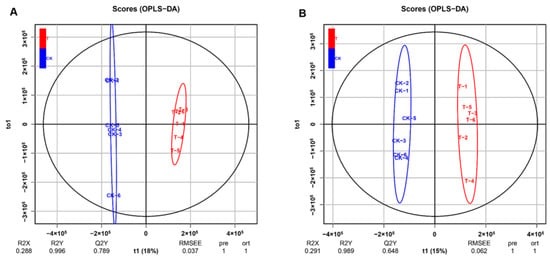
Figure 5.
Multivariate statistical analysis in positive and negative ion mode. (A) Orthogonal projections to latent structures-discriminant analysis (OPLS-DA) in positive mode between CK-H and T-H groups. (B) OPLS-DA in negative mode between CK-H and T-H groups.
LC-MS analysis revealed that in the positive ion model, 113 metabolites (50 upregulated and 63 downregulated metabolites) were significantly different in EHP-infected samples in comparison with the control (VIP > 1, p ≤ 0.05). On the other hand, in the negative ion model, 31 differential metabolites were identified (18 upregulated and 13 downregulated metabolites) (Table 2). These differential metabolites were involved in 77 metabolic pathways (Table S4). According to the enrichment analysis of KEGG pathways, they were mainly involved in the degradation of aromatic compounds, AMP-activated protein kinase (AMPK) signaling, C5-branched dibasic acid metabolism and thiamine metabolism (Figure 6A). Overall, the heatmap of significant differential metabolites showed obvious increases/decreases after EHP infection (Figure 6B).

Table 2.
Statistics of differential metabolites.
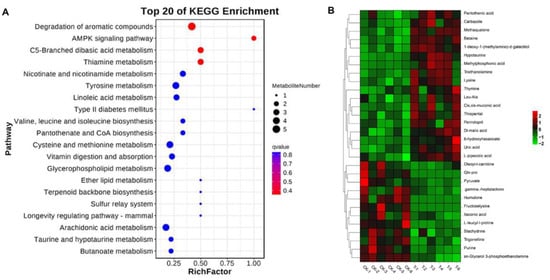
Figure 6.
Identification of significantly differential metabolites in Palaemon carinicauda after EHP infection. (A) Enriched metabolic pathways associated with EHP infection. (B) Heatmap of 30 significantly differential metabolites after EHP infection (VIP scores of the top 15 metabolites in positive and negative ion mode).
4. Discussion
EHP currently represents a major pathogen of shrimp culture in Asia [2], especially for species such as the Pacific white leg shrimp, the black tiger shrimp and the ridgetail white prawn, which are among the main hosts of EHP. The ridgetail white prawn is an economically valuable cultured shrimp species in East China as well as a widely used model organism in crustacean research [12,13,14]. Thus, this species was selected in this study to investigate changes in gene expression and metabolism during the early stage of EHP infection.
The hepatopancreas is an important organ in crustaceans due to its key role in energy metabolism, and after infecting this organ, EHP causes HPM, a disease characterized by slowed growth. However, based on practical experience and previous experiments [8], it is now known that slowed growth is not visible until the second month of infection. Previous transcriptomic and proteomic studies comparing healthy shrimps and those showing slowed growth revealed that the energy metabolism and immune genes/proteins expression were associated with EHP infection [9,10,11]. The results of this study, based on -omics analyses, also suggested that similar pathways, especially metabolic ones, were altered after EHP infection (Figure 2). In particular, the fact that “Metabolic processes” accounted for most of the enriched GO terms indicated that, during the early stages of EHP infection, shrimp metabolism was significantly changed. Furthermore, the infection process also altered the expression pattern of genes in P. carinicauda’s hepatopancreas, with the greatest changes noted for carbohydrate and lipid metabolism (i.e., “starch and sucrose metabolism”, “glycolysis/gluconeogenesis” and “fatty acid metabolism”) (Figure 3). The results therefore showed that infection by EHP was likely to affect energy metabolism in shrimps.
Carbohydrates are important sources of energy for organisms, with glucose catabolism being the main way through which they obtain energy [10]. The current study found that the relative expression of many important enzyme-encoding genes such as PKM, PFKM, GPI and GAPDH was significantly downregulated 14 days after EHP infection. This was a clear indication that EHP infection affected energy metabolism in shrimps. Although a number of additional genes related to glucose metabolism were also downregulated in the hepatopancreas, those involved in lipid metabolism were actually upregulated. For example, results from the RNA-seq analysis showed that Acox1, ACAC, Scd2 and Fasn were significantly upregulated after EHP infection, with acyl-coenzyme A oxidase (Acox1) and acetyl-CoA carboxylase (ACAC) involved in the synthesis and β-oxidation of fatty acids, respectively, being key enzymes in lipid metabolism [23,24]. Hence, the significantly upregulated genes were indicative of the fact that EHP infection enhanced lipid metabolism during the early stages. This was in contrast to results regarding the latter stages of infection, whereby the expression of lipid metabolism-related genes was reported to be downregulated [11]. Moreover, based on the results, it was speculated that the interaction between the host and the pathogen was a dynamic process which could differ between the early and later stages of the infection. In addition, since EHP lacks mitochondria, it is unable to carry out oxidative phosphorylation and glycolysis to produce ATP; as a result, it depends on the host for energy [25,26]. During the early stages of infection, the need to produce ATP as an energy source is not only essential for normal cellular functions but also for EHP replication. In order to meet those energy demands, it is likely that lipid metabolism was induced for energy production. Scd2, encoding stearoyl-CoA desaturase (SCD) that is associated with the biosynthesis of unsaturated fatty acids, catalyzes the conversion of saturated fatty acyl-CoAs to unsaturated ones [27,28]. The activity of this enzyme affects the ratio of monosaturated fatty acid (MUFA) to saturated fatty acid (SFA), and this subsequently influences lipid metabolism and membrane fluidity [29]. While many factors can regulate SCD expression, this study showed that EHP infection was among the factors that significantly upregulated its gene (Scd2) to induce lipid metabolism and increase the cell membrane’s fluidity.
In addition to energy metabolism-related genes, those associated with detoxification and antioxidants, including glutathione S-transferases (GST), were also upregulated during the early stage of EHP infection (Figure 3). GSTs are important detoxifying enzymes which catalyze the conjugation of glutathione to electrophilic centers of lipophilic compounds to make it more soluble and help in its excretion from the cell [30]. The increased mRNA level of GST in the hepatopancreas indicated that the detoxification mechanism in shrimps was activated after EHP infection to prevent damage to proteins, lipids and DNA.
Metabolic analyses also revealed significant changes in the metabolic biomarkers of the AMPK pathway (Figure 6). The AMPK is an essential metabolic sensor that maintains an energy balance in response to stress [31]. A previous study has shown that infectious diseases could trigger AMPK signaling cascades which subsequently regulated various catabolic and anabolic processes in response to altered energy levels [32,33]. These processes eventually modulated the host’s responses, resulting in either enhanced pathogen survival or induced host defenses in a context-dependent manner. In line with the above transcriptome analyses, it might be plausible that EHP infection caused metabolic stress which triggered AMPK signaling to maintain energy homeostasis. Although there is not enough information on the relationship between EHP and shrimp in terms of energy metabolism, it is clear that the AMPK signaling pathway does play an important role during the early stages of EHP infection.
Based on the above analysis, we speculate that the disorder of energy metabolism might be the major cause of slowed growth in EHP-infected shrimp. Many genes related to glucose and lipid metabolism exhibited significant changes after EHP infection; in addition, the AMPK pathway that maintains energy homeostasis was also affected after EHP infection. Although transcriptomic and metabolomics can provide significant evidence in interpreting the mechanisms of growth retardation in EHP-infected shrimps, further studies are needed to clarify the effect of EHP infection on physiology in P. carinicauda. For example, an analysis of enzymes related to energy metabolism would be useful.
5. Conclusions
In this study, we investigated the molecular responses induced by EHP at the transcript and metabolite levels in the hepatopancreas of P. carinicauda. The results indicated that EHP infection could induce detoxification and antioxidant processes, as well as disturb the energy metabolism in P. carinicauda. Furthermore, EHP infection could trigger the AMPK signaling pathway to maintain energy balance during the early stages of infection. These results not only provide useful insights into the interactions between EHP and shrimps but also help to understand the mechanisms underlying the stunted growth of shrimps following EHP infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8020092/s1, Figure S1: GO annotation of the unigenes in the EHP infected shrimp transcriptome; Figure S2: The most enriched GO terms of the DEGs in the hepatopancreas after EHP infection; Table S1: Primers used for qRT-PCR in this study; Table S2: Statistics of assembly quality; Table S3: Information of differentially expressed genes; Table S4: The enriched pathways of differential metabolites after EHP infection.
Author Contributions
Conceptualization, G.H., W.W. and K.X.; methodology, G.H., K.X. and N.J.; investigation, J.X., C.W. and D.L.; writing—original draft preparation, G.H. and N.J.; writing—review and editing, G.H., K.X. and N.J.; supervision, G.H.; project administration, B.Y.; funding acquisition, G.H. and H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Jiangsu Province (#BK20191007), the Natural Science Foundation of China (#31900370), the Lianyungang 521 Talent Projects (#2021-1021), the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS (2021)124) and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Jiangsu Ocean University.
Informed Consent Statement
Not applicable.
Data Availability Statement
In this study, all data generated are included in this article and Supplementary Materials. Further enquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Seethalakshmi, P.S.; Rajeev, R.; Kiran, G.S.; Selvin, J. Shrimp disease management for sustainable aquaculture: Innovations from nanotechnology and biotechnology. Aquacult. Int. 2021, 29, 1591–1620. [Google Scholar] [CrossRef]
- Karthikeyan, K.; Sudhakaran, R. Experimental horizontal transmission of Enterocytozoon hepatopenaei in post-larvae of whiteleg shrimp, Litopenaeus vannamei. J. Fish. Dis. 2019, 42, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Chayaburakul, K.; Nash, G.; Pratanpipat, P.; Sriurairatana, S.; Withyachumnarnkul, B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Organ. 2004, 60, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Hu, J.; Shen, H.; Deng, G.; Gao, W.; Mu, H.; Zhang, Q.; Gao, H. Effect of Enterocytozoon hepatopenaei infection on the intestinal microflora of Exopalaemon carinicauda. Prog. Fish. Sci. 2022, 43, 75–83. [Google Scholar]
- Tang, K.F.J.; Pantoja, C.R.; Redman, R.M.; Han, J.E.; Tran, L.H.; Lightner, D.V. Development of in situ hybridization and PCR assays for the detection of Enterocytozoon hepatopenaei (EHP), a microsporidian parasite infecting penaeid shrimp. J. Invertebr. Pathol. 2015, 130, 37–41. [Google Scholar] [CrossRef]
- Tang, K.F.J.; Han, J.E.; Aranguren, L.F.; White-Noble, B.; Schmidt, M.M.; Piamsomboon, P.; Risdiana, E.; Hanggono, B. Dense populations of the microsporidian Enterocytozoon hepatopenaei (EHP) in feces of Penaeus vannamei exhibiting white feces syndrome and pathways of their transmission to healthy shrimp. J. Invertebr. Pathol. 2016, 140, 1–7. [Google Scholar] [CrossRef]
- Chaijarasphong, T.; Munkongwongsiri, N.; Stentiford, G.D.; Aldama-Cano, D.J.; Thansa, K.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. The shrimp microsporidian Enterocytozoon hepatopenaei (EHP): Biology, pathology, diagnostics and control. J. Invertebr. Pathol. 2021, 186, 107458. [Google Scholar] [CrossRef]
- Kumar, T.S.; Praveena, P.E.; Sivaramakrishnan, T.; Rajan, J.J.S.; Makesh, M.; Jithendran, K.P. Effect of Enterocytozoon hepatopenaei (EHP) infection on physiology, metabolism, immunity, and growth of Penaeus vannamei. Aquaculture 2022, 553, 738105. [Google Scholar] [CrossRef]
- Yang, L.-G.; Wang, Y.; Wang, Y.; Fang, W.-H.; Feng, G.-P.; Ying, N.; Zhou, J.-Y.; Li, X.-C. Transcriptome analysis of pacific white shrimp (Penaeus vannamei) intestines and hepatopancreas in response to Enterocytozoon hepatopenaei (EHP) infection. J. Invertebr. Pathol. 2021, 186, 107665. [Google Scholar] [CrossRef]
- Ning, M.; Wei, P.; Shen, H.; Wan, X.; Jin, M.; Li, X.; Shi, H.; Qiao, Y.; Jiang, G.; Gu, W. Proteomic and metabolomic responses in hepatopancreas of whiteleg shrimp Litopenaeus vannamei infected by microsporidian Enterocytozoon hepatopenaei. Fish. Shellfish. Immun. 2019, 87, 534–545. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, H.; Wang, J.; Zeng, S.; Wang, Y.; Mo, Z.; Dan, X.; Li, Y. Response signatures of Litopenaeus vannamei to natural Enterocytozoon hepatopenaei infection revealed by the integration of the microbiome and transcriptome. Aquaculture 2021, 542, 736885. [Google Scholar] [CrossRef]
- Li, J.; Ma, P.; Liu, P.; Chen, P.; Li, J. The roles of Na+/K+-ATPase α-subunit gene from the ridgetail white prawn Exopalaemon carinicauda in response to salinity stresses. Fish. Shellfish. Immun. 2015, 42, 264–271. [Google Scholar] [CrossRef]
- Xu, W.; Xie, J.; Shi, H.; Li, C. Hematodinium infections in cultured ridgetail white prawns, Exopalaemon carinicauda, in eastern China. Aquaculture 2010, 300, 25–31. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Gui, T.; Sun, Z.; Xiang, J. A copper-induced metallothionein gene from Exopalaemon carinicauda and its response to heavy metal ions. Int. J. Biol. Macromol. 2014, 70, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.-H.; Yu, J.-Y.; Wang, J.-J.; Li, T.; Chang, L.-R.; Fang, Y.; Yan, D.-C. Development of a PCR assay for the effective detection of Enterocytozoon hepatopenaei (EHP) and investigation of EHP prevalence in Shandong Province, China. J. Invertebr. Pathol. 2021, 184, 107653. [Google Scholar] [CrossRef]
- Aldama-Cano, D.J.; Sanguanrut, P.; Munkongwongsiri, N.; Ibarra-Gámez, J.C.; Itsathitphaisarn, O.; Vanichviriyakit, R.; Flegel, T.W.; Sritunyalucksana, K.; Thitamadee, S. Bioassay for spore polar tube extrusion of shrimp Enterocytozoon hepatopenaei (EHP). Aquaculture 2018, 490, 156–161. [Google Scholar] [CrossRef]
- Xiao, J.; Li, Q.-Y.; Tu, J.-P.; Chen, X.-L.; Chen, X.-H.; Liu, Q.-Y.; Liu, H.; Zhou, X.-Y.; Zhao, Y.-Z.; Wang, H.-L. Stress response and tolerance mechanisms of ammonia exposure based on transcriptomics and metabolomics in Litopenaeus vannamei. Ecotox. Environ. Safe. 2019, 180, 491–500. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Bio. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Zhang, P.; Hua, S.; Li, Y.; Zhang, S.; Liu, X.; Shi, T.; Wang, P.; Yan, B.; Li, J.; Gao, H. Expression of the cyclin-dependent kinase 2 gene (cdk2) influences ovarian development in the ridgetail white prawn, Exopalaemon carinicauda. Aquacult. Rep. 2022, 25, 101265. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, X.F.; Tian, M.X.; Sun, R.Q.; Zhang, M.L.; Zhou, L.S.; Jin, L.; Chen, L.L.; Zhou, W.J.; Duan, K.L.; Chen, Y.J. SIRT 5 inhibits peroxisomal ACOX 1 to prevent oxidative damage and is downregulated in liver cancer. Embo. Rep. 2018, 19, e45124. [Google Scholar] [CrossRef] [PubMed]
- Brownsey, R.W.; Boone, A.N.; Elliott, J.E.; Kulpa, J.E.; Lee, W.M. Regulation of acetyl-CoA carboxylase. Biochem. Soc. T. 2006, 34, 223–227. [Google Scholar] [CrossRef]
- Stentiford, G.D.; Bass, D.; Williams, B.A.P. Ultimate opportunists-the emergent Enterocytozoon group microsporidia. PLoS Pathog. 2019, 15, e1007668. [Google Scholar] [CrossRef]
- Wiredu Boakye, D.; Jaroenlak, P.; Prachumwat, A.; Williams, T.A.; Bateman, K.S.; Itsathitphaisarn, O.; Sritunyalucksana, K.; Paszkiewicz, K.H.; Moore, K.A.; Stentiford, G.D. Decay of the glycolytic pathway and adaptation to intranuclear parasitism within Enterocytozoonidae microsporidia. Environ. Microbiol. 2017, 19, 2077–2089. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, F.S.; Ozols, J. Stearoyl-CoA desaturase, a short-lived protein of endoplasmic reticulum with multiple control mechanisms. Prostag. Leukotr. Ess. 2003, 68, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, F.; Hosseini, V.; Alihemmati, A.; Shaaker, M.; Mosayyebi, G.; Darabi, M.; Mehdizadeh, A. The role of stearoyl-coenzyme a desaturase 1 in liver development, function, and pathogenesis. J. Ren. Hepat. Disord. 2019, 3, 15–22. [Google Scholar] [CrossRef]
- Flowers, M.T.; Ntambi, J.M. Stearoyl-CoA desaturase and its relation to high-carbohydrate diets and obesity. Biochim. Biophys. Acta 2009, 1791, 85–91. [Google Scholar] [CrossRef]
- Vontas, J.G.; Small, G.J.; Hemingway, J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 2001, 357, 65–72. [Google Scholar] [CrossRef]
- Jo, E.-K.; Silwal, P.; Yuk, J.-M. AMPK-targeted effector networks in mycobacterial infection. Front. Microbiol. 2019, 10, 520. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.; Silvestre, R.; Cordeiro-da-Silva, A.; Estaquier, J.; Foretz, M.; Viollet, B. AMP-activated protein kinase as a target for pathogens: Friends or foes? Curr. Drug Targets 2016, 17, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Prantner, D.; Perkins, D.J.; Vogel, S.N. AMP-activated kinase (AMPK) promotes innate immunity and antiviral defense through modulation of stimulator of interferon genes (STING) signaling. J. Biol. Chem. 2017, 292, 292–304. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).