Abstract
The challenges inherent in many newborn screening programmes for medium-chain acyl-CoA dehydrogenase deficiency could be overcome by better use of the available second-line tests. Application of “next generation” technologies could minimize many of the problems generated by the current approach to genetic analysis. (Comment on Maier EM. Int. J. Neonatal Screen. 2015, 1, 79–88)To the Editor
The success of a screening programme rests on the ability to define the condition in question and to optimize the balance between sensitivity and specificity. Newborn screening for medium-chain acyl-CoA dehydrogenase deficiency (MCADD) is based initially on the detection of increased octanoylcarnitine using tandem mass-spectometry. This is an unspecific marker and reliable second-tier tests are essential. Maier describes the difficulties that arise from using comprehensive analysis of the ACADM gene for this. Sensitivity is high but specificity low. Homozygosity for the c.985A>G mutation is the most common genotype in MCADD in North-West Europe, constituting 80% of clinically-diagnosed cases in England but only 53% of screening-detected cases [1]. In a wider survey [2] only six of the 36 ACADM gene variants detected in screening-positive cases had also been identified in the clinically-presenting disorder making the presence of two variant alleles unsuitable as the sole diagnostic criterion.
Alternative approaches are available. Derks et al. suggest that measurement of MCAD activity in leukocytes or lymphocytes using phenylpropionyl-CoA as substrate should be the diagnostic gold-standard [3]. However this approach is only practicable if presumptive positive cases can all be referred to a specialist centre with appropriate laboratory facilities. Urine travels better and it contains increased amounts of hexanoylglycine in MCAD deficiency. Hexanoylglycine was reported as “detectable” in 25 of 29 samples collected for confirmation of positive screening results irrespective of whether the genotype was classed as mild or severe [4]. Quantitative analysis by stable-isotope dilution gas chromatography-mass spectrometry is a better discriminant [5]. In the English pilot study [1] only one of 93 screening-positive babies with a genotype previously reported from clinically-affected cases had urinary hexanoylglycine <8 micromol/mmol creatinine. This contrasts with 42% of cases with two ACADM mutations and a genotype of “uncertain” significance (Figure 1).
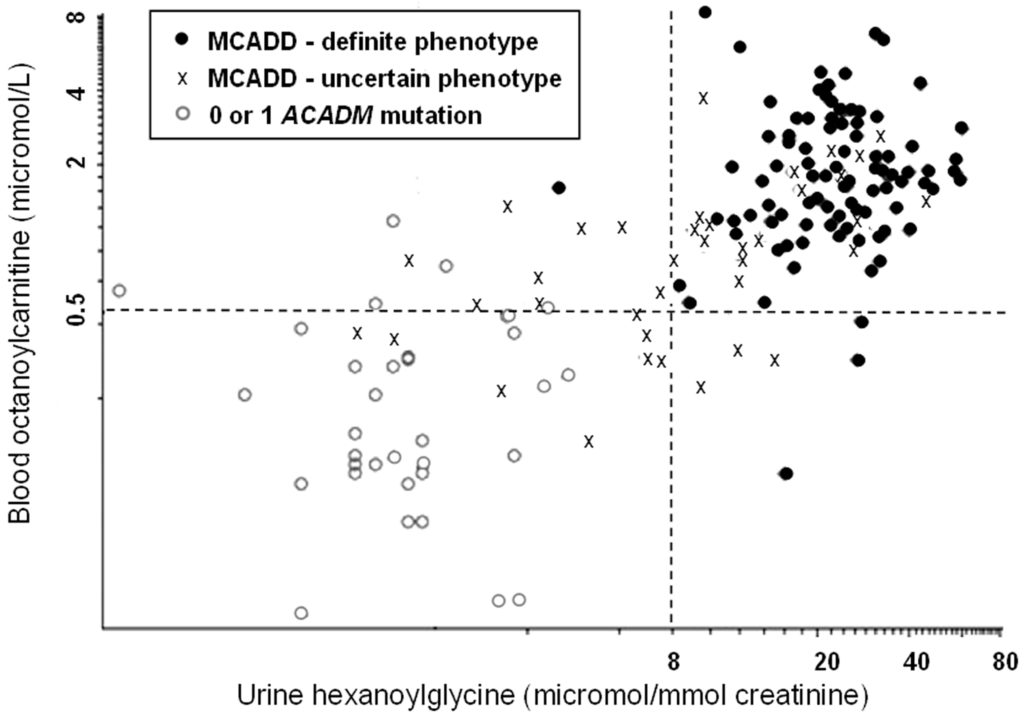
Figure 1.
Metabolites in samples taken at clinical evaluation following a presumptive-positive initial result (blood-spot octanoylcarnitine ≥0.5 micromol/L). Data points are taken from Oerton et al. [1]. The “cut-off” shown for hexanoylglycine is empirical; the upper limit of normal by the method used was 1.1 micromol/mmol creatinine [5].
Despite the difficulties of using the ACADM genotype alone to predict the risk of metabolic decompensation mutation analysis remains a valuable tool. If used selectively, testing specifically for c.985A>G and other “severe” mutations, it would lead directly to a definitive diagnosis in the majority of high-risk cases in North-Western Europe. The remainder would need to be classified according to their urinary hexanoylglycine excretion, preferably with more than one sample, and enzyme analysis in doubtful cases. However, even with selective mutation analysis, a significant number of carriers will be detected. Their parents will need to be informed and counselled appropriately. This is generally been regarded as an undesirable complication and in screening for cystic fibrosis a variety of strategies have been adopted to minimize or totally eliminate it. A generic solution might be “next generation” gene sequencing with a user-interface that reports only homozygotes or compound heterozygotes for the specified set of mutations.
Conflicts of Interest
The author has no conflict of interest.
Abbreviations
| MCAD | Medium-chain acyl-CoA dehydrogenase |
| MCADD | Medium-chain acyl-CoA dehydrogenase deficiency |
References
- Oerton, J.; Khalid, J.M.; Besley, G.; Dalton, R.N.; Downing, M.; Green, A.; Henderson, M.; Krywawych, S.; Leonard, J.; Andresen, B.S.; et al. Newborn screening for medium chain acyl-CoA dehydrogenase deficiency in England: Prevalence, predictive value and test validity based on 1.5 million screened babies. J. Med. Screen. 2011, 4, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, N.; Andresen, B.S.; Pedersen, C.B.; Olsen, R.K.J.; Corydon, T.J.; Bross, P. Mitochondrial fatty acid oxidation defects—remaining challenges. J. Inherit. Metab. Dis. 2008, 31, 643–657. [Google Scholar] [CrossRef] [PubMed]
- Derks, T.G.; Boer, T.S.; van Assen, A.; Bos, T.; Ruiter, J.; Waterham, H.R.; Niezen-Koning, K.E.; Wanders, R.J.A.; Rondeel, J.M.M.; Loeber, J.; et al. Neonatal screening for medium-chain acyl-CoA dehydrogenase (MCAD) deficiency in The Netherlands: The importance of enzyme analysis to ascertain true MCAD deficiency. J. Inherit. Metab. Dis. 2008, 31, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Gramar, G.; Haege, G.; Fang-Hoffmann, J.; Hoffmann, G.F.; Bartram, C.R.; Hinderhofer, K.; Burgard, P.; Lindner, M. Medium-chain acyl-CoA dehydrogenase deficiency: Evaluation of genotype-phenotype correlation in patients detected by newborn screening. JIMD Rep. 2015, 101–112. [Google Scholar]
- Downing, M.; Manning, N.J.; Dalton, R.N.; Krywawych, S.; Oerton, J. Detection of urinary hexanoylglycine in the diagnosis of MCAD deficiency from newborn screening. J. Inherit. Metab. Dis. 2008, 31, 550. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).