Structural Basis for the IgE-Binding Cross-Reacting Epitopic Peptides of Cup s 3, a PR-5 Thaumatin-like Protein Allergen from Common Cypress (Cupressus sempervirens) Pollen
Abstract
1. Introduction
2. Materials and Methods
2.1. Cypress Pollen Extract
2.2. Sera from Allergic Patients
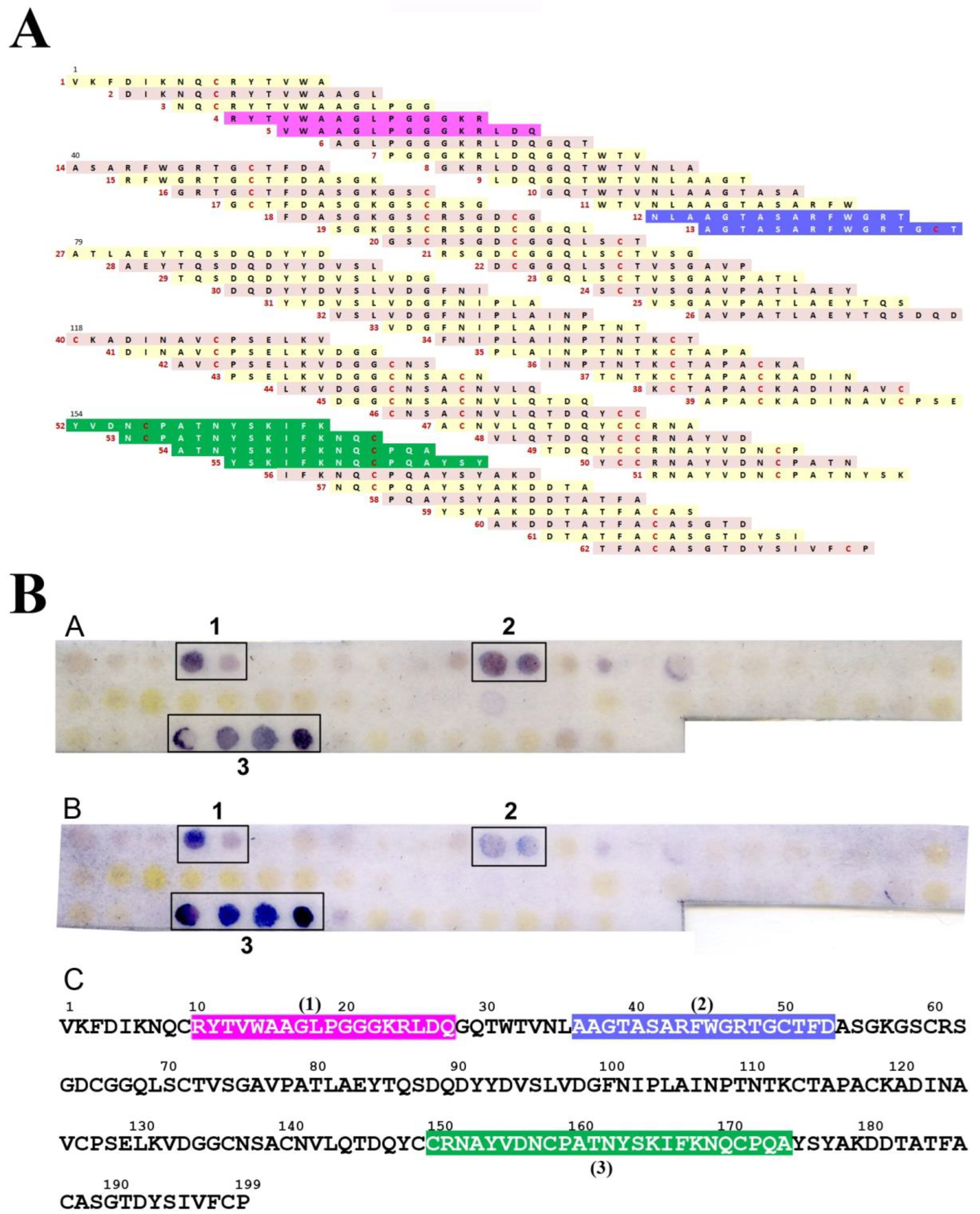
2.3. IgE-Binding Epitope Mapping
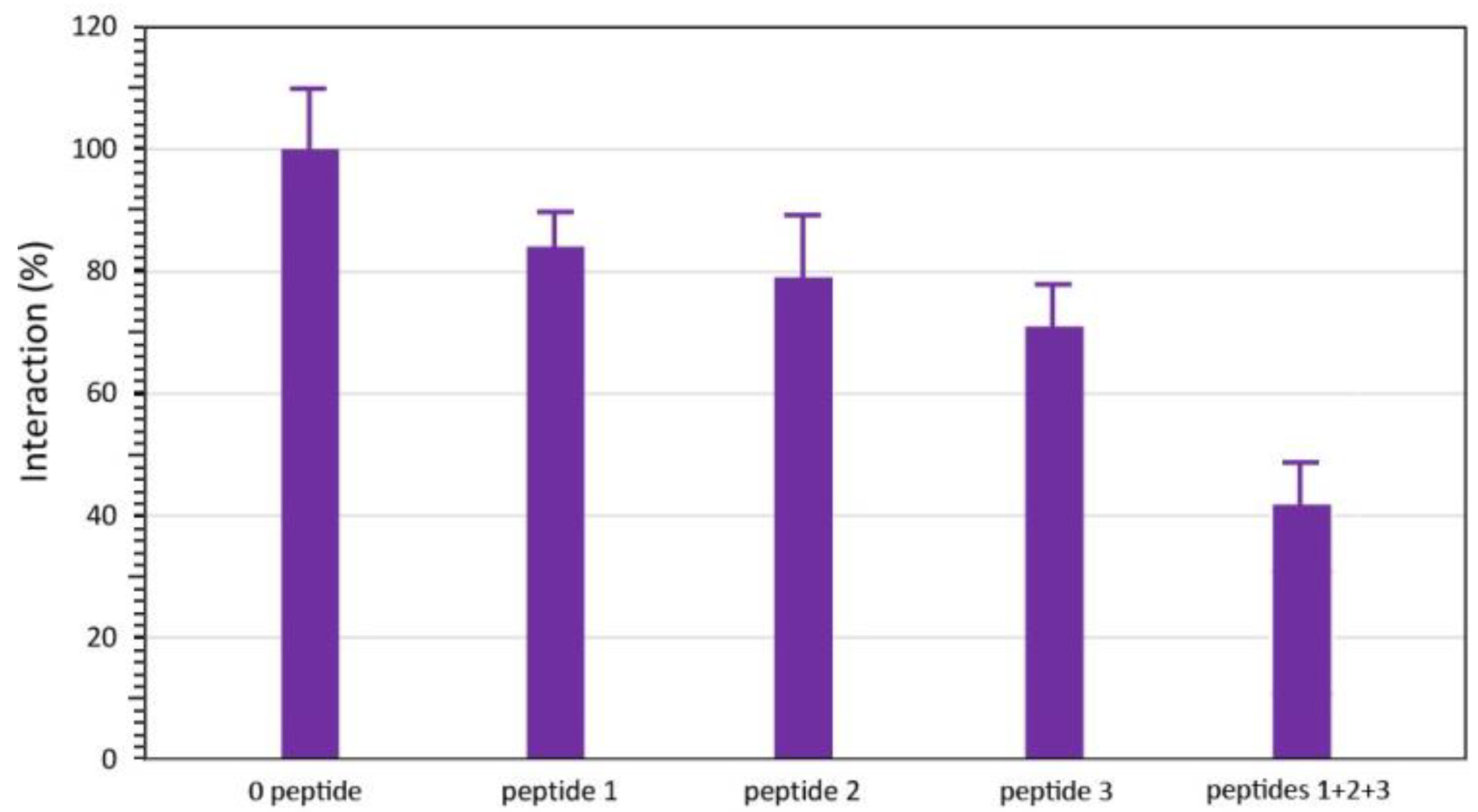
2.4. ELISA Inhibition Experiments
2.5. Bioinformatics
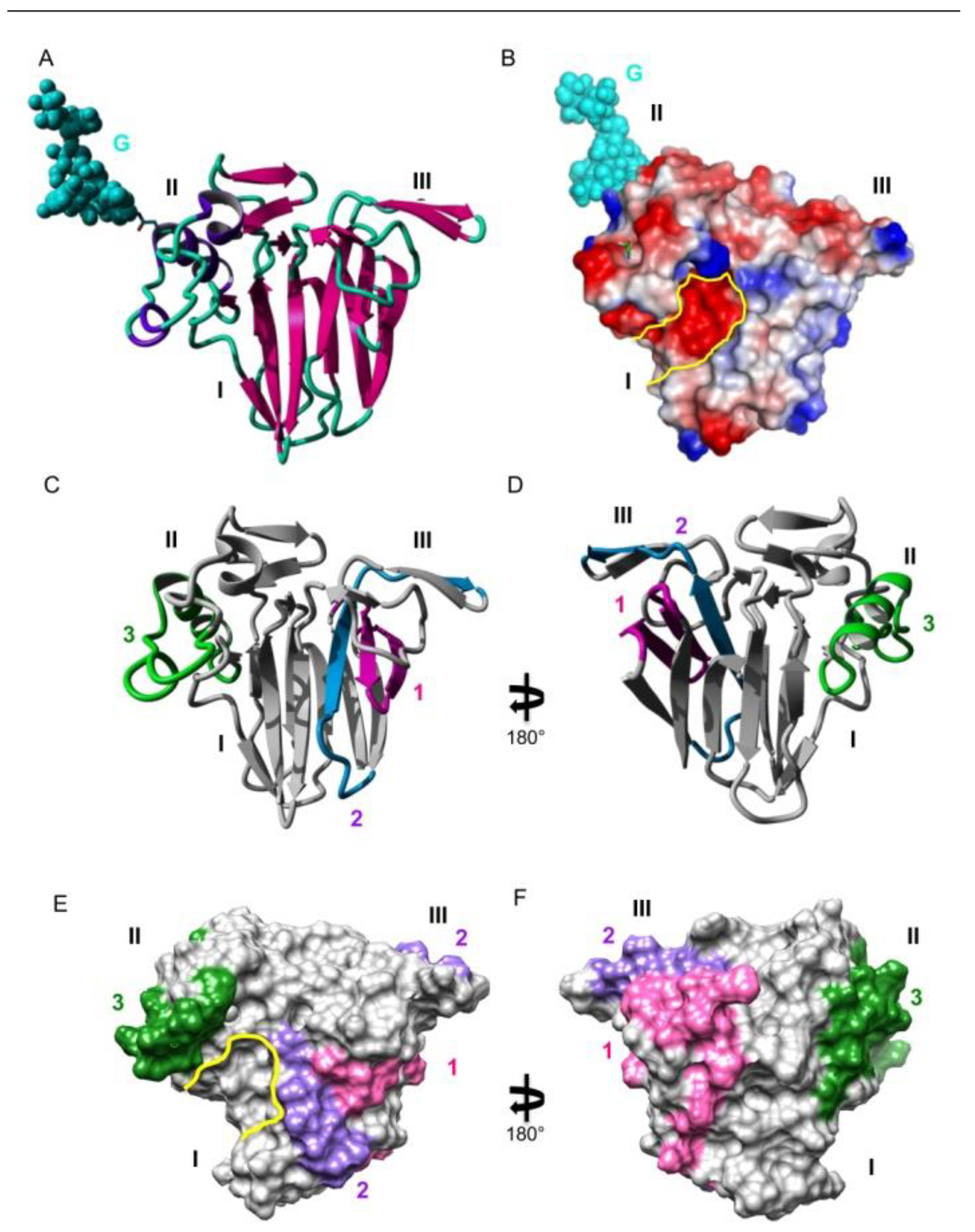
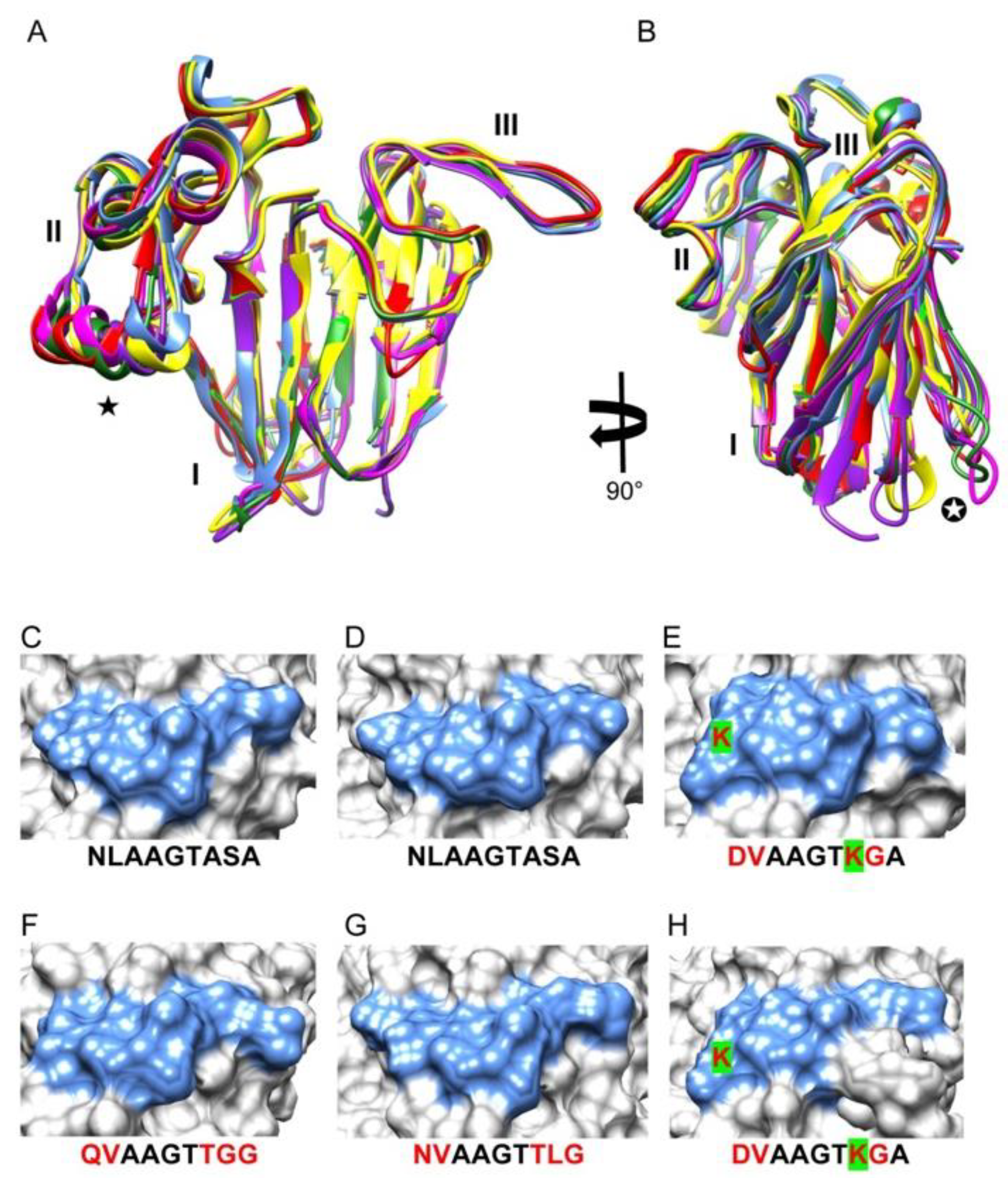
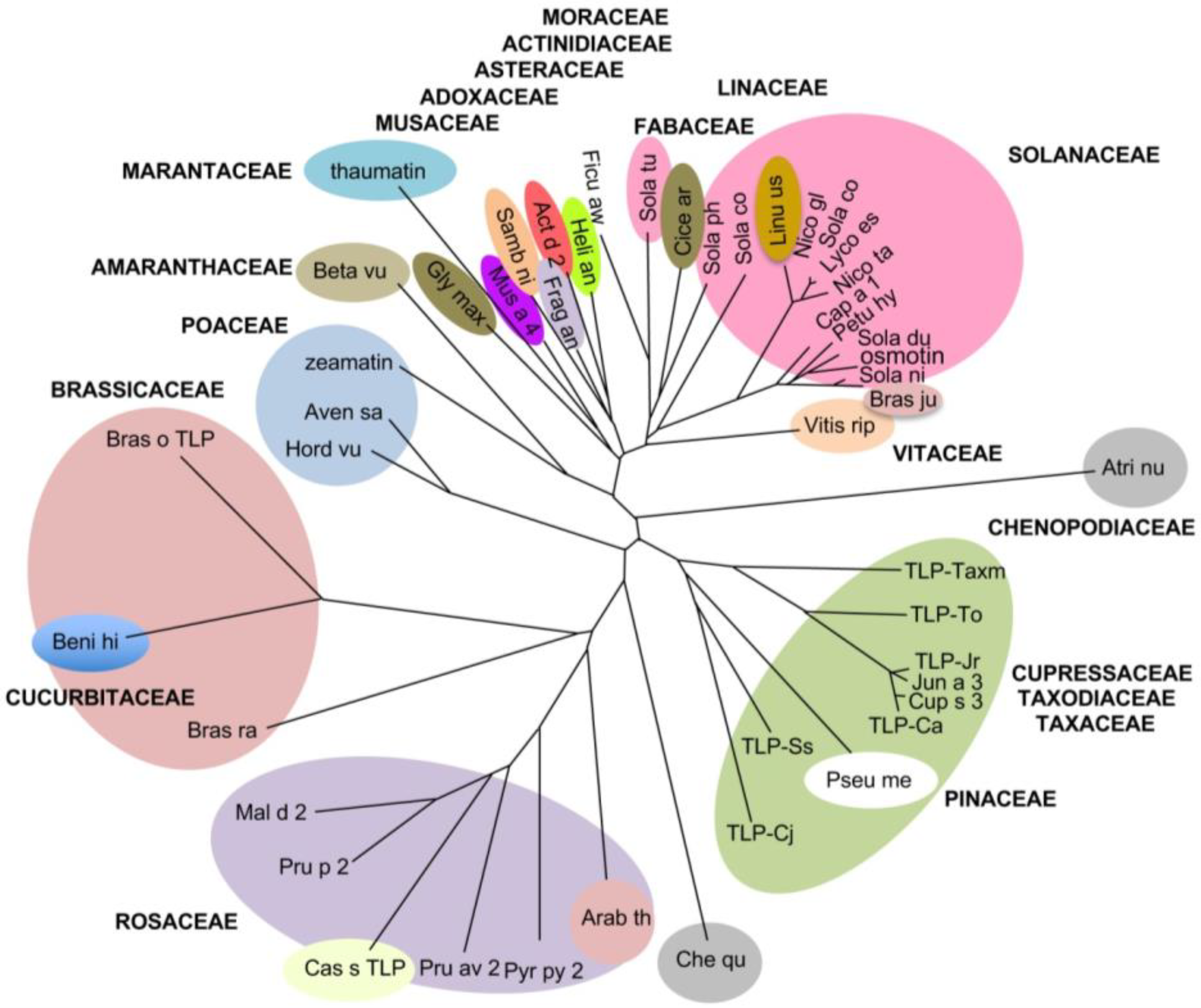
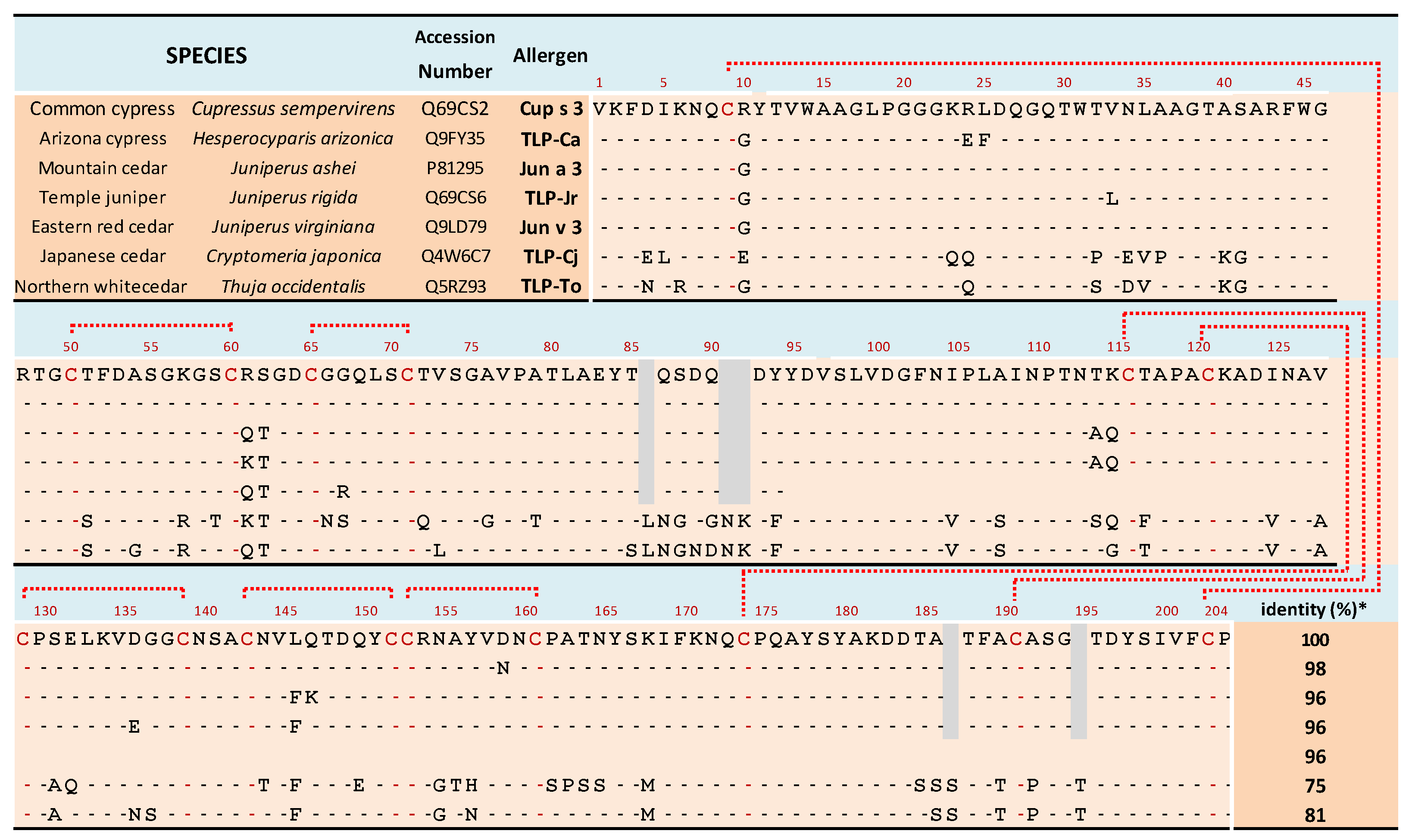
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- D’Amato, G.; Cecchi, L.; Bonini, S.; Nunes, C.; Annesi-Maesano, I.; Behrendt, H.; Liccardi, G.; Popov, T.; Van Cauwenberge, P. Allergenic pollen and pollen allergy in Europe. Allergy 2007, 62, 976–990. [Google Scholar] [CrossRef]
- Bibi, H.; Shoseyov, D.; Feigenbaum, D.; Nir, P.; Shiachi, R.; Scharff, S.; Peled, R. Comparison of positive allergy skin tests among asthmatic children from rural and urban areas living within small geographic area. Ann. Allergy Asthma Immunol. 2002, 88, 416–420. [Google Scholar] [CrossRef]
- Gonianakis, M.I.; Baritaki, M.A.; Neonakis, I.K.; Gonianakis, I.M.; Kypriotakis, Z.; Darivianaki, E.; Bouros, D.; Kontou-Filli, K. A 10-year aerobiological study (1994–2003) in the Mediterranean island of Crete, Greece: Trees, aerobiologic data, and botanical and clinical correlations. Allergy Asthma Proc. 2006, 27, 371–377. [Google Scholar] [CrossRef]
- Pérez-Badia, R.; Rapp, A.; Vaquero, C.; Fernández-González, F. Aerobiological study in east-central Iberian Peninsula: Pollen diversity and dynamics for major taxa. Ann. Agric. Environ. Med. 2011, 18, 99–111. [Google Scholar]
- Lamy, E.; Savournin, C.; Balansard, G. Ten years of cypress pollen counts in Marseille. Allergol. Immunopathol. 2001, 33, 103–104. [Google Scholar]
- Kishikawa, R.; Kotoh, E.; Oshikawa, C.; Soh, N.; Shimoda, T.; Saito, A.; Sahashi, N.; Enomoto, T.; Usami, A.; Teranishi, H.; et al. Longitudinal monitoring of tree airborne pollen in japan. Arerugi 2017, 66, 97–111. [Google Scholar] [PubMed]
- Pham, N.H.; Baldo, B.A.; Bass, D.J. Cypress pollen allergy. Identification of allergens and crossreactivity between divergent species. Clin. Exp. Allergy 1994, 24, 558–565. [Google Scholar] [CrossRef]
- Mohanty, R.P.; Buchheim, M.A.; Levetin, E. Molecular approaches for the analysis of airborne pollen: A case study of Juniperus pollen. Ann. Allergy Asthma Immunol. 2017, 118, 204–211.e202. [Google Scholar] [CrossRef] [PubMed]
- Guerin, B.; Kanny, G.; Terrasse, G.; Guyot, J.; Moneret-Vautrin, D. Allergic Rhinitis to Thuja Pollen. Int. Arch. Allergy Immunol. 1996, 110, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Iacovacci, P.; Afferni, C.; Barletta, B.; Tinghino, R.; Di Felice, G.; Pini, C.; Mari, A. Juniperus oxycedrus: A new allergenic pollen from the Cupressaceae family. J. Allergy Clin. Immunol. 1998, 101, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Sneller, M.R.; Hayes, H.D.; Pinnas, J.L. Pollen changes during five decades of urbanization in Tucson, Arizona. Ann. Allergy 1993, 71, 519–524. [Google Scholar] [PubMed]
- Charpin, D.; Calleja, M.; Pichot, C.; Penel, V.; Hugues, B.; Poncet, P. Cypress pollen allergy. Rev. Mal. Respir. 2013, 30, 868–878. [Google Scholar] [CrossRef]
- Charpin, D.; Pichot, C.; Belmonte, J.; Sutra, J.-P.; Zidkova, J.; Chanez, P.; Shahali, Y.; Sénéchal, H.; Poncet, P. Cypress Pollinosis: From Tree to Clinic. Clin. Rev. Allergy Immunol. 2019, 56, 174–195. [Google Scholar] [CrossRef]
- Cortegano, I.; Civantos, E.; Aceituno, E.; Del Moral, A.; López, E.; Lombardero, M.; Del Pozo, V.; Lahoz, C. Cloning and expression of a major allergen from Cupressus arizonica pollen, Cup a 3, a PR-5 protein expressed under polluted environment. Allergy 2004, 59, 485–490. [Google Scholar] [CrossRef]
- Sénéchal, H.; Šantrůček, J.; Melčová, M.; Svoboda, P.; Zídková, J.; Charpin, D.; Guilloux, L.; Shahali, Y.; Selva, M.-A.; Couderc, R.; et al. A new allergen family involved in pollen food-associated syndrome: Snakin/gibberellin-regulated proteins. J. Allergy Clin. Immunol. 2018, 141, 411–414.e4. [Google Scholar] [CrossRef]
- Iizuka, T.; Takei, M.; Saito, Y.; Rumi, F.; Zheng, J.; Lu, X.; Chafey, P.; Broussard, C.; Guilloux-Assalet, L.; Charpin, D.; et al. Gibberellin-regulated protein sensitization in Japanese cedar (Cryptomeria japonica) pollen allergic Japanese cohorts. Allergy 2021, 76, 2297–2302. [Google Scholar] [CrossRef]
- Ehrenberg, A.E.; Klingebiel, C.; Östling, J.; Larsson, H.; Mattsson, L.; Vitte, J.; Lidholm, J. Characterization of a 7 kDa pollen allergen belonging to the gibberellin-regulated protein family from three Cupressaceae species. Clin. Exp. Allergy 2020, 50, 964–972. [Google Scholar] [CrossRef]
- Sénéchal, H.; Keykhosravi, S.; Couderc, R.; Selva, M.-A.; Shahali, Y.; Aizawa, T.; Busnel, J.-M.; Arif, R.; Mercier, I.; Pham-Thi, N.; et al. Pollen/Fruit Syndrome: Clinical Relevance of the Cypress Pollen Allergenic Gibberellin-Regulated Protein. Allergy Asthma Immunol. Res. 2019, 11, 143–151. [Google Scholar] [CrossRef]
- Takei, M.; Nin, C.; Iizuka, T.; Pawlikowski, M.; Selva, M.-A.; Chantran, Y.; Nakajima, Y.; Zheng, J.; Aizawa, T.; Ebisawa, M.; et al. Capsicum Allergy: Involvement of Cap a 7, a New Clinically Relevant Gibberellin-Regulated Protein Cross-Reactive With Cry j 7, the Gibberellin-Regulated Protein From Japanese Cedar Pollen. Allergy Asthma Immunol. Res. 2022, 14, 328–338. [Google Scholar] [CrossRef]
- Panzani, R.; Yasueda, H.; Shimizu, T.; Shida, T. Cross-reactivity between the pollens of Cupressus sempervirens (common cypress) and of Cryptomeria japonica (Japanese cedar). Ann. Allergy 1986, 57, 26–30. [Google Scholar]
- Breiteneder, H. Thaumatin-like proteins—A new family of pollen and fruit allergens. Allergy 2004, 59, 479–481. [Google Scholar] [CrossRef]
- Ivanciuc, O.; Mathura, V.; Midoro-Horiuti, T.; Braun, W.; Goldblum, R.M.; Schein, C.H. Detecting Potential IgE-Reactive Sites on Food Proteins Using a Sequence and Structure Database, SDAP-Food. J. Agric. Food Chem. 2003, 51, 4830–4837. [Google Scholar] [CrossRef]
- Fujimura, T.; Futamura, N.; Midoro-Horiuti, T.; Togawa, A.; Goldblum, R.M.; Yasueda, H.; Saito, A.; Shinohara, K.; Masuda, K.; Kurata, K.; et al. Isolation and characterization of native Cry j 3 from Japanese cedar (Cryptomeria japonica) pollen. Allergy 2007, 62, 547–553. [Google Scholar] [CrossRef]
- Palacin, A.; Rivas, L.A.; Gómez-Casado, C.; Aguirre, J.; Tordesillas, L.; Bartra, J.; Blanco, C.; Carrillo, T.; Cuesta-Herranz, J.; Bonny, J.A.C.; et al. The Involvement of Thaumatin-Like Proteins in Plant Food Cross-Reactivity: A Multicenter Study Using a Specific Protein Microarray. PLoS ONE 2012, 7, e44088. [Google Scholar] [CrossRef]
- Midoro-Horiuti, T.; Goldblum, R.M.; Kurosky, A.; Wood, T.G.; Brooks, E.G. Variable Expression of Pathogenesis-Related Protein Allergen in Mountain Cedar (Juniperus ashei) Pollen. J. Immunol. 2000, 164, 2188–2192. [Google Scholar] [CrossRef]
- Togawa, A.; Panzani, R.C.; Garza, M.A.; Kishikawa, R.; Goldblum, R.M.; Midoro-Horiuti, T. Identification of Italian cypress (Cupressus sempervirens) pollen allergen Cup s 3 using homology and cross-reactivity. Ann. Allergy Asthma Immunol. 2006, 97, 336–342. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1987, 150, 76–85. [Google Scholar] [CrossRef]
- Barre, A.; Peumans, W.J.; Menu-Bouaouiche, L.; Van Damme, E.J.M.; May, G.D.; Herrera, A.F.; Van Leuven, F.; Rougé, P. Purification and structural analysis of an abundant thaumatin-like protein from ripe banana fruit. Planta 2000, 211, 791–799. [Google Scholar] [CrossRef]
- Tastet, C.; Lescuyer, P.; Diemer, H.; Luche, S.; Van Dorsselaer, A.; Rabilloud, T. A versatile electrophoresis system for the analysis of high- and low-molecular-weight proteins. Electrophoresis 2003, 24, 1787–1794. [Google Scholar] [CrossRef]
- Barre, A.; Sénéchal, H.; Nguyen, C.; Granier, C.; Rougé, P.; Poncet, P. Identification of Potential IgE-Binding Epitopes Contributing to the Cross-Reactivity of the Major Cupressaceae Pectate-Lyase Pollen Allergens (Group 1). Allergies 2022, 2, 106–118. [Google Scholar] [CrossRef]
- Frank, R. Spot-synthesis: An easy technique for the positionally addressable paralel chemical synthesis on a membrane support. Tetrehedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Laune, D.; Molina, F.; Ferrières, G.; Villard, S.; Bès, C.; Rieunier, F.; Chardès, T.; Granier, C. Application of the Spot method to the identification of peptides and amino acids from the antibody paratope that contribute to antigen binding. J. Immunol. Methods 2002, 267, 53–70. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Leone, P.; Menu-Bouaouiche, L.; Peumans, W.J.; Payan, F.; Barre, A.; Roussel, A.; Van Damme, E.; Rougé, P. Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-Å. Biochimie 2006, 88, 45–52. [Google Scholar] [CrossRef]
- Marangon, M.; Van Sluyter, S.C.; Waters, E.J.; Menz, R.I. Structure of Haze Forming Proteins in White Wines: Vitis vinifera Thaumatin-Like Proteins. PLoS ONE 2014, 9, e113757. [Google Scholar] [CrossRef] [PubMed]
- Batalia, M.A.; Monzingo, A.F.; Ernst, S.; Roberts, W.; Robertus, J.D. The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family. Nat. Struct. Biol. 1996, 3, 19–23. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Menu-Bouaouiche, L.; Vriet, C.; Peumans, W.J.; Barre, A.; Van Damme, E.J.M.; Rougé, P. A molecular basis for the endo-beta 1,3-glucanase activity of the thaumatin-like proteins from edible fruits. Biochimie 2003, 85, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Bistoni, O.; Emiliani, C.; Agea, E.; Russano, A.M.; Mencarelli, S.; Orlacchio, A.; Spinozzi, F. Biochemical and immunological characterization of pollen-derived beta-galactosidase reveals a new cross-reactive class of allergens among Mediterranean trees. Int. Arch. Allergy Immunol. 2005, 136, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Shahali, Y.; Sutra, J.-P.; Charpin, D.; Mari, A.; Guilloux, L.; Sénéchal, H.; Poncet, P. Differential IgE sensitization to cypress pollen associated to a basic allergen of 14 kDa. FEBS J. 2012, 279, 1445–1455. [Google Scholar] [CrossRef]
- Soman, K.V.; Midoro-Horiuti, T.; Ferreon, J.; Goldblum, R.M.; Brooks, E.G.; Kurosky, A.; Braun, W.; Schein, C.H. Homology Modeling and Characterization of IgE Binding Epitopes of Mountain Cedar Allergen Jun a 3. Biophys. J. 2000, 79, 161–169. [Google Scholar] [CrossRef]
- Afferni, C.; Iacovacci, P.; Barletta, B.; Di Felice, G.; Tinghino, R.; Mari, A.; Pini, C. Role of carbohydrate moieties in IgE binding to allergenic components of Cupressus arizonica pollen extract. Clin. Exp. Allergy 1999, 29, 1087–1094. [Google Scholar] [CrossRef]
- Barletta, B.; Tinghino, R.; Corinti, S.; Afferni, C.; Iacovacci, P.; Mari, A.; Pini, C.; Di Felice, G. Arizona cypress (Cupressus arizonica) pollen allergens. Identification of crossreactive periodate-resistant and sensitive epitopes with monoclonal antibodies. Allergy 1998, 53, 586–593. [Google Scholar] [CrossRef]
- Canini, A.; Giovinazzi, J.; Iacovacci, P.; Pini, C.; Caiola, M.G. Localisation of a carbohydrate epitope recognised by human IgE in pollen of Cupressaceae. J. Plant Res. 2004, 117, 147–153. [Google Scholar] [CrossRef]
- Iacovacci, P.; Afferni, C.; Butteroni, C.; Pironi, L.; Puggioni, E.M.R.; Orlandi, A.; Barletta, B.; Tinghino, R.; Ariano, R.; Panzani, R.C.; et al. Comparison between the native glycosylated and the recombinant Cup a1 allergen: Role of carbohydrates in the histamine release from basophils. Clin. Exp. Allergy 2002, 32, 1620–1627. [Google Scholar] [CrossRef]
- Shahali, Y.; Sutra, J.; Hilger, C.; Swiontek, K.; Haddad, I.; Vinh, J.; Guilloux, L.; Charpin, D.; Sénéchal, H.; Poncet, P. Identification of a polygalacturonase (Cup s 2) as the major CCD-bearing allergen in Cupressus sempervirens pollen. Allergy 2017, 72, 1806–1810. [Google Scholar] [CrossRef] [PubMed]
| Subjects | Sex/Age | Allergic History | Specific IgE |
|---|---|---|---|
| (kU/mL) | |||
| 1 | F/5 | * CY,DERF,PAR | 0.19 |
| 2 | M/4 | CY,DERP,PAR,DAC,PN,SIN,SHR | 0.44 |
| 3 | F/29 | CY,DERP/F,PAR | 0.70 |
| 4 | M/11 | CY,DERP/F,CAT,DOG | 1.10 |
| 5 | M/5 | CY,OLI,PAR | 1.27 |
| 6 | F/42 | CY,DERP/F,CAT | 1.39 |
| 7 | F/43 | CY,PAR,DAC | 1.98 |
| 8 | F/16 | CY,PAR | 4.49 |
| 9 | F/46 | CY,DERP,PAR,DAC | 4.92 |
| 10 | M/33 | CY,DERP,PAR,DAC,ALT | 6.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Sénéchal, H.; Nguyen, C.; Granier, C.; Poncet, P.; Rougé, P. Structural Basis for the IgE-Binding Cross-Reacting Epitopic Peptides of Cup s 3, a PR-5 Thaumatin-like Protein Allergen from Common Cypress (Cupressus sempervirens) Pollen. Allergies 2023, 3, 11-24. https://doi.org/10.3390/allergies3010002
Barre A, Sénéchal H, Nguyen C, Granier C, Poncet P, Rougé P. Structural Basis for the IgE-Binding Cross-Reacting Epitopic Peptides of Cup s 3, a PR-5 Thaumatin-like Protein Allergen from Common Cypress (Cupressus sempervirens) Pollen. Allergies. 2023; 3(1):11-24. https://doi.org/10.3390/allergies3010002
Chicago/Turabian StyleBarre, Annick, Hélène Sénéchal, Christophe Nguyen, Claude Granier, Pascal Poncet, and Pierre Rougé. 2023. "Structural Basis for the IgE-Binding Cross-Reacting Epitopic Peptides of Cup s 3, a PR-5 Thaumatin-like Protein Allergen from Common Cypress (Cupressus sempervirens) Pollen" Allergies 3, no. 1: 11-24. https://doi.org/10.3390/allergies3010002
APA StyleBarre, A., Sénéchal, H., Nguyen, C., Granier, C., Poncet, P., & Rougé, P. (2023). Structural Basis for the IgE-Binding Cross-Reacting Epitopic Peptides of Cup s 3, a PR-5 Thaumatin-like Protein Allergen from Common Cypress (Cupressus sempervirens) Pollen. Allergies, 3(1), 11-24. https://doi.org/10.3390/allergies3010002








