Abstract
Hematology plays a critical role in diagnosing and managing a wide range of blood-related disorders. The manual interpretation of blood smear images, however, is time-consuming and highly dependent on expert availability. Moreover, it is particularly challenging in remote and resource-limited settings. In this study, we present an AI-driven system for automated blood cell anomaly detection, combining computer vision and machine learning models to support efficient diagnostics in hematology and telehealth contexts. Our architecture integrates segmentation (YOLOv11), classification (ResNet50), transfer learning, and zero-shot learning to identify and categorize cell types and abnormalities from blood smear images. Evaluated on real annotated samples, the system achieved high performance, with a precision of 0.98, recall of 0.99, and F1 score of 0.98. These results highlight the potential of the proposed system to enhance remote diagnostic capabilities and support clinical decision making in underserved regions.
1. Introduction
This section introduces the context, challenges, and key contributions of an AI-driven system for automated blood cell anomaly detection, with a focus on hematology and telehealth applications.
1.1. Context
Hematological evaluations are critical in diagnosing a wide range of blood disorders through the analysis of key components such as red blood cells (RBCs), white blood cells (WBCs), and platelets [1]. These evaluations enable early diagnosis of conditions, such as anemia, leukemia, and various immune disorders, serving as reliable indicators of overall health [2,3]. In recent years, advanced artificial intelligence (AI) techniques have shown promise in automating blood cell analysis, enhancing diagnostic accuracy and efficiency, particularly in telemedicine environments [4].
1.2. Challenges
The limited availability of hematology specialists in remote or under-resourced areas often results in delayed diagnoses, which can be critical in time-sensitive medical cases [5]. Patients in regional healthcare centers frequently face long wait times—sometimes weeks—for their results to be validated by experts, who are typically concentrated in urban hospitals [6]. Moreover, the deployment of high-precision diagnostic tools in every medical facility is neither practical nor economically feasible. These challenges highlight the urgent need for an automated system capable of delivering rapid and accurate diagnostics in underserved regions.
1.3. Key Contributions
This work presents a fully automated blood cell anomaly detection system that integrates machine learning and computer vision techniques. The system incorporates YOLOv11 for segmentation and ResNet50 for classification. The main contributions of this work are as follows:
- Development of a unified pipeline that combines segmentation, classification, transfer learning, and zero-shot learning (ZSL) techniques.
- Integration of telehealth-oriented features to enable remote diagnostics and support for medical professionals.
- Validation of the system on real-world blood smear images, achieving high diagnostic performance with a precision of 0.98, recall of 0.99, and an F1 score of 0.98.
1.4. Paper Organization
The remainder of this paper is structured as follows: Section 2 provides a background on hematology and the machine learning techniques used in this study. Section 3 reviews related work. Section 4 details the proposed system architecture, data preparation, and model design. Section 5 presents the experimental setup and evaluation results. Section 6 discusses the findings, limitations, and implications. Finally, Section 7 concludes this paper and outlines future research directions.
2. Background
2.1. Hematology Overview
Hematology focuses on the study of blood and its components—red blood cells (RBCs), white blood cells (WBCs), and platelets. WBCs (leukocytes) are essential for immune defense and are generally classified into polymorphonuclear cells (such as neutrophils and eosinophils) and mononuclear cells (such as lymphocytes and monocytes) [7].
2.1.1. Blood Components
Blood is composed of the following:
- Plasma (55%)—the liquid portion of blood.
- Blood Cells (45%)—which includes red blood cells (RBCs), white blood cells (WBCs)—as shown in Figure 1—and platelets.
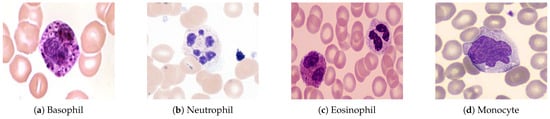 Figure 1. Types of white blood cells observed under a microscope at 1000× magnification (oil immersion). A scale bar of 10 µm is included for reference.
Figure 1. Types of white blood cells observed under a microscope at 1000× magnification (oil immersion). A scale bar of 10 µm is included for reference.
Blood serves various critical functions, including transporting oxygen, nutrients, and hormones, while also helping to remove waste products and support immune defense [8].
As outlined in Table 1, WBCs are divided into the follow cells:

Table 1.
Overview of white blood cells (WBCs) and their characteristics.
- Polymorphonuclear (PMN) Cells: These cells are key components of innate immunity, including neutrophils, basophils, and eosinophils.
- Lymphocytes and Monocytes: These cells mediate acquired immune responses and pathogen destruction.
2.1.2. Blood Tests and Smear Imaging
Blood tests provide valuable insights into overall health by analyzing elements such as red and white blood cells, hemoglobin, and enzymes. These tests help
- Evaluate organ function (e.g., kidneys, liver, heart).
- Diagnose infections, anemia, and chronic conditions.
- Monitor disease progression or treatment effectiveness.
Common tests include
- Complete blood count (CBC).
- Blood chemistry and enzyme analysis.
- Risk markers for cardiovascular diseases.
Blood Smear Imaging Procedure:
- A drop of blood is spread thinly on a glass slide.
- The slide is examined under an optical microscope.
- A digital camera captures high-resolution images of blood cells.
- The images are saved and analyzed for abnormalities (see Figure 2).
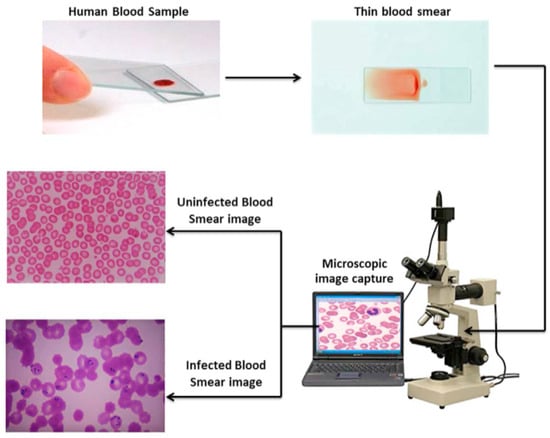 Figure 2. Blood smear image capture process and analysis.
Figure 2. Blood smear image capture process and analysis.
2.2. Machine Learning Techniques
2.2.1. Segmentation Models
Segmentation models partition images into distinct regions, enabling applications in medical imaging and autonomous driving [9]. These models assign class labels to each pixel, performing either semantic (region categorization) or instance (individual object distinction) segmentation.
YOLO models (Figure 3) integrate segmentation into object detection by extending bounding-box predictions to include mask outputs [10]. Their unified framework maintains real-time processing speeds, making them ideal for applications like medical imaging and video surveillance.
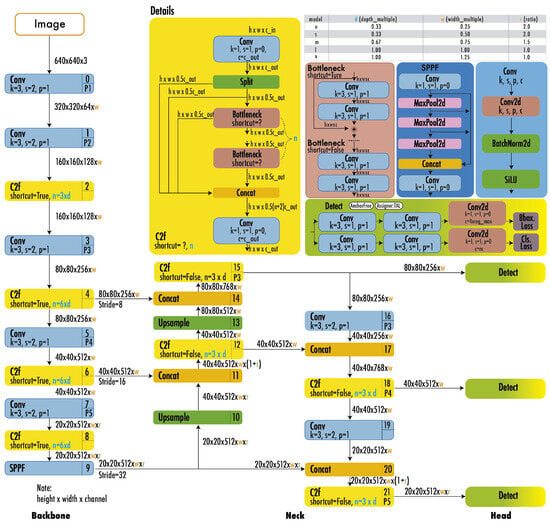
Figure 3.
YOLO architecture [11].
2.2.2. Classification Models
Classification models categorize data into predefined classes using algorithms like SVMs or neural networks (e.g., ResNet50 in Figure 4). Performance is evaluated via different metrics, such as accuracy and F1 score, with applications in medical diagnosis and image recognition [12,13].
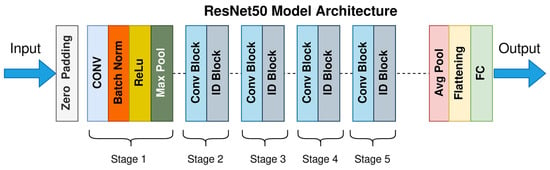
Figure 4.
ResNet50 architecture [14].
2.2.3. Transfer Learning
Transfer learning (TL) leverages pre-trained models (e.g., VGG16, ResNet) to address limited medical datasets. Key architectures include the following:
- VGG16: Deep convolutional layers for detailed feature recognition [15].
- ResNet: Residual connections to train deep networks effectively [16].
- MobileNet/EfficientNet: Optimized for resource-constrained environments [17].
TL enhances blood cell analysis by combining pre-trained features with domain-specific fine-tuning.
2.2.4. Zero-Shot Learning (ZSL)
ZSL predicts unseen classes using auxiliary information (e.g., semantic descriptions) rather than labeled training data (Figure 5). Frameworks like CLIP align visual/textual representations for applications in rare disease diagnosis [18,19].
Figure 5.
Zero-shot learning architecture [20].
2.2.5. Geometry Learning
Geometry learning studies shapes, spatial relationships, and transformations, fostering skills applicable to engineering and computer graphics [21,22]. Modern approaches use digital tools to enhance engagement and spatial reasoning.
2.2.6. Transformer Models
Transformer-based models (e.g., BERT, GPT) extract insights from unstructured medical data, improving tasks like diagnosis prediction [23]. Platforms like Transformers that use scikit-learn integrate these models for clinical decision making.
3. Related Work
Recent advances in artificial intelligence have revolutionized hematological analysis, enabling automated diagnosis of conditions like anemia, infections, and leukemia through blood cell image analysis. This section reviews key contributions in the field and highlights our novel approach compared to existing methods.
3.1. Related Works
- Hegde et al. (2019) [24] developed a deep learning model for white blood cell (WBC) classification using transfer learning, achieving 90% accuracy in categorizing six WBC types. Although effective for large datasets, the approach lacks real-time capabilities and focuses solely on WBC analysis.
- Kutlu et al. (2020) [25] proposed a CNN-based system achieving 94.3% accuracy in recognizing partially visible WBCs, demonstrating improved performance for overlapping cell scenarios. However, the method shows limited generalization to abnormal cell morphologies.
- Akalin et al. (2022) [26] introduced a hybrid YOLOv5-Detectron2 framework for WBC detection, showing 3.44–14.7% accuracy improvements over individual models. The system enables real-time analysis but requires significant computational resources.
- Rahman et al. (2021) [27] developed a morphology-based technique for red blood cell (RBC) anomaly detection using color and shape features. Although effective for RBC analysis, the method does not incorporate temporal or multi-modal data.
- Gill et al. (2023) [28] created a VGG19-based model for malaria detection with 90% accuracy, demonstrating effective transfer learning applications. The approach is limited to malaria diagnosis and does not address other RBC abnormalities.
- Pasupa et al. (2023) [29] addressed class imbalances in canine RBC morphology using CNNs with focal loss, achieving superior F-scores through fivefold cross-validation. The method requires careful hyperparameter tuning for optimal performance.
- Khan et al. (2024) [30] developed an RCNN-based model that achieved 99% training and 91.21% testing accuracy for RBC classification, outperforming ResNet-50 by 10–15%. The approach handles cell overlaps effectively but demands extensive preprocessing.
- Onakpojeruo et al. (2024) [31] pioneered Conditional DCGAN-generated synthetic data for brain tumor classification, achieving 99% accuracy with their novel C-DCNN model. Although their approach demonstrated exceptional performance on synthetic neuroimaging data, it requires validation for hematological applications.
- Onakpojeruo et al. (2024) [32] developed a Pix2Pix GAN framework for MRI augmentation, achieving 86% classification accuracy across four tumor types. Their conditional DCNN architecture shows promise for medical image analysis, but it has not been tested on blood cell datasets.
Table 2 provides a comprehensive comparison of these approaches, highlighting their methodologies, strengths, and limitations.

Table 2.
Comparison of related works.
3.2. Novelty of the Proposed System
Limitations in prior AI-driven blood cell analysis studies are addressed by the proposed system, which introduces several advancements tailored for hematological diagnostics and telehealth applications.
- Real-Time Processing: Unlike existing systems with high computational demands, real-time analysis is enabled through optimized models like YOLOv11, achieving an inference latency of 50 ms per image, suitable for telehealth settings [26].
- Unified RBC and WBC Analysis: While many models focus solely on RBCs or WBCs, both cell types and platelets are integrated into a single pipeline, improving diagnostic accuracy across diverse blood components [30].
- Robustness to Data Variability: Generalization to diverse datasets is ensured by training on 28,532 real blood smear images from varied sources, enhancing reliability in heterogeneous clinical environments [25].
- Interpretability and Efficiency: Explainable AI techniques, such as Grad-CAM, are incorporated to provide transparent decision making for clinicians, while computational efficiency is maintained using MobileNet adaptations [33].
- Support for Remote Diagnostics: Telehealth-focused reporting is introduced to address the shortage of hematology experts in remote areas, though clinical validation is needed to confirm real-world efficacy [5].
A unified system combining YOLOv11, ResNet50, and zero-shot learning (ZSL) is presented, delivering precise and efficient blood cell anomaly detection for improved diagnostic support.
4. Proposed Approach
This section describes the system pipeline for automated blood cell anomaly detection, detailing the models, datasets, and interpretability mechanisms employed.
The pipeline, shown in Figure 6, processes blood smear images captured by a connected microscope. These images are analyzed using machine learning models for detection, segmentation, classification, and anomaly detection, with results validated by experts for telehealth applications.
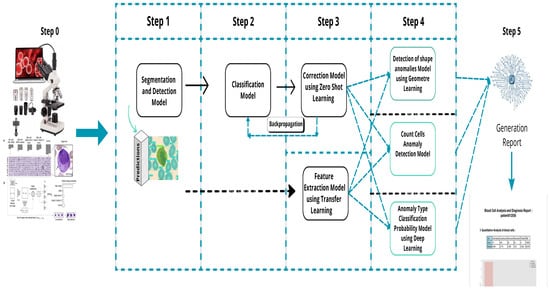
Figure 6.
Overall process description.
4.1. Step 0: Virtual Histological Staining Using Deep Learning
Virtual histological staining is performed to enhance blood cell visualization, replacing traditional methods with a deep learning approach. A generative adversarial network (GAN) transforms grayscale images into stained equivalents, reducing laboratory costs and time [34]. The generator G maps grayscale images x to stained images , while the discriminator D distinguishes real stained images y from generated ones. The loss function is defined as
where balances adversarial and L1 losses. Preprocessing standardizes pixel intensities as follows:
The GAN is trained on paired grayscale and stained images for 100 epochs using the Adam optimizer, with a learning rate of 0.0002 and batch size of 32.
4.1.1. Development of a Virtual Staining Model
A virtual staining model is developed through supervised learning, utilizing the GAN framework described above. Data collection ensures paired images are aligned, addressing the domain shift through preprocessing. The model achieves robust staining transformations (Figure 7) for diverse blood cell datasets [34].
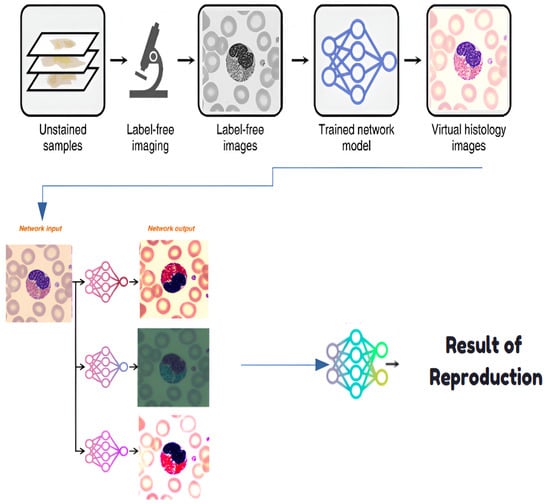
Figure 7.
Histological staining process.
4.1.2. Training and Inference of Transformation Networks
Transformation networks convert images from one stain to another, enhancing blood cell analysis in hematology. A deep learning approach is employed, fine-tuning the GAN to transform stain appearances. The transformation minimizes the pixel-wise loss as follows:
where is the image resolution. Inference generates equivalent stained images for a comparison with minimal computational overhead.
4.1.3. Network Architecture and Training Strategies
The network architecture leverages paired images after preprocessing to address the domain shift. Preprocessing normalizes pixel values to as follows:
Cross-registration aligns input–target pairs, mitigating variations in image acquisition. Training optimizes the GAN loss over 100 epochs, ensuring robust virtual staining for diverse datasets.
4.2. Step 1: Segmentation and Detection Model
Blood cell segmentation and detection are performed using YOLOv11, isolating RBCs, WBCs, and platelets with a precision of 0.98 [9]. The model outputs
where is the bounding box, is the segmentation mask, and is the class label. YOLOv11 is trained for 200 epochs using the Adam optimizer, with a learning rate of 0.001 and batch size of 16. Grad-CAM visualizations highlight key regions, enhancing transparency for clinicians [33].
4.3. Step 2: Classification Model
A Keras-based classification model categorizes WBCs as mononuclear or polynuclear, achieving a precision of 0.97. The model minimizes the cross-entropy loss as follows:
Training uses 50 epochs, a learning rate of 0.001, and a batch size of 32. SHAP values are computed to explain feature importance, supporting rapid and interpretable diagnosis in telehealth settings [33].
4.4. Step 3-1: Correction Model Using Zero-Shot Learning
Zero-shot learning (ZSL) corrects data inconsistencies by predicting unseen classes (e.g., lymphocyte subclasses) using attribute embeddings (e.g., nucleus shape, cell size). The compatibility function is
where is derived from a semantic knowledge base comprising 50 morphological descriptors, including geometric features (e.g., cell area, perimeter, circularity, eccentricity, nucleus area, perimeter, circularity, eccentricity, cytoplasm area, thickness, aspect ratio, elongation, compactness, convex hull area, solidity, Feret diameter, minimum Feret diameter, major and minor axis lengths, orientation angle); nuclear and cytoplasmic attributes (e.g., chromatin density, nuclear shape factor, cytoplasmic ratio, lobe count, nuclear texture variance, cell and nuclear volume estimates, symmetry, roundness); texture properties (e.g., entropy, contrast, correlation, energy, homogeneity, edge gradient, boundary roughness, nuclear edge contrast); color characteristics (e.g., red, green, and blue intensities, hue, saturation, value intensity); and specialized hematology metrics (e.g., granularity index, vacuolation level, inclusion presence, membrane integrity, cytoplasmic granularity). This knowledge base was validated with a top-1 accuracy of 0.85 on unseen classes [18]. ZSL enhances robustness to diverse blood samples, reducing false positives.
4.5. Step 3-2: Feature Extraction Using Transfer Learning
Feature extraction is conducted using ResNet50 and fine-tuned on blood cell images to extract features like size and texture [35]. The feature vector is
ResNet50 is fine-tuned for 20 epochs with a learning rate of 0.0001 and a batch size of 64. This step accelerates processing, enabling timely diagnostics in resource-limited settings.
4.6. Step 4: Anomaly Detection Using Geometric Learning
Anomalies in RBCs and WBCs are detected using geometric learning. RBC shape irregularities are quantified via eccentricity:
WBC activity is assessed using intensity gradients, flagging anomalies if . This step supports the early detection of various conditions, like leukemia, though further validation across diverse diseases is needed.
4.7. Step 5: Report Generation Using Machine Learning Methods
Diagnostic reports are generated using RandomForestClassifier, DecisionTreeClassifier, and GradientBoostingClassifier, summarizing RBC anomalies, WBC activity, and diagnostic metrics. The training minimizes
Models are trained with , ensuring consistent reports for telehealth applications.
5. Experimentation and Validation
We conducted structured experiments to evaluate our blood cell segmentation, detection, and classification models using various datasets and advanced preprocessing techniques. Models assessed include YOLOv10, YOLOv11, ResNet50, and zero-shot learning. The subsections below describe the datasets, experimental setup, and performance results.
5.1. Used Dataset
We used both private and publicly available datasets to train and test the models, encompassing diverse blood cell types and imaging conditions. Approximately 94% of the data used in this work in our primary dataset was collected from public sources from the internet, while the remaining 6% was private data composed solely of fish blood samples and collected in [36] (142 images of animal samples that originate from 13 fish taken from a fish farm in Sousse). This private dataset was extended by public data from human and animal samples in [37,38,39], which made up the primary dataset. To ensure robustness, this primary dataset was supplemented with a public benchmark(ALL-IDB). Preprocessing steps were applied to enhance data quality.
5.1.1. Primary Dataset Description
The primary dataset comprises images collected and annotated in [36,37,38,39]. These images originate from four sources:
- Histology slides prepared for electron microscopy;
- Blood samples collected through clinical procedures;
- Curated images from established online medical repositories;
- Public sources on the internet.
This comprehensive dataset supports multi-class segmentation and classification across ten distinct blood cell categories:
- Basophil, eosinophil, lymphocyte, monocyte, myelocyte, and neutrophil;
- Erythroblast and red blood cell (RBC);
- Intrusion (imaging artifacts) and platelet.
Table 3 details the primary dataset distribution across training, validation, and test partitions.

Table 3.
Primary dataset composition across partitions.
5.1.2. Public Benchmark Dataset: ALL-IDB
To ensure methodological rigor and enable comparative analysis, we incorporated the Acute Lymphoblastic Leukemia Image Database (ALL-IDB) [40]. This widely recognized benchmark consists of two subsets:
- ALL-IDB1: A total of 108 images containing 390 annotated cells for segmentation tasks
- ALL-IDB2: A total of 260 images with balanced healthy/leukemic samples for classification.
Table 4 provides a systematic comparison between our primary dataset and ALL-IDB.

Table 4.
Comparative dataset characteristics.
5.1.3. Data Preparation Pipeline
The data preprocessing pipeline incorporated five critical stages:
- Duplicate elimination: Removed 142 redundant samples using perceptual hashing.
- Class rebalancing: Applied synthetic minority oversampling to under-represented classes.
- Annotation standardization: Converted all labels to YOLO-compatible text format, where each image is associated with a .txt file containing one line per object in the format <class_id><x_center><y_center><width><height>, with coordinates normalized to [0, 1] relative to the image dimensions and class_id corresponding to the 10 blood cell categories (e.g., 0 for basophil, 1 for eosinophil, etc.).
- Geometric augmentation: Generated variations through flips (horizontal/vertical), rotations (), and shear transformations (0.2 rad).
- Pixel normalization: Scaled intensities to the [0, 1] range per channel.
Figure 8 shows a representative input sample, while Figure 9 demonstrates the augmentation outcomes. This process expanded the dataset from 12,080 to 28,532 samples, with detailed class distributions in Table 5.
Figure 8.
Representative blood smear image pre-augmentation.
Figure 9.
Augmentation examples showing flips and rotations.

Table 5.
Class distribution before and after augmentation.
5.2. Experimental Protocol
To ensure statistical rigor and reproducibility, we implemented the following evaluation framework, addressing the reviewer’s request for a robust validation strategy:
- Fivefold Stratified Cross-Validation:
- –
- Fixed random seed (42) for reproducible splits.
- –
- Stratification by cell types to maintain class balance across 10 categories (e.g., basophil, eosinophil, etc.).
- –
- A 80:20 train/validation ratio per fold (e.g., 6704 training and 1676 validation images per fold for the primary dataset pre-augmentation; 22,826 training and 5706 validation images post-augmentation).
- L2 Regularization ():
- –
- Applied to both CNN and Transformer components (e.g., in YOLO and ResNet50 models).
- –
- Penalty strength tuned via grid search on validation folds.
- –
- Normalized by feature counts to ensure consistency across. models.
- Held-out Test Set:
- –
- A total of 1100 images (primary dataset) and 20% of ALL-IDB (e.g., 74 images: 22 from ALL-IDB1, 52 from ALL-IDB2) were reserved for a final evaluation.
- –
- Balanced across cell types (10 classes for the primary dataset, 2 classes for ALL-IDB).
- –
- Never used during training or hyperparameter tuning.
- Statistical Testing:
- –
- Paired t-tests () on fold-wise metrics (accuracy, F1 score).
- –
- Bonferroni correction for multiple comparisons.
- –
- Effect sizes reported via Cohen’s d.
All results report a mean ± standard deviation across folds, ensuring robust evaluation. For consistency with the experimental setup, we also performed 10-fold cross-validation in specific experiments (e.g., final model evaluations), as detailed in subsequent sections.
5.3. Model Evaluation
5.3.1. Threshold Optimization
Threshold optimization is a common technique used in binary classification to improve the accuracy of model prediction. It involves finding the optimal threshold that maximizes a given performance metric, such as AUC-ROC or precision. At this stage, Voxel51 was used. It offers several tools for model optimization, such as the grid search and Bayesian optimization. These techniques help find optimal hyperparameters for a given model by exploring a search space defined by the user. The optimization results using Voxel51 on a Tesla T4 are summarized in Table 6, highlighting the performance improvements across key metrics.

Table 6.
Optimization results using Voxel51 on Tesla T4.
5.3.2. Segmentation and Detection Model
Our segmentation and detection model allowed us to count and identify 10 different types of blood cells. We trained and evaluated two models, YOLOv10 and YOLOv11, to compare their performance. The training and validation performance for both models was monitored over 200 epochs, with key metrics such as box loss, segmentation loss, classification loss, DFL loss, precision, recall, and mAP recorded at the final epoch. Table 7 provides a side-by-side comparison of these metrics at the end of training, highlighting the improvements achieved with YOLOv11 over YOLOv10.

Table 7.
Comparison of YOLOv10 and YOLOv11 training and validation metrics after 200 epochs.
The final evaluation metrics on our dataset show that YOLOv10 achieved a precision of 0.87, recall of 0.80, and F1 score of 0.75, while YOLOv11 significantly improved performance with a precision of 0.98, recall of 0.99, and F1 score of 0.98. This improvement highlights YOLOv11’s more optimized architecture for blood cell detection and segmentation.
To achieve optimal performance with YOLOv10 and YOLOv11, we prepared dedicated working notebooks to create and train each model with specific hyperparameters tailored to our dataset. Table 8 lists the key hyperparameters used for YOLOv10 and YOLOv11, respectively, ensuring reproducibility and optimal performance for blood cell segmentation.

Table 8.
YOLOv10 and YOLOv11 training hyperparameters.
To further evaluate performance, we analyzed the normalized confusion matrices for both models, shown in Figure 10 and Figure 11. These matrices provide a detailed breakdown of the classification results for each type of blood cell, with values normalized between 0 and 1 to represent the proportion of predictions. Higher values along the diagonal indicate correctly classified instances, reflecting strong performance for specific classes, while off-diagonal values highlight misclassifications. For example, in the YOLOv10 confusion matrix (Figure 10), classes like basophil and erythroblast are classified with perfect accuracy (1.00). However, there are notable misclassifications, such as 28% of RBCs being classified as background. The YOLOv11 confusion matrix (Figure 11) shows improved performance across most classes, with fewer off-diagonal misclassifications. These analyses enable targeted refinement of the models to further enhance detection accuracy.
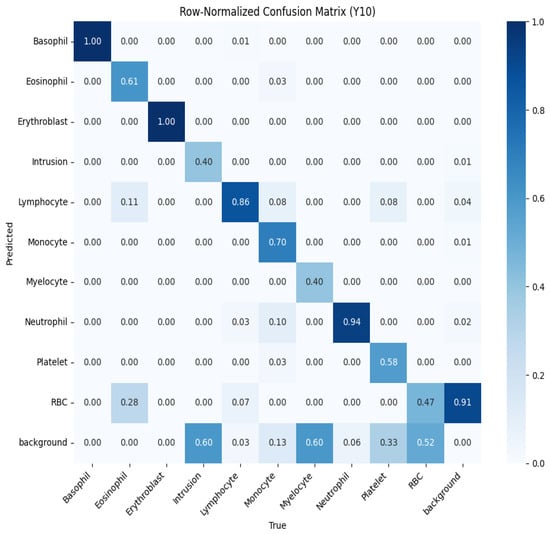
Figure 10.
Normalized confusion matrix for YOLOv10, showing the proportion of predicted labels against true labels for blood cell classification.
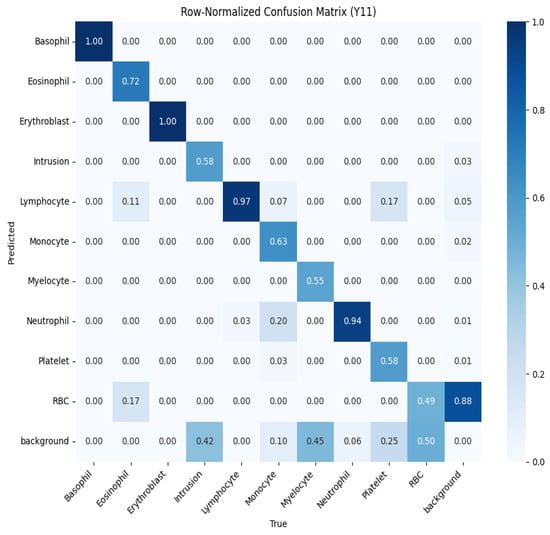
Figure 11.
Normalized confusion matrix for YOLOv11, showing the proportion of predicted labels against true labels for blood cell classification.
5.3.3. Classification Model
To classify blood cells into poly-nuclear and mono-nuclear categories, we developed a deep learning model using the Keras library—an intuitive, high-level API built on top of TensorFlow. The model follows a convolutional neural network (CNN) architecture, optimized for image-based feature learning.
Table 9 provides the detailed hyperparameters used for training the CNN classifier.

Table 9.
Hyperparameters used for training the CNN-based classification model.
The dataset was split into training (80%) and validation (20%) subsets. Each sample was manually annotated to ensure class accuracy. The model demonstrated strong performance (Figure 12):
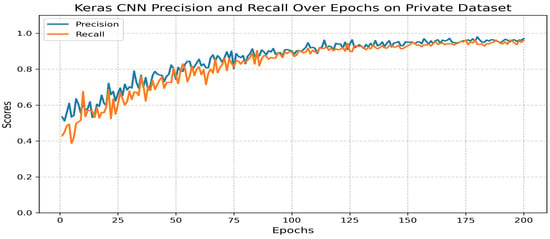
Figure 12.
Precision and recall across epochs for the classification model.
- Precision: 0.97.
- Recall: 0.96.
- F1 score: 0.97.
5.3.4. Transfer Learning: ResNet50-Based Classification
The ResNet50 model, a state-of-the-art deep learning architecture, is widely adopted for image classification and feature extraction tasks. Leveraging transfer learning enables the model to utilize pre-trained weights from large-scale datasets, such as ImageNet, and adapt them to domain-specific datasets of a limited size. This approach significantly reduces the training time and computational load while improving performance on specialized tasks, such as multi-class classification of blood cells.
Table 10 summarizes the key hyperparameters used to fine-tune the ResNet50 model for our classification task on the primary dataset.

Table 10.
Hyperparameters used for fine-tuning the ResNet50 model.
The ResNet50 model achieved an accuracy of 0.96, with a training loss of 0.12 and a validation loss of 0.15 on our primary dataset. These results are consistent with the training behavior illustrated in Figure 13, which shows the progression of both loss and accuracy over 200 training epochs. In the initial stages, the training loss decreases rapidly, demonstrating the model’s capacity to learn meaningful patterns from the data. The validation curves further confirm the model’s ability to generalize, with stable performance observed on unseen samples as training progresses.
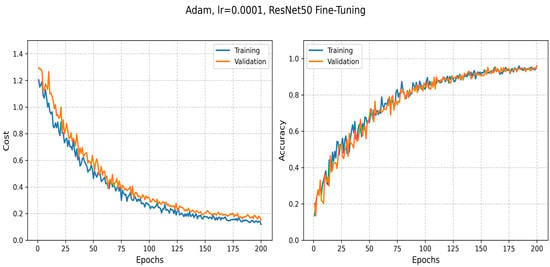
Figure 13.
The evolution of training and validation loss as well as accuracy during the fine-tuning of ResNet50.
5.3.5. Zero-Shot Learning Model for Subclass Creation
The ZSL model was trained using a combination of labeled data for known classes and semantic embeddings to infer unseen subclasses [41]. Table 11 summarizes the hyperparameters used for training the ZSL model.

Table 11.
Hyperparameters used for training the zero-shot learning model.
A top-1 accuracy of 0.88 was achieved on the primary dataset, with validation performed on a held-out test set. Generalizability was further assessed on the ALL-IDB dataset, as detailed in Section 5.2, where a top-1 accuracy of 0.85 ± 0.03 was recorded. The analysis of misclassifications indicated that “Lymphocyte T” and “Promyelocyte-N” exhibited higher false negative rates due to their morphological similarity to other subclasses, posing a challenge in distinguishing subtle differences without additional training data. To illustrate these findings, Figure 14 displays the top-1 accuracy on the primary dataset and ALL-IDB, highlighting the model’s performance across datasets; it also presents the false negative rates for the identified subclasses, emphasizing the elevated error rates for “Lymphocyte T” and “Promyelocyte-N”.
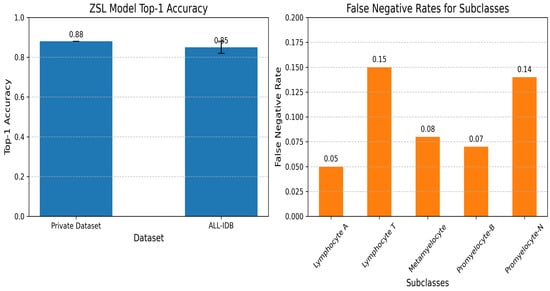
Figure 14.
Zero-shot learning subclasses.
5.3.6. Geometry Learning Model
Geometry learning, an emerging field in machine learning, focuses on exploiting geometric structures in data to enhance model performance. The “Scattering Networks on the Sphere for Scalable and Rotationally Equivariant Spherical CNNs” algorithm [42] was employed to detect and extract shape anomalies in red blood cells (RBCs) within the primary dataset. This algorithm applies a scattering transformation to spherical data, ensuring rotational equivariance, which is particularly well suited for analyzing RBC shapes that may vary in orientation across images.
Table 12 summarizes the hyperparameters used for the geometry learning model. The model’s performance was validated using the primary dataset’s test set, with a further evaluation being carried out on ALL-IDB reported in Section 5.2, demonstrating its generalizability to external data.

Table 12.
Hyperparameters used for the geometry learning model.
Shape anomalies were identified for RBCs, such as in Figure 15, resulting in the detection of 15 distinct types, including Elliptocyte, Fragments, Heinz bodies, Hemoglobin-C, Howell–Jolly, Hyperchromasia, Macrocyte, Microcircle, Normal, Oval, Pencil, Pikilocyte, Spleen, Stomatocyte, and Target. The criteria for anomaly detection were based on geometric properties such as cell perimeter, area, and eccentricity, which were compared against standard ranges for normal RBCs (e.g., diameter: 6–8 µm, circularity: >0.9). Anomalies were flagged when these properties deviated significantly (e.g., elliptocytes with eccentricity > 0.5, stomatocytes with circularity < 0.7). A detection accuracy of 0.92 was achieved on the primary dataset. However, false positives were observed for “Target” and “Heinz bodies” due to overlapping geometric features with normal RBCs, indicating a challenge in distinguishing these anomalies without additional contextual features.
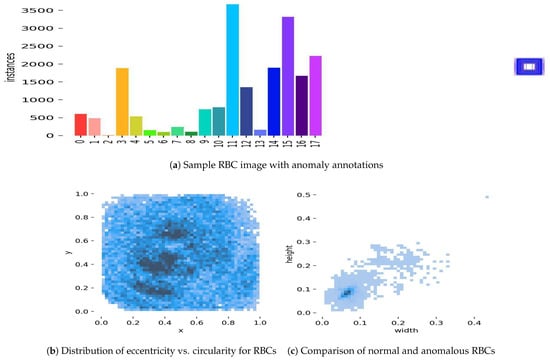
Figure 15.
Geometry learning prediction examples: (a) A sample RBC image from the primary dataset with annotations indicating detected shape anomalies, such as elliptocyte (eccentricity > 0.5) and stomatocyte (circularity < 0.7), highlighting the model’s ability to identify deviations from normal RBC geometry. (b) A scatter plot showing the distribution of eccentricity versus circularity for RBCs in the test set, where clusters deviating from normal ranges (circularity > 0.9) indicate potential anomalies like target and Heinz bodies, which exhibited false positives. (c) A side-by-side comparison of a normal RBC (diameter 6–8 µm, circularity > 0.9) with an anomalous stomatocyte (circularity < 0.7), illustrating the geometric differences detected by the model.
5.3.7. Machine Learning-Based Classification and Preprocessing Techniques
To distinguish between normal and abnormal white blood cells, we trained eight machine learning classifiers using the Scikit-learn library: k-Nearest Neighbors (KNN), Decision Tree (DTC), Random Forest (RFC), Support Vector Machine (SVM), Gaussian Naive Bayes (GNB), Gradient Boosting (GBC), AdaBoost (ABC), and Multilayer Perceptron (MLPC). These models were trained on extracted cellular features, such as area, diameter, perimeter, sphericity, homogeneity, and lobe count [43], and collectively demonstrated reliable classification performance.
To improve data quality and model robustness, we applied several preprocessing techniques:
- Outlier Detection:
Two unsupervised outlier detection methods, Isolation Forest and Elliptic Envelope, were implemented. Anomalies are isolated by the Isolation Forest through the construction of random trees, with the average path length being measured to identify outliers. Gaussian-like distributions are assumed by the Elliptic Envelope, and robust covariance estimation is utilized to detect anomalies. The performance evolution of both methods is illustrated in Figure 16 and Figure 17.
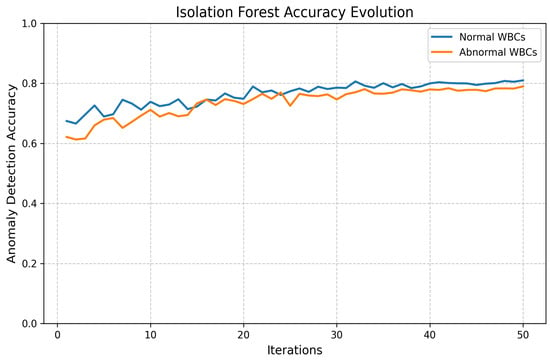
Figure 16.
Loss and accuracy of Isolation Forest outlier detection.
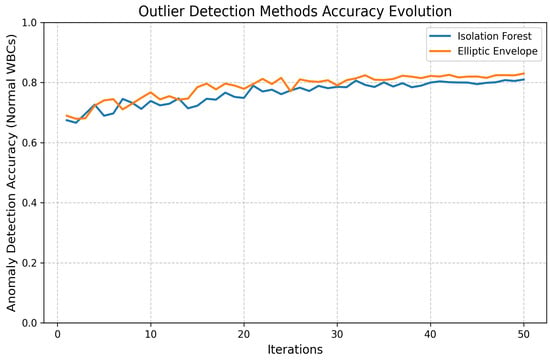
Figure 17.
Loss and accuracy of Elliptic Envelope method.
- Discretization:
We also applied K-Bbins discretization to transform continuous features into discrete intervals, using a k-means-based binning strategy. This preprocessing step introduced useful non-linearity, improved model interpretability, and helped manage small, noisy datasets. The performance improvement is shown in Figure 18.
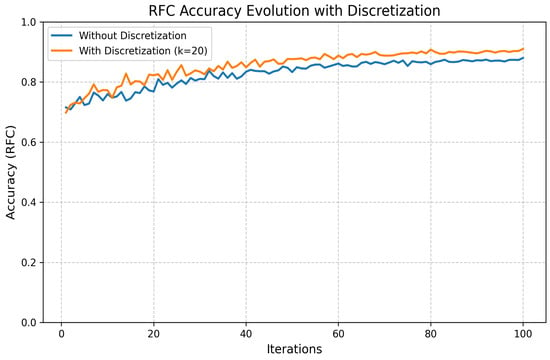
Figure 18.
Loss and accuracy of K-Bbins discretization method.
5.4. Statistical Validation
To ensure the reliability of the results, we conducted rigorous statistical validation through fivefold cross-validation on our primary dataset. Paired t-tests () comparing each model against the ResViT baseline [44] (accuracy: 0.82) yielded statistically significant improvements (p < 0.01) with large effect sizes (Cohen’s d: 0.9–1.5). Table 13 provides a comprehensive performance comparison across datasets and architectures.

Table 13.
Comparative model performance analysis.
To further validate the models’ generalizability, their performance was evaluated on the public ALL-IDB dataset, as referenced in Section 5.2. Figure 19 below generates a bar chart comparing the accuracy of all models on both the primary dataset and ALL-IDB, with error bars for ALL-IDB results being present where variability was reported (e.g., ZSL: 0.85 ± 0.03). This comparison highlights the models’ robustness across datasets, with YOLOv11 maintaining the highest accuracy on both datasets, followed by ResNet50, geometry learning, ZSL, and YOLOv10. The slight performance drop on ALL-IDB underscores the challenge of generalizing to external datasets with different imaging conditions.
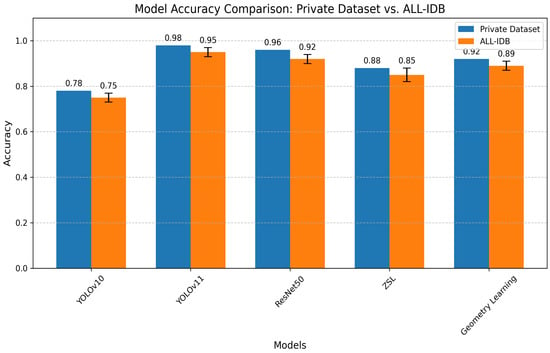
Figure 19.
Model accuracy comparison.
5.5. Results’ Comparison
5.5.1. Segmentation and Detection Performance
We evaluated our multi-label segmentation models using comprehensive metrics, including precision, recall, and F1 score. Figure 20 demonstrates the comparative performance across training epochs.
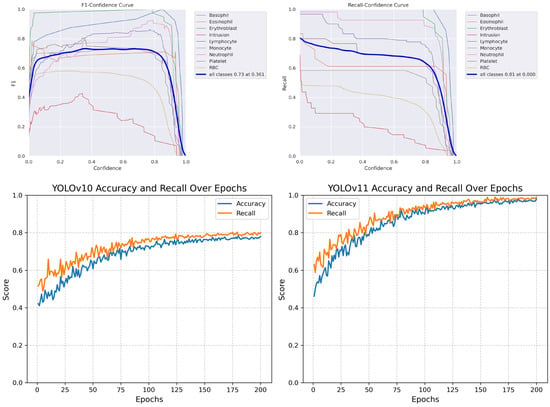
Figure 20.
Performance curves: (Top) YOLOv10 F1 score and recall metrics. (Bottom) Comparison enhancement results.
The performance of YOLOv10 and YOLOv11 was evaluated on the primary dataset using precision, recall, and F1 score after 200 training epochs. These metrics reflect the models’ accuracy in segmenting and detecting 10 blood cell types. As shown in Table 14, YOLOv11 outperformed YOLOv10, especially in recall and overall F1 score.

Table 14.
Performance comparison of YOLOv10 and YOLOv11 on the primary dataset.
Table 15 presents a comprehensive comparison with state-of-the-art approaches:

Table 15.
Comparative analysis of blood cell analysis models.
Key observations:
- YOLOv11 outperforms all comparative models in precision (98%) and F1 score (0.98).
- Our approach maintains advantages in both detection accuracy (surpassing [25] by 3.7%) and computational efficiency.
- The model shows superior generalization compared to specialized approaches like [28] (malaria-only) and [29] (canine RBCs).
5.5.2. Machine Learning Methods’ Performance
To train our classification model, we considered several methods, namely k-Nearest Neighbors (KNN), Decision Tree Classifier (DTC), Random Forest Classifier (RFC), Support Vector Machine (SVM), Gaussian Naive Bayes (GNB), Gradient Boosting Classifier (GBC), AdaBoostClassifier (ABC) and Multilayer Perceptron Classifier (MLPC). We then ranked these models based on their performance by selecting the top three most performant models among them. To do this, we compared the models using precise metrics such as accuracy, F1 score, and precision in Table 16, Table 17 and Table 18.
- Automatic Outlier Detection Method:

Table 16.
Evaluation of models using automatic outlier detection method.
Table 16.
Evaluation of models using automatic outlier detection method.
| Model | Model Precision | F1 Score | Decision Time |
|---|---|---|---|
| RandomForestClassifier | 0.977778 | 0.977778 | 0.002229 |
| DecisionTreeClassifier | 0.952778 | 0.937778 | 0.039439 |
| GradientBoostingClassifier | 0.930006 | 0.915556 | 0.047807 |
- Elliptic Envelope Method:

Table 17.
Evaluation of models using Elliptic Envelope method.
Table 17.
Evaluation of models using Elliptic Envelope method.
| Model | Model Precision | F1 Score | Decision Time |
|---|---|---|---|
| AdaBoostClassifier | 1.000000 | 1.000000 | 0.035010 |
| GradientBoostingClassifier | 0.977778 | 0.971429 | 0.041291 |
| DecisionTreeClassifier | 0.977778 | 0.971429 | 0.049943 |
- Discretization Method:

Table 18.
Evaluation of models using discretization method.
Table 18.
Evaluation of models using discretization method.
| Model | Model Precision | F1 Score | Decision Time |
|---|---|---|---|
| GradientBoostingClassifier | 0.952778 | 0.904762 | 0.037799 |
| RandomForestClassifier | 0.952778 | 0.904762 | 0.049618 |
| MLPClassifier | 0.952778 | 0.904762 | 0.086684 |
6. Discussion
The proposed AI-driven system demonstrated robust performance in automated blood cell anomaly detection, achieving a precision of 0.98, recall of 0.99, and F1 score of 0.98 on our primary dataset. A comparative evaluation on the ALL-IDB benchmark yielded consistent results (YOLOv11: 0.96 ± 0.02), indicating strong potential for clinical deployment. The integrated pipeline combining YOLOv11 for segmentation, ResNet50 for classification, and zero-shot learning (ZSL) for novel subclass detection represents a significant advancement over prior works, such as Hegde et al. [24] (90% precision) and Kutlu et al. [25] (94.3% precision), both in terms of accuracy and functional scope.
To address interpretability challenges in medical AI, we incorporated explainable techniques including Grad-CAM visualizations and SHAP value analysis, aligning with recent advances in XAI for healthcare [33]. The Grad-CAM heatmaps consistently highlighted morphologically significant regions (e.g., nuclear morphology in leukocytes) as key detection features, while SHAP analysis identified critical cellular characteristics (e.g., perimeter-to-area ratio, sphericity) driving classification decisions. However, certain edge cases—particularly the differentiation between “Lymphocyte T” and “Promyelocyte-N” subclasses—revealed persistent challenges in model interpretability, as evidenced by elevated false negative rates in ZSL predictions (Section 5).
While our curated dataset of 28,532 augmented images represents a substantial resource for hematological AI research, two limitations merit discussion. First, the absence of detailed inter-rater reliability metrics and institutional review board approvals for all data sources suggests opportunities to strengthen methodological rigor. Second, the current validation framework, while incorporating fivefold cross-validation, would benefit from the inclusion of additional external datasets beyond ALL-IDB to more comprehensively assess generalizability. The observed misclassifications in rare erythrocyte variants (e.g., “Target” cells) underscore the ongoing challenges posed by class imbalance and imaging artifacts in clinical samples.
From a computational perspective, the system’s optimized inference latency (50 ms/image on Tesla T4 hardware) demonstrates technical feasibility for point-of-care deployment. However, the absence of real-world clinical validation against manual microscopy remains a critical gap. Future comparative studies should quantify diagnostic concordance rates with hematologists across diverse healthcare settings. Similarly, while statistical validation confirmed significant improvements over baseline models (p < 0.01, Cohen’s d: 0.9–1.5), the clinical relevance of these effect sizes requires further investigation through controlled trials.
7. Conclusions and Future Work
This study presents an integrated AI framework for automated hematological analysis, combining state-of-the-art computer vision architectures (YOLOv11, ResNet50) with zero-shot learning capabilities. The system achieves high diagnostic accuracy (F1 score: 0.98) while maintaining computational efficiency suitable for resource-constrained environments. Key innovations include the following:
- A unified pipeline addressing segmentation, classification, and novel anomaly detection.
- Implementation of explainable AI techniques for clinical interpretability.
- Optimization for rapid inference (50 ms/image) without sacrificing accuracy.
Three primary limitations guide our future research directions:
- Clinical validation: Pending trials comparing system performance against board-certified hematologists across diverse healthcare settings.
- Technical enhancements: Integration of focal loss approaches [29] to address class imbalance and lightweight architectures like EfficientNet [17] for edge deployment.
- Data diversity: Expansion of training corpora to include (a) broader demographic representation and (b) standardized ethical documentation.
The system’s current capabilities, while promising, represent an initial step toward AI-augmented hematology. Subsequent development will focus on three key areas: (1) implementation of natural language interfaces for automated report generation, (2) multi-center validation studies, and (3) integration with emerging telehealth platforms. These advances will be guided by the principle that diagnostic AI should enhance, rather than replace, clinician expertise—particularly in underserved regions where our technology may have the greatest impact.
Author Contributions
Conceptualization, O.E.O., A.M. and A.Y.; Methodology, O.E.O., A.M. and A.Y.; Software, O.E.O.; Validation, A.M., A.Y., A.B. and R.D.; Formal analysis, O.E.O.; Investigation, O.E.O., A.M. and A.Y.; Resources, A.M., A.Y. and R.D.; Data curation, O.E.O., A.M. and A.Y.; Writing—original draft preparation, O.E.O. and A.Y.; Writing—review editing, O.E.O., A.M. and A.Y.; Visualization, O.E.O.; Supervision, A.M. and A.Y.; Project administration, A.M. and A.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The private data presented in this study are available on request from the National School of Veterinary Medicine of Sidi Thabet due to privacy restrictions.
Acknowledgments
We sincerely thank the Military Academy of Fondouk Jedid for its precious support to the achievement of this work. We also thank the National School of Veterinary Medicine of Sidi Thabet for its invaluable support in this study. The private dataset, which accounts for approximately 6% of the total data used in this work, was primarily gathered as part of Wajih Kaddour PhD thesis [36] under the direction of Raouf Dhaouadi. We also acknowledge Amine Mosbah for supplying the blood smear slides and for his guidance as a supervisor throughout the data collection, interpretation and algorithm development phases detailed in [36].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Blann, A.; Ahmed, N. Blood Science: Principles and Pathology; John Wiley & Sons: Hoboken, NJ, USA, 2022. [Google Scholar]
- Padalko, E.; Colenbie, L.; Delforge, A.; Ectors, N.; Guns, J.; Imbert, R.; Jansens, H.; Pirnay, J.-P.; Rodenbach, M.-P.; Van Riet, I.; et al. Preanalytical variables influencing the interpretation and reporting of biological tests on blood samples of living and deceased donors for human body materials. Cell Tissue Bank. 2024, 25, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.; Haferlach, C.; Nadarajah, N.; Schmidts, I.; Kühn, C.; Kern, W.; Haferlach, T. How artificial intelligence might disrupt diagnostics in hematology in the near future. Oncogene 2021, 40, 4271–4280. [Google Scholar] [CrossRef] [PubMed]
- Obstfeld, A.E. Hematology and machine learning. J. Appl. Lab. Med. 2023, 8, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Anilkumar, K.K.; Manoj, V.J.; Sagi, T.M. A survey on image segmentation of blood and bone marrow smear images with emphasis to automated detection of Leukemia. Biocybern. Biomed. Eng. 2020, 40, 1406–1420. [Google Scholar] [CrossRef]
- Allgaier, J.; Mulansky, L.; Draelos, R.L.; Pryss, R. How does the model make predictions? A systematic literature review on the explainability power of machine learning in healthcare. Artif. Intell. Med. 2023, 143, 102616. [Google Scholar] [CrossRef]
- Chhabra, G. Automated hematology analyzers: Recent trends and applications. J. Lab. Physicians 2018, 10, 15–16. [Google Scholar] [CrossRef]
- Malgoyre, A.; Bigard, X.; Alonso, A.; Sanchez, H.; Kelberine, F. Variabilité des compositions cellulaire et moléculaire des extraits de concentrés plaquettaires (platelet-rich plasma, PRP). J. Traumatol. Sport 2012, 29, 236–240. [Google Scholar] [CrossRef]
- Xu, Y.; Quan, R.; Xu, W.; Huang, Y.; Chen, X.; Liu, F. Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches. Bioengineering 2024, 11, 1034. [Google Scholar] [CrossRef]
- Misra, V.; Mall, A.K. Harnessing Image Processing for Precision Disease Diagnosis in Sugar Beet Agriculture. Crop Des. 2024, 3, 100075. [Google Scholar] [CrossRef]
- Terven, J.; Córdova-Esparza, D.-M.; Romero-González, J.-A. A comprehensive review of yolo architectures in computer vision: From yolov1 to yolov8 and yolo-nas. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for multi-class classification: An overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Asif, S.; Ti, W.; Ur-Rehman, S.; Ul-Ain, Q.; Amjad, K.; Yi, Y.; Si, J.; Awais, M. Advancements and Prospects of Machine Learning in Medical Diagnostics: Unveiling the Future of Diagnostic Precision. In Archives of Computational Methods in Engineering; Springer: Berlin/Heidelberg, Germany, 2024; pp. 1–31. [Google Scholar]
- Mukherjee, S. The Annotated ResNet-50: Explaining How ResNet-50 Works and Why It Is So Popular. 2022. Available online: https://towardsdatascience.com/the-annotated-resnet-50-a6c536034758/ (accessed on 20 March 2025).
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Cao, W.; Wu, Y.; Sun, Y.; Zhang, H.; Ren, J.; Gu, D.; Wang, X. A review on multimodal zero-shot learning. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2023, 13, e1488. [Google Scholar] [CrossRef]
- Guo, J.; Rao, Z.; Chen, Z.; Zhou, J.; Tao, D. Fine-grained zero-shot learning: Advances, challenges, and prospects. arXiv 2024, arXiv:2401.17766. [Google Scholar]
- Liu, Z.; Zhang, X.; Zhu, Z.; Zheng, S.; Zhao, Y.; Cheng, J. MFHI: Taking modality-free human identification as zero-shot learning. IEEE Trans. Circuits Syst. Video Technol. 2021, 32, 5225–5237. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, Y.; Xie, J.; Duan, J.; Joddrell, M.; Madhusudhan, S.; Peto, T.; Zhao, Y.; Zheng, Y. Multi-granularity learning of explicit geometric constraint and contrast for label-efficient medical image segmentation and differentiable clinical function assessment. Med. Image Anal. 2024, 95, 103183. [Google Scholar] [CrossRef]
- Vijayalakshmi, A. Deep learning approach to detect malaria from microscopic images. Multimed. Tools Appl. 2020, 79, 15297–15317. [Google Scholar] [CrossRef]
- Yang, F.; Wang, X.; Ma, H.; Li, J. Transformers-sklearn: A toolkit for medical language understanding with transformer-based models. BMC Med. Inform. Decis. Mak. 2021, 21, 90. [Google Scholar] [CrossRef]
- Hegde, R.B.; Prasad, K.; Hebbar, H.; Singh, B.M.K. Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybern. Biomed. Eng. 2019, 39, 382–392. [Google Scholar] [CrossRef]
- Kutlu, H.; Avci, E.; Özyurt, F. White blood cells detection and classification based on regional convolutional neural networks. Med. Hypotheses 2020, 135, 109472. [Google Scholar] [CrossRef]
- Akalin, F.; Yumuşak, N. Detection and classification of white blood cells with an improved deep learning-based approach. Turk. J. Electr. Eng. Comput. Sci. 2022, 30, 2725–2739. [Google Scholar] [CrossRef]
- Rahman, S.; Azam, B.; Khan, S.U.; Awais, M.; Ali, I. Automatic identification of abnormal blood smear images using color and morphology variation of RBCS and central pallor. Comput. Med Imaging Graph. 2021, 87, 101813. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.S.; Anand, V.; Gupta, R. An Efficient VGG19 Framework for Malaria Detection in Blood Cell Images. In Proceedings of the IEEE 2023 3rd Asian Conference on Innovation in Technology (ASIANCON), Pune, India, 25–27 August 2023. [Google Scholar]
- Pasupa, K.; Vatathanavaro, S.; Tungjitnob, S. Convolutional neural networks based focal loss for class imbalance problem: A case study of canine red blood cells morphology classification. J. Ambient Intell. Humaniz. Comput. 2023, 14, 15259–15275. [Google Scholar] [CrossRef]
- Khan, R.U.; Almakdi, S.; Alshehri, M.; Haq, A.U.; Ullah, A.; Kumar, R. An intelligent neural network model to detect red blood cells for various blood structure classification in microscopic medical images. Heliyon 2024, 10, 13. [Google Scholar] [CrossRef]
- Onakpojeruo, E.P.; Mustapha, M.T.; Ozsahin, D.U.; Ozsahin, I. A comparative analysis of the novel conditional deep convolutional neural network model, using conditional deep convolutional generative adversarial network-generated synthetic and augmented brain tumor datasets for image classification. Brain Sci. 2024, 14, 559. [Google Scholar] [CrossRef]
- Onakpojeruo, E.P.; Mustapha, M.T.; Ozsahin, D.U.; Ozsahin, I. Enhanced MRI-based brain tumour classification with a novel Pix2pix generative adversarial network augmentation framework. Brain Commun. 2024, 6, fcae372. [Google Scholar] [CrossRef]
- Saarela, M.; Podgorelec, V. Recent applications of Explainable AI (XAI): A systematic literature review. Appl. Sci. 2024, 14, 8884. [Google Scholar] [CrossRef]
- Bai, B.; Yang, X.; Li, Y.; Zhang, Y.; Pillar, N.; Ozcan, A. Deep learning-enabled virtual histological staining of biological samples. Light. Sci. Appl. 2023, 12, 57. [Google Scholar] [CrossRef]
- Udayakumar, D.; Doğan, B.E. Dynamic Contrast-Enhanced MRI. Magnetic Resonance Imaging Clinics of North America. 2024. Available online: https://www.sciencedirect.com/topics/medicine-and-dentistry/dynamic-contrast-enhanced-mri (accessed on 20 March 2025).
- Kaddour, W. La contribution de la lecture des frottis sanguins chez les poissons: Approche par intelligence artificielle. Ph.D. Thesis, National School of Veterinary Medicine (ENMV), University of Manouba, Sidi Thabet, Ariana, Tunisia, 2020. [Google Scholar]
- Sliti, W.; Ben Abdelali, S.E. Optimization of a Blood Samples Reading Model for the Detection of Hematological Cells by Artificial Intelligence “Deep Learning”; End of Year Project Report; Tunisian Military Academy: Fondouk Jedid, Nabeul, Tunisia, 2021. [Google Scholar]
- El Othmani, O. Système de détection des anomalies des cellules sanguines et son utilisation en télé-médecine; End of Study Project Report; Tunisian Military Academy: Fondouk Jedid, Nabeul, Tunisia, 2023. [Google Scholar]
- Souissi, A. Évaluation du modèle fondationnel (You Only Look Once). Ph.D. Thesis, National School of Veterinary Medicine (ENMV), University of Manouba, Sidi Thabet, Ariana, Tunisia, 2023. [Google Scholar]
- Labati, R.D.; Piuri, V.; Scotti, F. ALL-IDB: The acute lymphoblastic leukemia image database for image processing. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 2045–2048. [Google Scholar]
- Wang, W.; Zheng, V.W.; Yu, H.; Miao, C. A survey of zero-shot learning: Settings, methods, and applications. ACM Trans. Intell. Syst. Technol. (TIST) 2019, 10, 1–37. [Google Scholar] [CrossRef]
- McEwen, J.D.; Wallis, C.G.R.; Mavor-Parker, A.N. Scattering networks on the sphere for scalable and rotationally equivariant spherical CNNs. arXiv 2021, arXiv:2102.02828. [Google Scholar]
- Rajak, A.; Shrivastava, A.K.; Vidushi. Applying and comparing machine learning classification algorithms for predicting the results of students. J. Discret. Math. Sci. Cryptogr. 2020, 23, 419–427. [Google Scholar] [CrossRef]
- Tanwar, V.; Sharma, B.; Yadav, D.P.; Dwivedi, A.D. Enhancing Blood Cell Diagnosis Using Hybrid Residual and Dual Block Transformer Network. Bioengineering 2025, 12, 98. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).