Abstract
The accumulation of salt in arable lands is a source of significant abiotic stress, contributing to a 10% decline in the world’s total arable lands and threatening food productivity and the sustainability of agriculture. About 76 million hectares of productive land are estimated to have been affected by human-induced salinization such as extreme salt deposits in soil, which are mainly caused by the actions of humans. For instance, continued irrigation and the frequent use of chemical fertilizers need to be understood. To ensure food availability, it is essential to improve upon traditional farming methods using current technologies to facilitate the reclamation of saline-affected arable lands to achieve high and sustainable food production. This review details current innovative strategies such as the modification of metabolic pathways, manipulation of antioxidant pathways, genetic engineering, RNA interference technology, engineered nanoparticles, arbuscular mycorrhizal fungi (AMF), organic amendments, and trace elements for improving saline marginal lands. These strategies were identified to have contributed to the improvement of plants salinity tolerance in diverse ways. For instance, the accumulation of plant metabolites such as amino acids, sugars, polyols, organic acids, saponins, anthocyanins, polyphenols, and tannins detoxify plants and play crucial roles in mitigating the detrimental effects of oxidative damage posed by salinity stress. Multiple plant miRNAs encoding the up- and down-regulation of single- and multi-ion transporters have been engineered in plant species to enhance salt tolerance. Nanomaterials and plant root system colonized by arbuscular mycorrhizal increase water uptake, photosynthetic efficiency, and biomass allocation in plants exposed to saline stress by excluding 65 percent of the Na+ uptake and enhancing K+ uptake by 84.21 percent. Organic amendments and trace elements reduced salinity concentrations by 22 percent and improved growth by up to 84 percent in maize subjected to salinity stress. This study also discusses how researchers can use these strategies to improve plants growth, development, and survival in saline soil conditions to enhance the productivity and sustainability of agriculture. The strategies discussed in this study have also proven to be promising approaches for developing salinity stress tolerance strategies for plants to increase agricultural productivity and sustainability.
1. Introduction
The issue of food security has been worsening in recent times due to the continuous increase in the global population [1]. The global population, currently estimated to be 7 billion, is expected to reach 10 billion within the next 50 years [1]. Feeding an ever-increasing global population has become a significant issue. The high rate of population growth, coupled with the decline of agricultural lands, are the most important contributing factors impending agriculture productivity and sustainability [2]. Consequently, enhancing agricultural productivity represents the sole viable strategy for attaining future food security [3]. Salinity, high temperatures, UVC radiation, and drought are significant environmental stresses that have been reported to significantly negatively impact agricultural productivity [4,5,6,7,8,9]. The total amount of salt dissolved in water is referred to as salinity. Salt deposition in arable lands, predominantly sodium and chloride ions, can result in saline soil conditions of arable lands. Salinity can be classified into sodic (alkaline) and saline soils [10]. Sodic soils are characterized by soils of a poor structure, commonly observed in arid and semi-arid areas. These soils exhibit high Na+ concentrations at the exchangeable site of the clay particles in most soils, with a pH above 8.5 and an exchangeable sodium percentage (ESP) greater than 15 [10]. In contrast, saline soils are typically found in arid areas, estuaries, and coastal fringes, where the electrical conductivity (EC) is greater than 4 dS/m, corresponding to approximately 40 mM NaCl [10,11]. Saline soils exhibit an ESP of less than 15 and a lower pH than sodic soils [10]. Salinity harms plant growth and development, causing osmotic stress, ionic toxicity, hormonal imbalances, and a reduction in nutrient uptake and mobilization [12,13]. Salinity also enhances the accumulation of reactive carbonyl species such as acrolein, 4-hydroxy-(E)-2-nominal (HNE), and 4-hydroxy-(E)-2-hexenal (HHE), which are downstream products of reactive oxygen species (ROS) [14,15]. The generation of reactive carbonyl species in plant organs can lead to slowed growth, senescence, wilting, drying, and even plant death [7,14,15]. These findings are supported by numerous reports documenting the agricultural output reduction resulting from these environmental factors. As reported by Saghafi et al., about 800 million hectares, representing 6% of the world’s total land area, are affected by saline stress conditions, and this percentage continues to increase [2]. This evidence indicates that soil salinity stress represents a significant problem that can inhibit agricultural plants’ growth, development, and productivity [16]. Therefore, it has become of paramount importance to gain insight into the mechanisms regulating plant tolerance to saline soil stress conditions.
Plants use strategies involving physiology, biochemical, and molecular mechanisms to respond to different levels of salinity. Antioxidant enzyme activity, the synthesis of osmoprotectants and antioxidant compounds, plant ion uptake and transport, polyamine and nitric oxide synthesis, and hormones modulation serve as the principal strategies that plants use to mitigate salinity stress [17]. Plants have been classified into two categories based on their capacity to flourish and persist in saline soils: glycophytes and halophytes. The majority of plants classified as halophytes can survive in saline conditions. This is achieved through root and shoot salts exclusion, ions compartmentation in various organs, and the synthesis of compatible solutes that are adopted in salt stress mechanisms. Recently, it has been demonstrated that new ecotypes of Sarcocornia plants exhibit higher biomasses under nitrogen and salinity conditions [18]. However, plants classified as glycophytes constitute the bulk of agricultural crops, which are highly susceptible to salinity stress [19]. To enhance the resilience of farming plants to this challenge, it is essential to elucidate additional strategies that breeders can employ to improve the growth, development, and survival of plants in saline soil conditions, thereby promoting increased productivity and sustainability in agriculture. This review will discuss the current innovative plant strategies used to tolerate salt stress to increase and sustain productivity. Additionally, the prospects for future study of plant responses to saline soil conditions will be considered.
2. Sustainable Plant Strategies in Mitigating Salinity Stress Conditions
Saline stress has a considerable impact on plants and their environment, which has led to the development of various strategies and technologies to alleviate the harmful effects of stress on plant growth, development, and physiology [20]. Plant response to salinity stress is a complex phenomenon and this involves multiple physiological and biochemical processes, including various genes (Table 1). The increase in soil salt concentration negatively disturbs plant nutrient and water uptake, which results in the accumulation of toxic ions. Plants that are tolerant to salinity conditions maintain proper cellular ion homeostasis, which is essential for their growth, development, and survival. This homeostasis is attained by enhancing the regulation of ion uptake, vacuolar compartmentalization, and active ion extrusion to the external environment [21,22].

Table 1.
Ion transporter and their genes associated with plant resistance to salt stress.
The innovative approaches that scientists are investigating to improve plants exposed to salinity stress adaptation include the modification of metabolic pathways, manipulation of antioxidant pathways, advancements in genetic engineering on salinity stress, RNA interference technology, engineered nanoparticles to alleviate salinity, arbuscular mycorrhizal fungi (AMF) for salt stress alleviation, the application of organic amendments, and trace elements for improving plant salt stress (Figure 1). These approaches facilitate the allocation of greater resources toward developing and enhancing plant salinity tolerance mechanisms, which will be discussed in detail in the subsequent sections [22]. This enhances our understanding of how most agriculture crops can be improved to adapt to salinity stress conditions; for instance, employing the genetic variability of numerous agricultural crops in different gene banks to develop stress-tolerant crops. These approaches offer a sustainable method of mitigating salt stress.
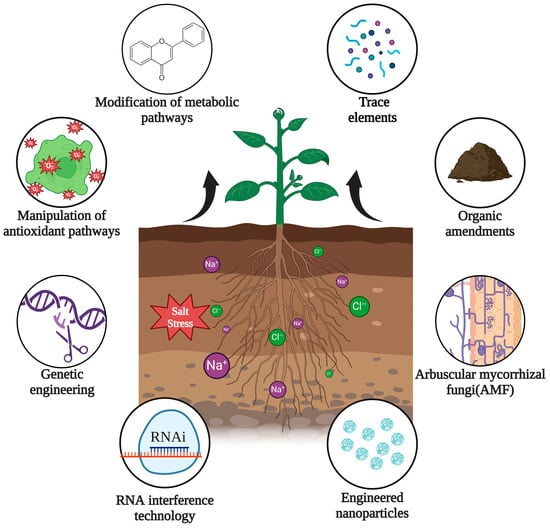
Figure 1.
Innovative approaches to improve plants exposed to salinity stress. Created using BioRender.com.
2.1. Modification of Plant Primary Metabolite Response to Salinity Stress
Primary metabolic processes represent a fundamental aspect of normal plant cell function. Such processes relate to numerous physiological and biochemical processes, including gas exchange and chlorophyll fluorescence. The energies that are involved in these processes and other precursors are necessary for synthesizing the compounds essential for various plant developmental processes [39,40]. Primary metabolites encompass polyols, sugars, amino acids, and organic acids. It has been demonstrated that primary metabolites serve as osmolytes, osmoprotectants, compatible solutes, and detoxification agents in plants subjected to salinity stress [17,41]. These compounds are small, non-toxic, hydrophilic molecules that do not affect cellular functions, even at high concentrations [42]. Kulkani et al. subjected Sesuvium portulacastrum to a 250 mM NaCl stress treatment for 8 and 24 h and reported the detection of 53 and 62 primary metabolites in the roots and shoots, respectively [43]. They pointed out that the high score of the cumulative pathways of both the roots and shoots was aspartate–alanine–glutamate and concluded that the analysis of metabolites and their salt responses pathways in halophytes will enhance salinity stress tolerance in plants. Increased concentrations of compatible solutes in the cytoplasm balance the decreased water potential, which is normally related to vacuole Na+ accumulation [44]. Additionally, primary metabolites can mitigate the repressing ability of elevated ion levels of enzyme activities without affecting protein structure or function. For instance, these compounds might act as antioxidants by scavenging free radicals, and they can assist membrane stability in response to salinity stress [44,45]. The most prevalent compatible solutes in plants are polyhydroxy compounds (e.g., sucrose, oligosaccharides, and polyhydric alcohols), as well as nitrogen-containing solutes (e.g., amino acids, quaternary ammonium compounds, and polyamines) [45,46,47]. These solutes have the ability to stabilize the enzymatic activities and structures of the cells and act as ROS scavengers, which serve as an important mechanism for alleviating the harmful effects of saline stress on plants. The metabolomic approaches employed when studying plants exposed to saline conditions and the changes observed among salt-sensitive and tolerant plants revealed distinct patterns. These patterns included two main categories: differences in the magnitude of metabolites within tolerant and sensitive plants, and different plants exhibiting both conserved and divergent metabolite responses to salinity stress [44]. Research conducted under control conditions demonstrated that Thellungiella halophila, a salt-tolerant plant, exhibited higher levels of sucrose, fructose, glucose, proline, inositol, galactinol, raffinose, citrate, malate, and succinate compared to Arabidopsis [48]. This indicates that a deep understanding of primary metabolites is important for enhancing the agricultural plant’s resilience to salt stress, thereby ensuring sustainability in production.
2.2. Plant Secondary Metabolite Modification in Response to Salinity Stress
Generally, secondary metabolites are not a basic requirement for plant cell functioning but are involved in the crucial role of protecting plants exposed to salinity conditions. Plant species use a considerably different level of secondary metabolites when exposed to the environment [49,50,51]. A significant proportion of these secondary metabolites have been grouped into three main categories (Figure 2), including nitrogen-containing compounds (e.g., alkaloids and glucosinolates), terpenes (terpenoids—e.g., isoprene-C5, monoterpenes-C10, sesquiterpenes-C15, diterpenes-C20, and polyterpenoids-C5xn), and phenolic compounds (e.g., phenylpropanoids and flavonoids) [52,53,54].
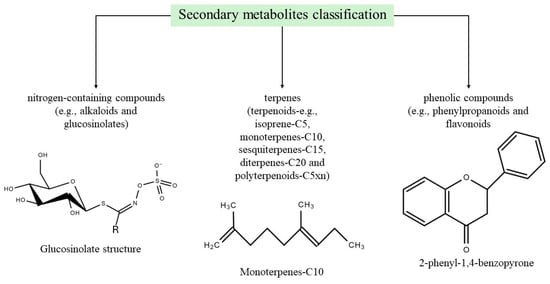
Figure 2.
Schematic representation of main plant secondary metabolite groups.
Plant secondary metabolite (SM) concentrations vary when exposed to salt stress-induced osmotic stress and ion toxicity [49]. Anthocyanins have been observed to amass in salt-tolerant plants and decrease in sensitive plants in response to saline soil stress conditions [49,55]. Research has demonstrated that endogenous jasmonic acid and polyphenols accumulate in tomato cultivars under salt stress conditions [55,56]. Several findings have shown a correlation between peppers’ total phenolic content and plant polyamine accumulation in response to salinity [57]. A salt-tolerant quinoa plant subjected to elevated salinity conditions exhibited a pronounced accumulation of saponin levels compared to the control plants [58,59]. Furthermore, a correlation was demonstrated between saponins and plant salinity tolerance, with a notable increase in triterpenoid concentrations observed in the shoots and roots of Kandeliacandel and Bruguiera gymnorrhiza under high-salinity conditions. In addition to saponins, other secondary metabolites have been observed to have accumulated at high levels, instigating salinity stress tolerance by enhancing antioxidant functions [49,60,61]. Saponins have also been found to be overexpressed in the shoots and roots of Medicago sativa and Medicago arborea, respectively, indicating its critical role in response to salinity stress [61]. Flavonoids, polyphenols, tannins, and anthocyanins are secondary metabolites known to render antioxidant activity and accumulate in response to salt stress [49,60]. Such alterations are conducive to a diminution of perturbational effects on the production of crops.
3. Manipulation of Plant Antioxidant Pathways Under Saline Conditions
Plants have evolved a complex antioxidant system to prevent cellular damage caused by salinity stress (Figure 3). The primary components of this system are carotenoids, ascorbate, glutathione, and tocopherols, in addition to the following enzymes: superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPX), and peroxidases (Figure 3) [62]. Furthermore, enzymes involved in the ascorbate–glutathione cycle, such as ascorbate peroxidase (APX) and glutathione reductase (GR), have been found to lessen the adverse effects of salinity stress (Figure 3) [63].
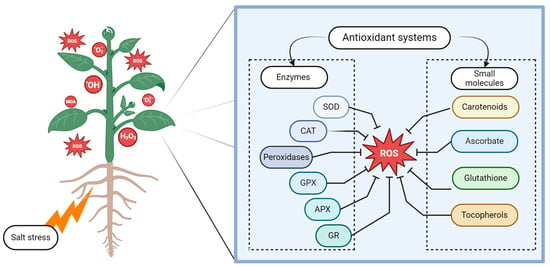
Figure 3.
Plant antioxidant protection components against reactive oxygen species induced by salt stress. MDA—malondialdehyde; ROS—reactive oxygen species; SOD—superoxide dismutase; CAT—catalase; GPX—glutathione peroxidase, APX—ascorbate peroxidase; GR—glutathione reductase; blunt-ended lines (T) inhibiting effects. Created using BioRender.com.
The manipulation of antioxidant enzymes and small antioxidant molecules in chloroplasts is essential to ensure the continuous productivity of agricultural plants exposed to salinity stress. Numerous studies have shown that salt-tolerant plants exhibit high peroxidase activity and enhanced antioxidant enzymatic activity and antioxidant contents [64]. For example, Sarker et al. showcased a salt-tolerant variety of Amaranthus tricolor (VA14) that exhibited a high level of superoxide dismutase (SOD), ascorbate, and APX, in support of the detoxification of reactive oxygen species (ROS) [65]. The investigation by Hussain et al. on the salt tolerance of contrasting wheat cultivars revealed that low malondialdehyde deposition in saline stress-tolerant varieties was associated with low membrane lipid peroxidation [66,67]. The upregulation of the peroxidase gene GsPRX9 and enhanced antioxidant activity in wild-type soybeans has been shown to enhance salt tolerance [68]. Seedlings of barley showed a significant effect on the percent germination and enzymatic activity changes, coupled with the roles of antioxidants and expressed genes in responding to salt stress [69]. The findings demonstrated that numerous enzymes, molecules, and pigments play crucial roles in mitigating the detrimental effects of oxidative damage, thereby enhancing plants’ salinity tolerance [70]. Deeper knowledge of antioxidant enzyme regulation and their production is crucial for developing transgenic plants with modified levels of antioxidant enzymes, because elevated antioxidant production in response to specific stress could lead to tolerance against multiple stressors.
4. Advances in Plant Genetic Engineering to Withstand Salinity Stress
Under salinity stress conditions, agricultural cultivars exhibited differential gene regulation with diverse functions (Figure 4) [71]. These regulatory processes result in the activation of different developmental and physiological mechanisms that are involved in stress-associated growth and metabolic alterations [71]. To improve plant saline stress tolerance, different approaches have been developed in advanced plant genetic engineering. These approaches aim to regulate the activity of enzymes involved in functional metabolites, antioxidant enzymes, transporters synthesis, and enzymes for membrane lipid biosynthesis [72,73]. The encoded proteins include transporters and channels for ions, enzymes associated with the biosynthesis of osmolytes, antioxidant systems, and protective proteins such as late embryogenesis abundant (LEA) proteins. These proteins play a pivotal role in the exhibition of salt stress-sensing and signal transduction pathways [74]. Substantial recent findings have revealed the significance of transcription factors (TFs) belonging to the TF families of ERF/AP2, bZIP, MYB, MYC, NAC, WRKY, and zinc-finger proteins as regulatory elements in the modulation of salinity stress conditions [75,76]. Genes encoding single- and multi-ion transporters, derived from multiple sources, have been engineered in numerous plant species to enhance their salt tolerance. The overexpression of AtNHX1 and related NHX proteins has been demonstrated to enhance salinity stress tolerance in several plants, including brassica, wheat, cotton, tobacco, tomato, and soybeans [77,78,79,80,81,82]. Over the years, halophytes have served as a primary source of genes to enhance salinity stress tolerance in numerous plant cultivars. For instance, the expression of a vacuolar H+-ATPase subunit c1 (SaVHAc1) gene from the halophyte grass Spartina alterniflora led to an improved rice salinity stress tolerance [83]. An associated study on a genome-wide of 350 barley genotypes at seed germination annotated 19 loci including less than 50 molecular markers associated with salt stress tolerance [84]. Salt tolerance-related genes within species can be generated through the process of mutation or genetic engineering. Salt tolerance-related genes within species can be generated through the process of mutation or genetic engineering. Consequently, this is used as donors in genetic engineering integration into plant breeding programs has targeted developing plants tolerant to saline soil conditions, representing a significant and valuable advancement.
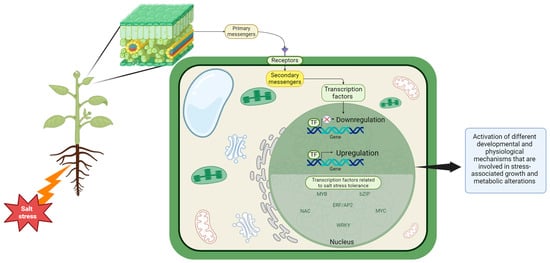
Figure 4.
The mechanism of regulation of gene expression via transcription factors as a response to salt stress, that agricultural cultivars exhibited in differential gene regulation with diverse functions in response to salinity stress conditions; TF—transcription factors; ⊗—suppression of the gene expression. Created in BioRender.com.
RNA Interference Technology for Enhancing Plants’ Saline Soil Stress Tolerance
When subjected to salinity stress conditions, the up and down-regulation of transcript plant genes plays a critical role in post-transcriptional genes in plants grown in saline soil conditions [85]. For instance, Arabidopsis thaliana, Oryza sativa, Phaseolous vulgaris, Populus trichocarpa, and other plant species have been investigated for the importance of microRNA (miRNA) in response to saline soil stress (Figure 5) [86]. A research work on Arabidopsis revealed the differential regulation of several miRNAs in salt-stressed tissue [87]. In response to salinity condition, several miRNAs were upregulated in Arabidopsis, including miR156, miR158, miR159, miR165, miR167, miR168, miR169, miR171, miR319, miR393, miR394, miR396, and miR397, while miR398 was down-regulated, thus indicating adaptive response of miRNAs under salt stresses. In P. vulgaris miRS1 and miR159.2 was also up-regulated under salt stress [88]. Up-regulation of miR530a, miR1445, miR1446a-e, miR1447, miR171l-n and down-regulation of miR482.2 and miR1450 were also observed in P. trichocarpa plants under salt stress conditions. These findings indicate the crucial role that miRNAs play in environmental stress conditions—particularly saline soil stress conditions—and it can be an important tool to create cultivars that are tolerant to salt stress conditions [87].
The targets of the sulfurylase and ASP1 genes are expressed in salt-induced soybean lines under sulfate starvation conditions and are regulated by miR395. This suggests that miR395 may play a role in nonspecific salt-stressed pathways, such as those involved in energy supply maintenance [89]. The homologous artichoke cca-miR397 and 399 are members of a laccase gene family that has also been found to participate in salt stress response [89]. In artichokes, reduced expression of miR397 in the roots during salt stress has been observed, leading to enhanced expression of laccase [89]. A multi-copper-containing glycoprotein laccase is present in plants and contributes to lignin formation. The upregulation of the laccase gene is enhanced by a high concentration of NaCl in tomato and maize roots. The AGROUNAUTAE1 (AGO1) gene, which encodes for the RNA slicer enzyme in the miRNA pathway, is regulated by miR168 [90,91,92,93]. It can be observed that both AGO1 and miR168 are important in maintaining an equilibrium between the target miRNA and their targeted gene. Furthermore, miR168 has been identified in maize subjected to salt stress [94]. In a recent study on rice, Wan et al. proposed that miR168 directly regulates AGO1-dependent gene regulation, which explains the differences in the salt stress response of STTM168 rice roots and that of the non-transgenic controls [95]. It has been previously proposed that miR168 may interfere with salt stress responses by downregulating its target gene AGO1, thereby affecting the activities of other miRNAs. Furthermore, it has been demonstrated that miR319, which acts positively to enhance salt response mechanisms, inhibits its target genes at the post-transcriptional level. The group identified the following genes as differentially expressed between the STTM168 and non-transgenic control plants: LOC_Os02g03840.2, OsRCI25 (LOC_Os03g17790.1), LOC_Os07g07270.1, LOC_Os02g09480.1, and LOC_Os03g50540 [96]. The researchers observed that STTM168 improved rice salt stress tolerance and concluded that the miR168-AGO1 cascade may be functionally conserved in plant salt stress adaptation. The results demonstrate that plant genes can be up- or down-regulated, indicating that RNA interference (RNAi) technology can be employed to enhance agricultural plants’ resilience to salinity stress. Importantly, investigation and identification of miRNA-mediated gene regulation under salt stress will improve our understanding of the complex regulatory networks involved in salt stress. A deeper understanding of miRNA during salt stress will create new possibilities to enhance plant tolerance under salt stress.
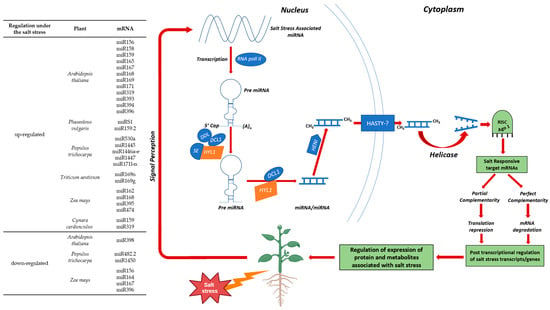
Figure 5.
Mechanisms and regulation of various miRNAs in response to salt stress [86]. HASTY-?—assumed exporter of the plant miRNAs [97].
5. Engineered Nanoparticles and Nanomaterials to Improve the Resilience of Plants Exposed to Salinity Stress
Recently, nanotechnology has been highlighted for its innovative approach to improve agricultural crop resilience in harmful environmental conditions, including salinity stress [98,99]. Engineered nanoparticles are a highly promising avenue for mitigating salinity stress, a phenomenon that presents a significant challenge to agricultural productivity. Engineered nanoparticles applied in agriculture are ultra-small particles with distinctive physicochemical properties, which can improve agricultural efficacy like fertilizers, pesticides, and plant growth regulators [99]. Engineered nanoparticles have been developed to play specific functions in assisting the growth and development of plants in facing salinity stress, including improvements in physiological processes and biochemical reactions, as shown in Figure 6. Furthermore, these nanoparticles have been demonstrated to regulate saline soil stress tolerance in wheat via alterations in abscisic acid (ABA) concentrations, ion homeostasis, and defense systems [100].
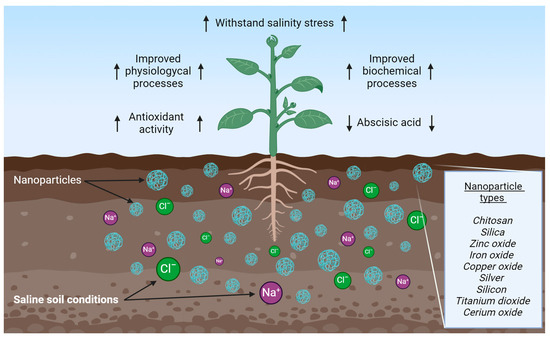
Figure 6.
Effects of the nanoparticle usage for plants under salt stress. Arrows illustrate ↑ enhancing and ↓ reducing effects. Created using BioRender.com.
Engineered nanoparticle types have been made to mitigate the adversity effects of salinity stress on plant species, with varying results. These include chitosan (Ch) nanoparticles, silica (SiO2) nanoparticles, zinc oxide (ZnO) nanoparticles, iron oxide (IO) nanoparticles, copper oxide (CuO) nanoparticles, silver (Ag) nanoparticles, silicon (Si) nanoparticles, and titanium dioxide (TiO2) nanoparticles [22,101]. For example, TiO₂ nanoparticles have been demonstrated to improve salt-stressed tomato plants’ growth and photosynthetic efficiency [22,101]. Similarly, Ag nanoparticles have revealed improvements in the tolerance of wheat cultivars under high salinity, increasing growth [98,100,101,102]. Additionally, ZnO nanoparticles increased the survival of maize cultivars grown in saline soils [102]. In contrast, CuO nanoparticles have been observed to hurt soybean plants’ growth under saline conditions [103]. Furthermore, cerium oxide (CeO2) nanoparticles resulted in stunted growth of maize plants grown in saline soils [104]. The application of engineered nanoparticles in agricultural systems is known to improve water retention, increase antioxidant activity, and regulate ion transport and uptake in plants under salt stress conditions [22].
Carbon-based nanomaterials (CBNs) have also demonstrated efficacy in promoting plant growth and alleviating abiotic stress, particularly salt stress, in a range of crops, including bioenergy plants as engineered nanoparticles [105,106,107]. It has been shown that CBNs not only play a role in improving salt stress tolerance but can also increase seed germination rates and biomass production [99]. Additionally, nanomaterials have been demonstrated to enhance water uptake, photosynthetic efficiency, and antioxidant activities while reducing oxidative stress markers at lower concentrations [108]. CBNs can safeguard plants by mimicking antioxidative enzymes and mitigating damage caused by ROS [109]. However, higher concentrations of nanomaterials may induce phytotoxicity and increase ROS generation [109]. These affect the physiology of plants, reducing growth and development and eventually leading to the death in most plants. The application of nanomaterials in agriculture presents both opportunities and challenges, necessitating further research to elucidate their behavior and effects on plants at the molecular and subcellular levels under various agroecosystem conditions [110]. Therefore, there is a need to highlight the research to understand the challenges that come with the use of nanoparticles [109].
6. Arbuscular Mycorrhizal Fungi (AMF) and Plant Growth-Promoting Bacteria (PGPB) Mediate the Growth of Plants Exposed to Salinity Stress
Plant root systems are mostly colonized by arbuscular mycorrhizal, which influence growth and development, improving the resilience of plants exposed to saline stress conditions (Figure 7). Numerous findings have described the role of AMF in improving plant defenses against salinity stress. These findings have demonstrated that increases in nutrient uptake, osmoregulator accumulation, improvements in photosynthetic rate, and increases in water use efficiency are associated with the mitigation of saline soil stress conditions by AMF [111,112,113].
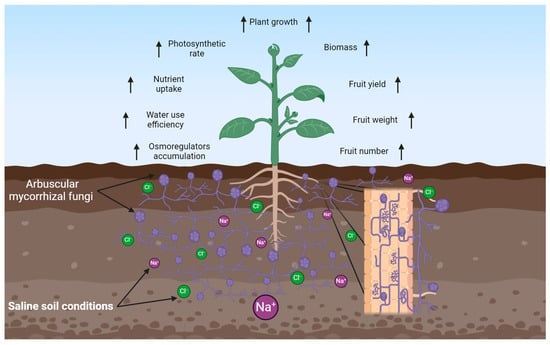
Figure 7.
Effects of arbuscular mycorrhizal fungi on plants grown in salt saline soil. Arrows (↑) illustrate positive effects. Created using BioRender.com.
Several mechanisms have also been suggested for AMF symbiosis in alleviating salt stress in plant hosts. Among them are plant growth and biomass, biochemical, physiological, molecular, and ultra-structural changes [114]. Plant growth and biomass allocation are hindered by salinity stress conditions. This may be because of the increase in the osmotic potential of the salt-affected soils, which hinders nutrient uptake by plants. However, AMF root colonization has been reported to result in increased nutrient uptake of the host plant, improving growth and biomass allocation. Higher shoot and root dry masses were recorded in mycorrhizal-inoculated Acacia nilotica seedlings than in uninoculated mycorrhizal seedlings [115]. A mycorrhizal tomato plant also reported increased shoot and root dry mass, fresh fruit yield, fruit mass, and number of fruits compared to control plants [116]. The improved growth of AMF-colonized plants has been moderately ascribed to improved nutrient uptake mediated by AMF; particularly improved phosphorous nutrition [117]. Together, these findings revealed the benefits of symbiosis in the alleviation of salinity stress on agricultural plant cultivars.
In addition to AMF, plant growth-promoting bacteria (PGPB) also have great potential to mitigate salt stress in plants. These bacteria enhance plant growth and salt stress tolerance through different PGPB-mediated mechanisms such as the production of phytohormones, 1-aminocyclopropane-1-carboxylic acid deaminase, and exopolysaccharides [14,118]. PGPB induces hormones such as auxin, cytokinin, and gibberellin, while hormones such as ethylene can be reduced by ACC deaminase. Ethylene is known to be involved in growth and developmental processes; however, higher concentrations of ethylene can be detrimental and inhibit plant growth. PGPB regulates ethylene levels in plants via ACC deaminase, thus inducing plant tolerance under salt stress [119]. PGPB can also modulate antioxidant defense systems, maintain ion homeostasis, and induce salt-responsive genes in plants [118]. For example, Klebsiella sp. SBP-8 has been demonstrated to enhance wheat growth under salt stress conditions by increasing Na+ exclusion (65%) and K+ uptake (84.21%) [120]. Moreover, the SBP-8 strain demonstrated increased 1-aminocyclopropane-1-carboxylatedeaminase activity by up to 6% under salt stress conditions, indicating its ability to survive and interact with plants in saline environments [120]. Thus, bacteria containing ACCD with salt tolerance could be beneficial in saline environments, providing positive effects on plants. Additionally, PGPB have been demonstrated to enhance nutrient mobilization, nitrogen fixation, and phosphate solubilization [121]. Employing PGPB as a biological instrument for the alleviation of salt stress offers a cost-effective and sustainable methodology for the enhancement of crop yield in saline environments [14,121].
7. Organic Amendments Moderate Plant Growth and Biomass Allocation under Salinity Stress
The recent use of various types of organic matter alterations to promote the growth of plants exposed to salt stress has demonstrated a decrease in the oxidative and osmotic stress of agricultural plants exposed to saline conditions. This reduction has been attributed to increased microbial activities [122,123]. This was achieved by enhancing the disposal of energy-rich C-containing compounds in the organic amendments, allowing soil microorganisms to biosynthesize osmolytes to mitigate the effects of osmotic pressure from high-salinity conditions [124]. Organic soil amendments have been shown to offer considerable benefits for saline soil ecosystems when used in conjunction with compost [125]. It has been demonstrated that incorporating organic matter into saline soils can intensify the dissolution percentage of calcite (CaCO3) by facilitating the fast generation of carbonic acid, thereby hastening the binding of small particles within the matrix. This results in the formation of substantial aggregates, which remain stable in the aqueous environment. A wide range of organic amendments for soil, including manure composites and biochar, have been subjected to rigorous investigation to enhance the physical and chemical properties of saline–alkali soil, thereby lowering salinity, pH, and mitigating salinity stress conditions (Figure 8) [126]. For example, recent studies have indicated that biochar can result in notable differences in water content and movement within different layers and depths of the soil [127]. Organic matter-rich amendments incorporated into the soil have been demonstrated to enhance soil aggregation and improve the water-holding capacity, thereby hindering the impacts of salt stress on productivity [128,129]. For example, the use of biochar has been demonstrated to increase soil water content while reducing salinity concentrations by up to 22% [130,131]. Importantly, biochar increased the availability of soil potassium (K) concentrations by approximately 89% and foliar K concentrations up to 25% [131]. It is known that biochar possesses the ability to bind potentially toxic salt ions (Na+) at different scales [132]. This binding occurs due to the biochar’s inherent absorptive properties, which are further augmented by its enormously high porosity, cation exchange capacity, and large surface area [132]. Moreover, it has been shown that organic amendments can enhance 48, 39, and 84% of the root fresh weight in maize plants under salt stress [133]. The incorporation of organic amendments into marginal soils has been demonstrated to enhance the growth and development of agricultural crops, particularly those exposed to salinity stress conditions, thereby ensuring the sustainability of production. Thus, it is important to apply biochar to reduce salinity and support sustainable food production under changing climate conditions.
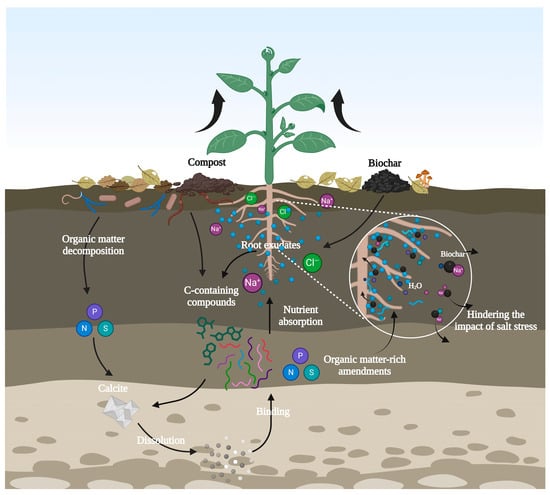
Figure 8.
Effects of organic amendments on plants growing in salt saline soil. Created using BioRender.com.
8. Trace Elements Used in Salinity Stress Alleviation
Trace elements are mostly required at low levels for plant survival and growth. Chlorine, boron, copper, ion, magnesium, manganese, zinc, etc. are some common examples of trace elements. Applying micronutrients enhances the biosynthesis and synthesis of genes, compatible solutes, and multiple enzymatic and nonenzymatic antioxidants [134]. These efficiently scavenge toxic biomolecules like ROS and methylglyoxal (MG), and they accumulate ophenolics and flavonoids thus mitigating salt stress. The trace elements promote the exportation of accumulated Na+ ions from the plant cells through induced upregulation of transporter and stress-associated osmotic stress-responsive genes, contributing to plant salt tolerance (Figure 9). Multiple salinity-related genes, including those encoding dehydration-responsive element-binding proteins 1, 2, and 3 (DREB1, 2, 3), as well as APX, SOD, CAT, and genes involved in silicon transport like leLsi-1, -2, and -3, were observed to be upregulated in salt-stressed tomato seedlings under Si treatment [135]. Exogenous plant micronutrient treatment supplements metabolic pathways and promotes growth [134,136]. For example, Zn-treated plants showed higher heights, dry weights, and fresh weights under 70 mM NaCl conditions [134]. Molybdenum reduced oxidative damage to plant tissues when crested wheatgrass was subjected to saline soil stress conditions by raising the activities of the three molybdenum-containing enzymes: nitrate reductase, aldehyde oxidase, and xanthine dehydrogenase [137,138]. Evidence has shown that applying selenium to parsley plants alleviated salinity stress by enhancing PSII function and decreasing Na content in the shoot through the binding of Na to the root cell wall [139]. Al-Zahrani et al. exogenously applied zinc to the seedlings of Vigna radiata and reported salinity stress tolerance [140]. Despite several positive correlations between trace elements and salt tolerance, this relationship is not completely known and still needs further investigation. This field requires investigation at the molecular and epigenomic levels. Furthermore, optimal application of trace elements will help to avoid unnecessary crop losses. In this regard, it is very important to conduct experiments to identify the exact dose, duration, and application procedure for different species and cultivars.
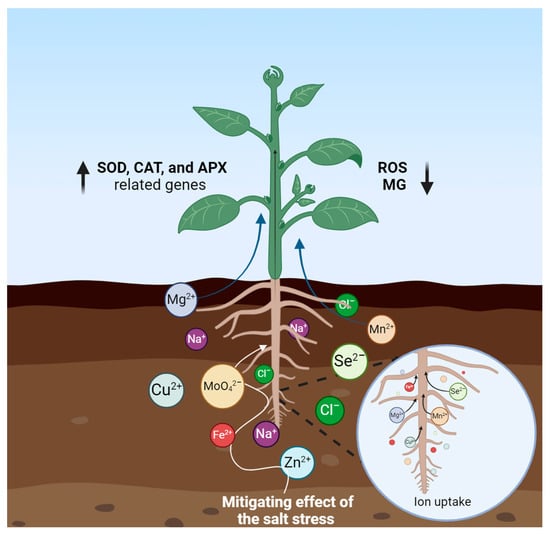
Figure 9.
Effects of the trace elements mitigating salt stress. Bold arrows illustrate ↑ enhancing and ↓ reducing effects. Thin arrows demonstrate ion uptake. Created using BioRender.com.
9. Concluding Remarks and Future Perspectives
This review highlights the innovative strategies applied to agricultural crop plants to alleviate salinity stress, displaying their potential to ensure sustainable agricultural productivity. The use of metabolomics approaches in the total inclusivity of cellular metabolite profiling provides a high-level approach to examining the influence of metabolite changes on a wider scale. State-of-the-art tools and procedures, including transcriptomics, proteomics, and metabolomics, have been employed regularly to comprehend the cellular processes, genetic control, and signaling networks involved in plant exposure to salinity stress. Plant-associated microorganisms as biofertilizers promote plant productivity and salinity-affected soils. These organisms can initiate the osmotic reaction, growth hormones, and nutritional elements, working as biocontrol agents by inducing specific plant genes. Organic matter and amendments can be relied on to amend saline soil conditions by enhancing the soil’s physical and chemical conditions. Recently, nanotechnology has been used for specific nutrient availability purposes and the conservation of soil fertility. Trace elements, applied in the right quantity, have also been highlighted to improve the tolerance of crops. There is a need for research scientists to conduct field experiments regarding salinity, particularly regarding land evaluation and opportunities for salinity management in the agroecosystem to promote land productivity. The future perspective of this study lies in the introduction and improvement of the discussed developmental innovative strategies in future research and breeding programs. These efforts will significantly improve the tolerance of agricultural crops in response to saline stress, thereby increasing the productivity of degraded agricultural lands. Additionally, farmers need to be informed and motivated to adopt such developmental technological solutions to mitigate soil salinization, which will help to achieve food security targets.
Author Contributions
Conceptualization, Z.N.; investigation, Z.N. and M.S.; data curation, Z.N., M.S. and S.Z.; writing—original draft preparation, Z.N., M.S. and S.Z.; writing—review and editing, Z.N., M.S., S.Z., J.C., Y.C. and Z.W.; visualization, Z.N., Z.W. and M.S.; revising, Z.N., Z.W., M.S. and S.Z.; supervision, S.Z.; project administration, M.B., A.A. and R.U.; funding acquisition, S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan, grant number AP19679378.
Data Availability Statement
All the data are available within the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying Mechanics of Plant Growth Promoting Rhizobacteria (PGPR): A Review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Saghafi, D.; Delangiz, N.; Lajayer, B.A.; Ghorbanpour, M. An Overview on Improvement of Crop Productivity in Saline Soils by Halotolerant and Halophilic PGPRs. 3 Biotech 2019, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, A.; Varma, A.; Tuteja, N.; Choudhary, D.K. PGPR-Mediated Amelioration of Crops Under Salt Stress. In Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Choudhary, D.K., Varma, A., Tuteja, N., Eds.; Springer: Singapore, 2016; pp. 205–226. ISBN 978-981-10-2854-0. [Google Scholar]
- Yergaliyev, T.M.; Nurbekova, Z.; Mukiyanova, G.; Akbassova, A.; Sutula, M.; Zhangazin, S.; Bari, A.; Tleukulova, Z.; Shamekova, M.; Masalimov, Z.K.; et al. The Involvement of ROS Producing Aldehyde Oxidase in Plant Response to Tombusvirus Infection. Plant Physiol. Biochem. 2016, 109, 36–44. [Google Scholar] [CrossRef]
- Sutula, M.Y.; Akbassova, A.Z.; Yergaliyev, T.M.; Nurbekova, Z.A.; Mukiyanova, G.S.; Omarov, R.T. Endowing Plants with Tolerance to Virus Infection by Their Preliminary Treatment with Short Interfering RNAs. Russ. J. Plant Physiol. 2017, 64, 939–945. [Google Scholar] [CrossRef]
- Srivastava, S.; Brychkova, G.; Yarmolinsky, D.; Soltabayeva, A.; Samani, T.; Sagi, M. Aldehyde Oxidase 4 Plays a Critical Role in Delaying Silique Senescence by Catalyzing Aldehyde Detoxification. Plant Physiol. 2017, 173, 1977–1997. [Google Scholar] [CrossRef]
- Nurbekova, Z.; Srivastava, S.; Standing, D.; Kurmanbayeva, A.; Bekturova, A.; Soltabayeva, A.; Oshanova, D.; Turečková, V.; Strand, M.; Biswas, M.S.; et al. Arabidopsis Aldehyde Oxidase 3, Known to Oxidize Abscisic Aldehyde to Abscisic Acid, Protects Leaves from Aldehyde Toxicity. Plant J. 2021, 108, 1439–1455. [Google Scholar] [CrossRef]
- Kurmanbayeva, A.; Bekturova, A.; Soltabayeva, A.; Oshanova, D.; Nurbekova, Z.; Srivastava, S.; Tiwari, P.; Dubey, A.K.; Sagi, M. Active O-Acetylserine-(Thiol) Lyase A and B Confer Improved Selenium Resistance and Degrade l-Cys and l-SeCys in Arabidopsis. J. Exp. Bot. 2022, 73, 2525–2539. [Google Scholar] [CrossRef] [PubMed]
- Soltabayeva, A.; Bekturova, A.; Kurmanbayeva, A.; Oshanova, D.; Nurbekova, Z.; Srivastava, S.; Standing, D.; Sagi, M. Ureides Are Accumulated Similarly in Response to UV-C Irradiation and Wounding in Arabidopsis Leaves but Are Remobilized Differently during Recovery. J. Exp. Bot. 2022, 73, 1016–1032. [Google Scholar] [CrossRef]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity Tolerance Mechanisms in Glycophytes: An Overview with the Central Focus on Rice Plants. Rice 2012, 5, 11. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Aubakirova, K.; Satkanov, M.; Kulataeva, M.; Assylbekova, G.; Kambarbekova, A.; Alikulov, Z. Molybdoenzymes Isolated from S. glanis Liver Can Produce Nitric Oxide from Nitrates and Nitrites. Czech J. Anim. Sci. 2023, 68, 222–230. [Google Scholar] [CrossRef]
- Aitlessov, K.; Zhumabekova, B.; Sagyndykov, U.; Tuyakbayeva, A.; Bitkeyeva, A.; Bazarbaeva, K.Z.; Mukhtarov, A.; Nurbekova, Z.; Satkanov, M.; Kulatayeva, M.; et al. Foliar Fertilization with Molybdate and Nitrate Up-Regulated Activity of Nitrate Reductase in Lemon Balm Leaves. Horticulturae 2023, 9, 1325. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant Growth-Promoting Bacteria: Biological Tools for the Mitigation of Salinity Stress in Plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef] [PubMed]
- Sultana, M.S.; Sakurai, C.; Biswas, M.S.; Szabados, L.; Mano, J. Accumulation of Reactive Carbonyl Species in Roots as the Primary Cause of Salt Stress-Induced Growth Retardation of Arabidopsis thaliana. Physiol. Plant. 2024, 176, e14198. [Google Scholar] [CrossRef]
- Yan, K.; Shao, H.; Shao, C.; Chen, P.; Zhao, S.; Brestic, M.; Chen, X. Physiological Adaptive Mechanisms of Plants Grown in Saline Soil and Implications for Sustainable Saline Agriculture in Coastal Zone. Acta Physiol. Plant. 2013, 35, 2867–2878. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Sisay, T.A.; Nurbekova, Z.; Oshanova, D.; Dubey, A.K.; Khatri, K.; Mudgal, V.; Mudgal, A.; Neori, A.; Shpigel, M.; Srivastava, R.K.; et al. Effect of Salinity and Nitrogen Fertilization Levels on Growth Parameters of Sarcocornia fruticosa, Salicornia brachiata, and Arthrocnemum macrostachyum. Agronomy 2022, 12, 1749. [Google Scholar] [CrossRef]
- Saberi Riseh, R.; Ebrahimi-Zarandi, M.; Tamanadar, E.; Moradi Pour, M.; Thakur, V.K. Salinity Stress: Toward Sustainable Plant Strategies and Using Plant Growth-Promoting Rhizobacteria Encapsulation for Reducing It. Sustainability 2021, 13, 12758. [Google Scholar] [CrossRef]
- Mishra, A.K.; Das, R.; George Kerry, R.; Biswal, B.; Sinha, T.; Sharma, S.; Arora, P.; Kumar, M. Promising Management Strategies to Improve Crop Sustainability and to Amend Soil Salinity. Front. Environ. Sci. 2023, 10, 962581. [Google Scholar] [CrossRef]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium Transport in Plant Cells. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1465, 140–151. [Google Scholar] [CrossRef]
- Junedi, M.A.; Mukhopadhyay, R.; Manjari, K.S. Alleviating Salinity Stress in Crop Plants Using New Engineered Nanoparticles (ENPs). Plant Stress 2023, 9, 100184. [Google Scholar] [CrossRef]
- Oh, D.-H.; Lee, S.Y.; Bressan, R.A.; Yun, D.-J.; Bohnert, H.J. Intracellular Consequences of SOS1 Deficiency during Salt Stress. J. Exp. Bot. 2010, 61, 1205–1213. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, P.; Liu, Z.; Yu, B.; Shi, H. Soybean Na+/H+ Antiporter GmsSOS1 Enhances Antioxidant Enzyme Activity and Reduces Na+ Accumulation in Arabidopsis and Yeast Cells under Salt Stress. Acta Physiol. Plant. 2016, 39, 19. [Google Scholar] [CrossRef]
- Feki, K.; Quintero, F.J.; Khoudi, H.; Leidi, E.O.; Masmoudi, K.; Pardo, J.M.; Brini, F. A Constitutively Active Form of a Durum Wheat Na+/H+ Antiporter SOS1 Confers High Salt Tolerance to Transgenic Arabidopsis. Plant Cell Rep. 2014, 33, 277–288. [Google Scholar] [CrossRef]
- Gao, J.; Sun, J.; Cao, P.; Ren, L.; Liu, C.; Chen, S.; Chen, F.; Jiang, J. Variation in Tissue Na+ Content and the Activity of SOS1 Genes among Two Species and Two Related Genera of Chrysanthemum. BMC Plant Biol. 2016, 16, 98. [Google Scholar] [CrossRef]
- Amin, I.; Rasool, S.; Mir, M.A.; Wani, W.; Masoodi, K.Z.; Ahmad, P. Ion Homeostasis for Salinity Tolerance in Plants: A Molecular Approach. Physiol. Plant. 2021, 171, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cai, S.; Wang, G.; Wang, F.; Dong, F.; Mak, M.; Holford, P.; Ji, J.; Salih, A.; Zhou, M.; et al. Halophytic NHXs Confer Salt Tolerance by Altering Cytosolic and Vacuolar K+ and Na+ in Arabidopsis Root Cell. Plant Growth Regul. 2017, 82, 333–351. [Google Scholar] [CrossRef]
- Ye, C.-Y.; Zhang, H.-C.; Chen, J.-H.; Xia, X.-L.; Yin, W.-L. Molecular Characterization of Putative Vacuolar NHX-Type Na+/H+ Exchanger Genes from the Salt-Resistant Tree Populus euphratica. Physiol. Plant 2009, 137, 166–174. [Google Scholar] [CrossRef]
- Albaladejo, I.; Meco, V.; Plasencia, F.; Flores, F.B.; Bolarin, M.C.; Egea, I. Unravelling the Strategies Used by the Wild Tomato Species Solanum Pennellii to Confront Salt Stress: From Leaf Anatomical Adaptations to Molecular Responses. Environ. Exp. Bot. 2017, 135, 1–12. [Google Scholar] [CrossRef]
- Gu, S.; Han, S.; Abid, M.; Bai, D.; Lin, M.; Sun, L.; Qi, X.; Zhong, Y.; Fang, J. A High-K+ Affinity Transporter (HKT) from Actinidia valvata Is Involved in Salt Tolerance in Kiwifruit. Int. J. Mol. Sci. 2023, 24, 15737. [Google Scholar] [CrossRef]
- Irulappan, V.; Park, H.W.; Han, S.-Y.; Kim, M.-H.; Kim, J.S. Genome-Wide Identification of a Novel Na+ Transporter from Bienertia sinuspersici and Overexpression of BsHKT1;2 Improved Salt Tolerance in Brassica rapa. Front. Plant Sci. 2023, 14, 1302315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qi, C.; Li, C.; Huang, D.; Mao, H.; Lin, X. Overexpression of High Affinity K+ Transporter from Nitraria sibirica Enhanced Salt Tolerance of Transgenic Plants. Plant Sci. 2024, 342, 112052. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Wu, X.; Jiang, X.; Gong, B.; Gao, H. Types of Membrane Transporters and the Mechanisms of Interaction between Them and Reactive Oxygen Species in Plants. Antioxidants 2024, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Do, T.D.; Chen, H.; Hien, V.T.T.; Hamwieh, A.; Yamada, T.; Sato, T.; Yan, Y.; Cong, H.; Shono, M.; Suenaga, K.; et al. Ncl Synchronously Regulates Na+, K+ and Cl− in Soybean and Greatly Increases the Grain Yield in Saline Field Conditions. Sci. Rep. 2016, 6, 19147. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, Z.; Wei, J.; Zhao, Z.; Sun, D.; Cui, S. A Na+/Ca2+ Exchanger-like Protein (AtNCL) Involved in Salt Stress in Arabidopsis. J. Biol. Chem. 2012, 287, 44062–44070. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, G.; Gonzales, N.; Guo, Y.; Hu, H.; Park, S.; Zhao, J. Ca2+-Regulated and Diurnal Rhythm-Regulated Na+/Ca2+ Exchanger AtNCL Affects Flowering Time and Auxin Signalling in Arabidopsis. Plant Cell Environ. 2016, 39, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Tyagi, S.; Sharma, Y.; Madhu; Sharma, A.; Pandey, A.; Singh, K.; Upadhyay, S.K. Expression of TaNCL2-A Ameliorates Cadmium Toxicity by Increasing Calcium and Enzymatic Antioxidants Activities in Arabidopsis. Chemosphere 2023, 329, 138636. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for Plant Stress Response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Yancey, P.H. Organic Osmolytes as Compatible, Metabolic and Counteracting Cytoprotectants in High Osmolarity and Other Stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Kulkarni, J.; Sharma, S.; Sahoo, S.A.; Mishra, S.; Nikam, T.D.; Borde, M.; Penna, S.; Srivastava, A.K. Resilience in Primary Metabolism Contributes to Salt Stress Adaptation in Sesuvium portulacastrum (L.). Plant Growth Regul. 2022, 98, 385–398. [Google Scholar] [CrossRef]
- Widodo; Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic Responses to Salt Stress of Barley (Hordeum vulgare L.) Cultivars, Sahara and Clipper, Which Differ in Salinity Tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar] [CrossRef]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the Roles of Osmolyte Accumulation during Stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef]
- Kadyrbaev, M.K.; Golovatskaya, I.F.; Satkanov, M.Z. Features of Regenerants Morphogenesis and Metabolism in Vitro, Obtained from Different Fragments of Potato Shoots. Vestn. Tomsk. Gos. Univ. Biol. 2021, 55, 114–134. [Google Scholar] [CrossRef]
- Gong, Q.; Li, P.; Ma, S.; Indu Rupassara, S.; Bohnert, H.J. Salinity Stress Adaptation Competence in the Extremophile Thellungiella halophila in Comparison with Its Relative Arabidopsis thaliana. Plant J. 2005, 44, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Akula, R.; Ravishankar, G.A. Influence of Abiotic Stress Signals on Secondary Metabolites in Plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef]
- Verma, N.; Shukla, S. Impact of Various Factors Responsible for Fluctuation in Plant Secondary Metabolites. J. Appl. Res. Med. Aromat. Plants 2015, 2, 105–113. [Google Scholar] [CrossRef]
- Kusano, M.; Yang, Z.; Okazaki, Y.; Nakabayashi, R.; Fukushima, A.; Saito, K. Using Metabolomic Approaches to Explore Chemical Diversity in Rice. Mol. Plant 2015, 8, 58–67. [Google Scholar] [CrossRef]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A Central Role for Thiols in Plant Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [PubMed]
- Xin, F.; Yang, C.; Wei, Y.; Ma, Q.X.; Yang, L.; Chen, X.-Y. Genomics Grand for Diversified Plant Secondary Metabolites. Plant Divers. Res. 2011, 33, 53–64. [Google Scholar]
- Wink, M. Modes of Action of Herbal Medicines and Plant Secondary Metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt Tolerance and Salinity Effects on Plants: A Review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Muthukumarasamy, M.; Gupta, S.D.; Panneerselvam, R. Enhancement of Peroxidase, Polyphenol Oxidase and Superoxide Dismutase Activities by Triadimefon in NaCl Stressed Raphanus sativus L. Biol. Plant. 2000, 43, 317–320. [Google Scholar] [CrossRef]
- Navarro, J.M.; Flores, P.; Garrido, C.; Martinez, V. Changes in the Contents of Antioxidant Compounds in Pepper Fruits at Different Ripening Stages, as Affected by Salinity. Food Chem. 2006, 96, 66–73. [Google Scholar] [CrossRef]
- Pulvento, C.; Riccardi, M.; Lavini, A.; Iafelice, G.; Marconi, E.; d’Andria, R. Yield and Quality Characteristics of Quinoa Grown in Open Field Under Different Saline and Non-Saline Irrigation Regimes. J. Agron. Crop Sci. 2012, 198, 254–263. [Google Scholar] [CrossRef]
- Moses, T.; Papadopoulou, K.K.; Osbourn, A. Metabolic and Functional Diversity of Saponins, Biosynthetic Intermediates and Semi-Synthetic Derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. [Google Scholar] [CrossRef]
- Edreva, A.; Velikova, V.; Tsonev, T.; Dagnon, S.; Aktaş, L.; Gesheva, E. Stress-Protective Role of Secondary Metabolites: Diversity of Functions and Mechanisms. Gen. Appl. Plant. Physiol. 2008, 34, 67–78. [Google Scholar]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, E.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity Stress Alters the Secondary Metabolic Profile of M. Sativa, M. Arborea and Their Hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef]
- Mukhamejanova, A.; Alikulov, Z.; Shapekova, N.; Aubakirova, K.; Mukhtarov, A. The Effect of Antioxidants on Xanthine Oxidase Activity in Fresh Ovine Milk. Potravin. Slovak J. Food Sci. 2021, 15, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.A.; Jiménez, A.; Mullineaux, P.; Sevilia, F. Tolerance of Pea (Pisum sativum L.) to Long-Term Salt Stress Is Associated with Induction of Antioxidant Defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Demiral, T.; Türkan, İ. Comparative Lipid Peroxidation, Antioxidant Defense Systems and Proline Content in Roots of Two Rice Cultivars Differing in Salt Tolerance. Environ. Exp. Bot. 2005, 53, 247–257. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. The Response of Salinity Stress-Induced A. tricolor to Growth, Anatomy, Physiology, Non-Enzymatic and Enzymatic Antioxidants. Front. Plant Sci. 2020, 11, 559876. [Google Scholar] [CrossRef]
- Hussain, N.; Sohail, Y.; Shakeel, N.; Javed, M.; Bano, H.; Gul, H.S.; Zafar, Z.U.; Frahat Zaky Hassan, I.; Ghaffar, A.; Athar, H.-R.; et al. Role of Mineral Nutrients, Antioxidants, Osmotic Adjustment and PSII Stability in Salt Tolerance of Contrasting Wheat Genotypes. Sci. Rep. 2022, 12, 12677. [Google Scholar] [CrossRef]
- Ali Solangi, K.; Wu, Y.; Xing, D.; Ahmed Qureshi, W.; Hussain Tunio, M.; Ali Sheikh, S.; Shabbir, A. Can Electrophysiological Information Reflect the Response of Mangrove Species to Salt Stress? A Case Study of Rewatering and Sodium Nitroprusside Application. Plant Signal. Behav. 2022, 17, 2073420. [Google Scholar] [CrossRef]
- Jin, T.; Sun, Y.; Zhao, R.; Shan, Z.; Gai, J.; Li, Y. Overexpression of Peroxidase Gene GsPRX9 Confers Salt Tolerance in Soybean. Int. J. Mol. Sci. 2019, 20, 3745. [Google Scholar] [CrossRef]
- Derakhshani, Z.; Bhave, M.; Shah, R.M. Metabolic Contribution to Salinity Stress Response in Grains of Two Barley Cultivars with Contrasting Salt Tolerance. Environ. Exp. Bot. 2020, 179, 104229. [Google Scholar] [CrossRef]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef]
- Saradadevi, R.; Mukankusi, C.; Li, L.; Amongi, W.; Mbiu, J.P.; Raatz, B.; Ariza, D.; Beebe, S.; Varshney, R.K.; Huttner, E.; et al. Multivariate Genomic Analysis and Optimal Contributions Selection Predicts High Genetic Gains in Cooking Time, Iron, Zinc, and Grain Yield in Common Beans in East Africa. Plant Genome 2021, 14, e20156. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, S.; Xie, L.; Lu, Z.; Liu, M.; Han, X.; Qiao, G.; Jiang, J.; Zhuo, R.; Qiu, W.; et al. Overexpression of Cysteine Protease Gene from Salix Matsudana Enhances Salt Tolerance in Transgenic arabidopsis. Environ. Exp. Bot. 2018, 147, 53–62. [Google Scholar] [CrossRef]
- Iksat, N.; Masalimov, Z.; Omarov, R. Plant Virus Resistance Biotechnological Approaches: From Genes to the CRISPR/Cas Gene Editing System. J. Water Land Dev. 2023, 57, 147–158. [Google Scholar] [CrossRef]
- Mann, A.; Kumar, N.; Lata, C.; Kumar, A.; Kumar, A.; Meena, B.L. Functional Annotation of Differentially Expressed Genes under Salt Stress in Dichanthium annulatum. Plant Physiol. Rep. 2019, 24, 104–111. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Luo, X.; Zhu, X.; Kinuthia, K.B.; Nie, S.; Feng, H.; Li, C.; Liu, L. Transcriptome-Based Gene Expression Profiling Identifies Differentially Expressed Genes Critical for Salt Stress Response in Radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jin, X.; Huang, H.; Derebe, M.G.; Levin, E.J.; Kabaleeswaran, V.; Pan, Y.; Punta, M.; Love, J.; Weng, J.; et al. Crystal Structure of a Potassium Ion Transporter, TrkH. Nature 2011, 471, 336–340. [Google Scholar] [CrossRef]
- Gouiaa, S.; Khoudi, H. Co-Expression of Vacuolar Na+/H+ Antiporter and H+-Pyrophosphatase with an IRES-Mediated Dicistronic Vector Improves Salinity Tolerance and Enhances Potassium Biofortification of Tomato. Phytochemistry 2015, 117, 537–546. [Google Scholar] [CrossRef]
- He, X.; Huang, X.; Shen, Y.; Huang, Z. Wheat V-H+-ATPase Subunit Genes Significantly Affect Salt Tolerance in Arabidopsis thaliana. PLoS ONE 2014, 9, e86982. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing Seed Mass and Oil Content in Transgenic Arabidopsis by the Overexpression of Wri1-like Gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Wei, J.; Qiu, X.; Hu, R.; Kuppu, S.; Auld, D.; Blumwald, E.; Gaxiola, R.; Payton, P.; Zhang, H. Co-Overexpression of AVP1 and AtNHX1 in Cotton Further Improves Drought and Salt Tolerance in Transgenic Cotton Plants. Plant Mol. Biol. Rep. 2015, 33, 167–177. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Z.-Z.; Zhou, X.-F.; Yin, H.-B.; Li, X.; Xin, X.-F.; Hong, X.-H.; Zhu, J.-K.; Gong, Z. Overexpression of SOS (Salt Overly Sensitive) Genes Increases Salt Tolerance in Transgenic arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef]
- Baisakh, N.; RamanaRao, M.V.; Rajasekaran, K.; Subudhi, P.; Janda, J.; Galbraith, D.; Vanier, C.; Pereira, A. Enhanced Salt Stress Tolerance of Rice Plants Expressing a Vacuolar H+-ATPase Subunit C1 (SaVHAc1) Gene from the Halophyte Grass Spartina alterniflora Löisel. Plant Biotechnol. J. 2012, 10, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.; Hill, C.B.; Zhang, X.-Q.; Li, C. Genome-Wide Association Study of Salinity Tolerance During Germination in Barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef]
- Pradhan, A.; Naik, N.; Sahoo, K.K. RNAi Mediated Drought and Salinity Stress Tolerance in Plants. Am. J. Plant Sci. 2015, 6, 1990–2008. [Google Scholar] [CrossRef]
- Mangrauthia, S.K.; Agarwal, S.; Sailaja, B.; Madhav, M.S.; Voleti, S.R. MicroRNAs and Their Role in Salt Stress Response in Plants. In Salt Stress in Plants: Signalling, Omics and Adaptations; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2013; pp. 15–46. ISBN 978-1-4614-6108-1. [Google Scholar]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De la Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and Novel miRNAs in the Legume Phaseolus Vulgaris in Response to Stress. Plant Mol. Biol. 2009, 70, 385–401. [Google Scholar] [CrossRef]
- De Paola, D.; Cattonaro, F.; Pignone, D.; Sonnante, G. The miRNAome of Globe Artichoke: Conserved and Novel Micro RNAs and Target Analysis. BMC Genom. 2012, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Davis, E.J.; Ballif, J.; Liang, M.; Bushman, E.; Haroldsen, V.; Torabinejad, J.; Wu, Y. Mutant Identification and Characterization of the Laccase Gene Family in Arabidopsis. J. Exp. Bot. 2006, 57, 2563–2569. [Google Scholar] [CrossRef]
- Liang, M.; Haroldsen, V.; Cai, X.; Wu, Y. Expression of a Putative Laccase Gene, ZmLAC1, in Maize Primary Roots under Stress*. Plant Cell Environ. 2006, 29, 746–753. [Google Scholar] [CrossRef]
- Wei, J.-Z.; Tirajoh, A.; Effendy, J.; Plant, A.L. Characterization of Salt-Induced Changes in Gene Expression in Tomato (Lycopersicon esculentum) Roots and the Role Played by Abscisic Acid. Plant Sci. 2000, 159, 135–148. [Google Scholar] [CrossRef]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The Nuclear dsRNA Binding Protein HYL1 Is Required for MicroRNA Accumulation and Plant Development, but Not Posttranscriptional Transgene Silencing. Curr. Biol. 2004, 14, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential Expression of miRNAs in Response to Salt Stress in Maize Roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef]
- Wan, J.; Meng, S.; Wang, Q.; Zhao, J.; Qiu, X.; Wang, L.; Li, J.; Lin, Y.; Mu, L.; Dang, K.; et al. Suppression of microRNA168 Enhances Salt Tolerance in Rice (Oryza sativa L.). BMC Plant Biol. 2022, 22, 563. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive Expression of a miR319 Gene Alters Plant Development and Enhances Salt and Drought Tolerance in Transgenic Creeping Bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, X. HASTY moves to chromatin for miRNA production. Mol. Plant 2021, 14, 364–365. [Google Scholar] [CrossRef]
- Khan, I.; Raza, M.A.; Awan, S.A.; Shah, G.A.; Rizwan, M.; Ali, B.; Tariq, R.; Hassan, M.J.; Alyemeni, M.N.; Brestic, M.; et al. Amelioration of Salt Induced Toxicity in Pearl Millet by Seed Priming with Silver Nanoparticles (AgNPs): The Oxidative Damage, Antioxidant Enzymes and Ions Uptake Are Major Determinants of Salt Tolerant Capacity. Plant Physiol. Biochem. 2020, 156, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.; Lahiani, M.H.; Hicks, V.K.; Hudson, M.K.; Green, M.J.; Khodakovskaya, M. Effects of Carbon-Based Nanomaterials on Seed Germination, Biomass Accumulation and Salt Stress Response of Bioenergy Crops. PLoS ONE 2018, 13, e0202274. [Google Scholar] [CrossRef] [PubMed]
- Wahid, I.; Kumari, S.; Ahmad, R.; Hussain, S.J.; Alamri, S.; Siddiqui, M.H.; Khan, M.I.R. Silver Nanoparticle Regulates Salt Tolerance in Wheat through Changes in ABA Concentration, Ion Homeostasis, and Defense Systems. Biomolecules 2020, 10, 1506. [Google Scholar] [CrossRef] [PubMed]
- Moulahoum, H.; Ghorbanizamani, F. Navigating the Development of Silver Nanoparticles Based Food Analysis through the Power of Artificial Intelligence. Food Chem. 2024, 445, 138800. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Ahmad, A.; Alhammad, B.A.; Tola, E. Exogenous Application of Zinc Oxide Nanoparticles Improved Antioxidants, Photosynthetic, and Yield Traits in Salt-Stressed Maize. Agronomy 2023, 13, 2645. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Torabian, S. Nano-Silicon Alters Antioxidant Activities of Soybean Seedlings under Salt Toxicity. Protoplasma 2018, 255, 953–962. [Google Scholar] [CrossRef]
- Rossi, L.; Zhang, W.; Lombardini, L.; Ma, X. The Impact of Cerium Oxide Nanoparticles on the Salt Stress Responses of Brassica napus L. Environ. Pollut. 2016, 219, 28–36. [Google Scholar] [CrossRef]
- Bhati, A.; Gunture; Tripathi, K.M.; Singh, A.; Sarkar, S.; Sonkar, S.K. Exploration of Nano Carbons in Relevance to Plant Systems. New J. Chem. 2018, 42, 16411–16427. [Google Scholar] [CrossRef]
- Darwish, M.; Mohammadi, A. Functionalized Nanomaterial for Environmental Techniques. In Nanotechnology in Environmental Science; Vch Pub: Hoboken, NJ, USA, 2018; pp. 315–350. ISBN 978-3-527-80885-4. [Google Scholar]
- Adhikari, S.; Eswar, N.K.R.; Mishra, A.K.; Sarkar, D.; Madras, G. Functionally Active Nanomaterials for Environmental Remediation. In Nanotechnology in Environmental Science; Vch Pub: Hoboken, NJ, USA, 2018; pp. 293–314. ISBN 978-3-527-80885-4. [Google Scholar]
- Alabdallah, N.M.; Alluqmani, S.M. Emerging Pivotal Role of Carbon Nanomaterials in Abiotic Stress Tolerance in Plants: A Mini Review. Arab J. Basic Appl. Sci. 2023, 30, 463–471. [Google Scholar] [CrossRef]
- Khan, M.N.; Mobin, M.; Abbas, Z.K.; AlMutairi, K.A.; Siddiqui, Z.H. Role of Nanomaterials in Plants under Challenging Environments. Plant Physiol. Biochem. 2017, 110, 194–209. [Google Scholar] [CrossRef] [PubMed]
- Azameti, M.K.; Imoro, A.-W.M. Nanotechnology: A Promising Field in Enhancing Abiotic Stress Tolerance in Plants. Crop Des. 2023, 2, 100037. [Google Scholar] [CrossRef]
- Hoekstra, F.A.; Crowe, J.H.; Crowe, L.M.; Van Roekel, T.; Vermeer, E. Do Phospholipids and Sucrose Determine Membrane Phase Transitions in Dehydrating Pollen Species? Plant Cell Environ. 1992, 15, 601–606. [Google Scholar] [CrossRef]
- Ouziad, F.; Wilde, P.; Schmelzer, E.; Hildebrandt, U.; Bothe, H. Analysis of Expression of Aquaporins and Na+/H+ Transporters in Tomato Colonized by Arbuscular Mycorrhizal Fungi and Affected by Salt Stress. Environ. Exp. Bot. 2006, 57, 177–186. [Google Scholar] [CrossRef]
- Zuccarini, P. Mycorrhizal Infection Ameliorates Chlorophyll Content and Nutrient Uptake of Lettuce Exposed to Saline Irrigation. Plant Soil Environ. 2007, 53, 283–289. [Google Scholar] [CrossRef]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular Mycorrhizal Fungi in Alleviation of Salt Stress: A Review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef]
- Giri, B.; Mukerji, K.G. Mycorrhizal Inoculant Alleviates Salt Stress in Sesbania aegyptiaca and Sesbania grandiflora under Field Conditions: Evidence for Reduced Sodium and Improved Magnesium Uptake. Mycorrhiza 2004, 14, 307–312. [Google Scholar] [CrossRef]
- Al-Karaki, G.N. Nursery Inoculation of Tomato with Arbuscular Mycorrhizal Fungi and Subsequent Performance under Irrigation with Saline Water. Sci. Hortic. 2006, 109, 1–7. [Google Scholar] [CrossRef]
- Sharifi, M.; Ghorbanli, M.; Ebrahimzadeh, H. Improved Growth of Salinity-Stressed Soybean after Inoculation with Salt Pre-Treated Mycorrhizal Fungi. J. Plant Physiol. 2007, 164, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Mishra, R.; Rai, S.; Bano, A.; Pathak, N.; Fujita, M.; Kumar, M.; Hasanuzzaman, M. Mechanistic Insights of Plant Growth Promoting Bacteria Mediated Drought and Salt Stress Tolerance in Plants for Sustainable Agriculture. Int. J. Mol. Sci. 2022, 23, 3741. [Google Scholar] [CrossRef] [PubMed]
- Ansari, F.A.; Ahmad, I.; Pichtel, J. Growth Stimulation and Alleviation of Salinity Stress to Wheat by the Biofilm Forming Bacillus Pumilus Strain FAB10. Appl. Soil Ecol. 2019, 143, 45–54. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.; Jha, P.N. The Plant-Growth-Promoting Bacterium Klebsiella Sp. SBP-8 Confers Induced Systemic Tolerance in Wheat (Triticum aestivum) under Salt Stress. J. Plant Physiol. 2015, 184, 57–67. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; AL-Harrasi, A. Plant Growth Promoting Bacteria as an Alternative Strategy for Salt Tolerance in Plants: A Review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Khan, S.; Irshad, S.; Mehmood, K.; Hasnain, Z.; Nawaz, M.; Rais, A.; Gul, S.; Wahid, M.A.; Hashem, A.; Abd_Allah, E.F.; et al. Biochar Production and Characteristics, Its Impacts on Soil Health, Crop Production, and Yield Enhancement: A Review. Plants 2024, 13, 166. [Google Scholar] [CrossRef]
- Ahmed, A.; Kurian, J.; Raghavan, V. Biochar Influences on Agricultural Soils, Crop Production, and the Environment: A Review. Environ. Rev. 2016, 24, 495–502. [Google Scholar] [CrossRef]
- Wichern, F.; Islam, M.R.; Hemkemeyer, M.; Watson, C.; Joergensen, R.G. Organic Amendments Alleviate Salinity Effects on Soil Microorganisms and Mineralisation Processes in Aerobic and Anaerobic Paddy Rice Soils. Front. Sustain. Food Syst. 2020, 4, 30. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rengel, Z. Environmental Salinization Processes: Detection, Implications & Solutions. Sci. Total Environ. 2021, 754, 142432. [Google Scholar] [CrossRef] [PubMed]
- Andrade Foronda, D. Reclamation of a Saline-Sodic Soil with Organic Amendments and Leaching. Environ. Sci. Proc. 2022, 16, 56. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, M.; Chen, H.; Chen, Y.; Wang, L.; Wang, R. Organic Amendments Promote Saline-Alkali Soil Desalinization and Enhance Maize Growth. Front. Plant Sci. 2023, 14, 1177209. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Liu, G.; Jiang, S.; Guan, X.; Chen, J.; Yao, R.; Wang, X. Bio-Organic Fertilizer Combined with Different Amendments Improves Nutrient Enhancement and Salt Leaching in Saline Soil: A Soil Column Experiment. Water 2022, 14, 4084. [Google Scholar] [CrossRef]
- Hoque, M.N.; Imran, S.; Hannan, A.; Paul, N.C.; Mahamud, M.A.; Chakrobortty, J.; Sarker, P.; Irin, I.J.; Brestic, M.; Rhaman, M.S. Organic Amendments for Mitigation of Salinity Stress in Plants: A Review. Life 2022, 12, 1632. [Google Scholar] [CrossRef]
- Khaled, H.; Fawy, A.H. Effect of Different Levels of Humic Acids on the Nutrient Content, Plant Growth, and Soil Properties under Conditions of Salinity. Soil Water Res. 2011, 6, 21–29. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, J.; Yao, R.; Chen, X.; Wang, X. Biochar and Fulvic Acid Amendments Mitigate Negative Effects of Coastal Saline Soil and Improve Crop Yields in a Three Year Field Trial. Sci. Rep. 2020, 10, 8946. [Google Scholar] [CrossRef] [PubMed]
- Mona, S.; Bhateria, R.; Deepak, B.; Kiran, B.; Nisha, R. Biochar for Reclamation of Saline Soils. In Microorganisms in Saline Environments: Strategies and Functions; Giri, B., Varma, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 451–466. ISBN 978-3-030-18975-4. [Google Scholar]
- Irshad, I.; Anwar-ul-Haq, M.; Akhtar, J.; Maqsood, M. Effects of Different Organic Amendments on Maize (Zea mays L.) Growth in Salt Affected Soil. J. Glob. Innov. Agric. Sci. 2022, 10, 121–130. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Role of Beneficial Trace Elements in Salt Stress Tolerance of Plants. In Plant Nutrients and Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Oku, H., Nahar, K., Hawrylak-Nowak, B., Eds.; Springer: Singapore, 2018; pp. 377–390. ISBN 978-981-10-9044-8. [Google Scholar]
- Muneer, S.; Jeong, B.R. Proteomic Analysis of Salt-Stress Responsive Proteins in Roots of Tomato (Lycopersicon esculentum L.) Plants towards Silicon Efficiency. Plant Growth Regul. 2015, 77, 133–146. [Google Scholar] [CrossRef]
- Pilon-Smits, E.A.; Quinn, C.F.; Tapken, W.; Malagoli, M.; Schiavon, M. Physiological Functions of Beneficial Elements. Curr. Opin. Plant Biol. 2009, 12, 267–274. [Google Scholar] [CrossRef]
- Babenko, O.N.; Brychkova, G.; Sagi, M.; Alikulov, Z.A. Molybdenum Application Enhances Adaptation of Crested Wheatgrass to Salinity Stress. Acta Physiol. Plant. 2015, 37, 14. [Google Scholar] [CrossRef]
- Satkanov, M.; Tazhibay, D.; Zhumabekova, B.; Assylbekova, G.; Abdukarimov, N.; Nurbekova, Z.; Kulatayeva, M.; Aubakirova, K.; Alikulov, Z. Method for Assessing the Content of Molybdenum Enzymes in the Internal Organs of Fish. MethodsX 2024, 12, 102576. [Google Scholar] [CrossRef] [PubMed]
- Habibi, G. Selenium Ameliorates Salinity Stress in Petroselinum crispum by Modulation of Photosynthesis and by Reducing Shoot Na Accumulation. Russ. J. Plant Physiol. 2017, 64, 368–374. [Google Scholar] [CrossRef]
- Al-Zahrani, H.S.; Alharby, H.F.; Hakeem, K.R.; Rehman, R.U. Exogenous Application of Zinc to Mitigate the Salt Stress in Vigna radiata (L.) Wilczek—Evaluation of Physiological and Biochemical Processes. Plants 2021, 10, 1005. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).