Abstract
The adaptability of bermudagrass genotypes to high-pH saline–alkali conditions was investigated through a comprehensive evaluation of 38 genotypes during the seedling stage. For this purpose, two distinct treatments were established: exposure to saline–alkali solution composed of 45% NaCl, 5% Na2SO4, 5% NaHCO3, and 45% Na2CO3 (pH 10.0), and exposure to distilled water as control. On 6th day of treatment, eight physiological indicators were measured. Compared with the control, the net photosynthetic rates, leaf water content, and chlorophyll content of the test genotypes decreased under stress. In contrast, the soluble protein content, proline levels, malondialdehyde concentration, and conductivity exhibited an increase. The salt–alkali tolerance coefficients of each indicator ranged from 0.24 to 8.54, and the variable coefficient was from 9.77% to 62.82%. Based on the salt–alkali tolerance coefficients, the comprehensive evaluation value (D) and resistance coefficient (CSAC) for each genotype were calculated. Subsequently, 38 genotypes were classified into three salt–alkali tolerance clusters by hierarchical clustering analysis, with Cluster I consisting of 10 genotypes with the most salt–alkali tolerance, and Cluster II with intermediate tolerance. Cluster III was comprised of 18 genotypes showing the lowest tolerance. The predictive model for assessing salt–alkali tolerance in bermudagrass is (D) = −0.238 + 0.106 × SACChlb + 0.209 × SACRWC + 0.015 × SACPro + 0.284 × SACProtein + 0.051 × SACPn. Notably, Cluster I genotypes were more vigorous and showed lower damage under saline stress compared to Cluster III. Moreover, stepwise regression analysis pinpointed Chlb, RWC, and Pro as crucial indicators for evaluating salt–alkali tolerance in bermudagrass genotypes.
1. Introduction
Soil salinity is one of the major environmental factors that influence plant growth and development [1]. Globally, there are over 800 million hectares of saline–alkali soil. Due to population growth, industrialization, and environmental degradation, the issue of soil salinization is becoming increasingly severe. Elevated concentrations of Na+ and carbonate/HCO3− in saline–alkali soils increase soil pH and sodium absorption rates, decrease fertility, and cause the deterioration of soil structure [2]. The impacts of soil salinization on plants primarily encompass both salt stress (primarily NaCl and Na2SO4) and alkali stress (primarily NaHCO3 and Na2CO3) [3]. The concurrent occurrence of soil salinization and alkaline stress exerts a pronounced inhibitory effect on plant growth [4]. In light of this, studies have indicated that the screening and breeding of plant genotypes with strong salt–alkali tolerance may be an effective means of mitigating the adverse impacts of saline–alkaline stress. Notable progress has been achieved in oats (Avena sativa L.), alfalfa (Medicago sativa L.), and rice (Oryza sativa L.) [5,6,7].
Bermudagrass [Cynodon dactylon (L.) Pers.] is a warm-season turfgrass widely used for lawn construction due to its strong ecological adaptability and rich genetic variation [8]. Significant differences in stress resistance exist among bermudagrass ecotypes. The screening mainly focuses on drought and cold tolerance [9,10], whereas studies on salt–alkali tolerance are limited. Bermudagrass has been proven to grow well under mild saline–alkali stress (10 mM Na+ at pH 8.0) and exhibits a survival rate of 15% under severe saline–alkali stress (150 mM Na+ at pH 10.6) [11]. Xinjiang, China, being a major saline–alkali distribution region, harbors a diverse range of bermudagrass genotypes. An evaluation of different ecological types may facilitate obtaining exceptional stress-resistant genotypes.
In order to clarify the stress tolerance of various plant genotypes, morphological and physiological indicators serve as effective evaluation tools [12]. When exposed to saline–alkali conditions, plants suffer significant impacts on both photosynthesis and cell membrane structural integrity due to elevated pH levels. To quantify these impacts, some physiological parameters are considered as essential indicators, including the net photosynthetic rate (Pn), which reflects the overall efficiency of photosynthesis [13], and the contents of chlorophyll a (Chla) and chlorophyll b (Chlb), pigments crucial for light absorption and energy conversion during photosynthesis [14]. Moreover, relative water content (RWC) is a crucial parameter for understanding salt–alkali tolerance in plants, as it indicates the plant’s capacity to maintain water balance under stressful conditions [15]. Additionally, membrane damage and cell stability are assessed through measurements of relative electrolyte conductivity (REL) and malondialdehyde (MDA) levels. REL indicates membrane permeability and the degree of membrane damage, while MDA serves as a marker for lipid peroxidation, indicating oxidative stress and cellular damage [16]. The accumulation of osmotic adjustment substances, such as proline (Pro) and soluble proteins (Protein), play a critical role in mitigating the adverse effects of abiotic stress. These osmolytes accumulate in response to stress, aiding in reducing cellular damage by maintaining turgor pressure and protecting cellular structures, thereby enhancing plant tolerance to stressful environments [17]. By analyzing these parameters, a comprehensive evaluation of plant stress tolerance can be carried out, providing valuable insights into the mechanisms of stress adaptation for different genotypes.
The experimental materials for this study consisted of 38 bermudagrass genotypes originating from Xinjiang. These genotypes were subjected to high-pH saline–alkali stress, and the physiological parameters closely related to overall physiology, including Pn, Chla, Chlb, RWC, REL, MDA, Pro, and Protein, were measured. A comprehensive evaluation of the saline–alkali tolerance of these genotypes was subsequently carried out. This methodology facilitated the identification of resistant bermudagrass genotypes and the screening of salt–alkali tolerance indicators, laying a foundation for future research endeavors.
2. Materials and Methods
2.1. Experimental Materials and Growth Conditions
A total of 38 Xinjiang bermudagrass genotypes were propagated and maintained as individual plants (Table 1). The experiment was conducted in Xinjiang Agricultural University, located in Urumqi, Xinjiang. In June 2021, a turf plug with a diameter of 12 cm and a soil depth of 5 cm of each genotype was transplanted into plastic pots (20 cm diameter, 15 cm depth, containing a mixture of nutrient loamy soil and vermiculite in a 2:1 ratio (Table 2). Eight pots were transplanted for each genotype and maintained for 60 days with regular weeding and trimming. Quick-acting fertilizer (N:P2O5:K2O = 3:1:1) was applied every ten days.

Table 1.
Information on the test genotypes of bermudagrass.

Table 2.
Principal physical and chemical properties of nutrient soil mixed with vermiculite in pots.
2.2. Salt and Alkali Stress Treatment
A mixture of NaCl, Na2SO4, NaHCO3, and Na2CO3 was used to induce salt–alkali stress in plants, with a mass ratio of 9:1:1:9 and a pH of 10.0. The concentrations ranged from 0 to 150 mmol/L (pH 10.0), and each concentration treatment was repeated four times. Before the experiment started, all materials were fully irrigated (to reach the saturated water-holding capacity of the field) and trimmed, leaving a stump height of 4 cm. Subsequently, stress treatment was carried out gradually, with each pot irrigated with 100 mL of saline–alkali solution at concentrations of 0, 30, 60, 90, 120, and 150 mmol/L from the 1st to the 6th day, respectively. The same amount of clear water was used as a control.
2.3. Determination of Net Photosynthetic Rate
The photosynthetic rate (Pn) of the third fully expanded leaf from the top of bermudagrass plants was measured using the Ciras-2 portable photosynthesis instrument. Measurements were taken on a cloudless day between 9:30 and 13:00 across different treatment conditions. For each treatment, four leaves were selected, and each leaf was measured four times.
2.4. Determination of Chlorophyll Content
Chlorophyll a (Chla) and b (Chlb) content were measured with the 95% ethanol method by spectrophotometer Briefly, 0.2 g of bermudagrass leaves were weighed, ground with a mortar and pestled using 95% ethanol, diluted to 10 mL, and centrifuged. The supernatant was then transferred into a cuvette, and absorbance was measured at 649 nm and 665 nm, with 95% ethanol serving as the blank. The concentrations of Chla and Chlb were calculated using the formulas: , .
2.5. Relative Moisture Content Determination
The relative water content (RWC) of leaves was determined using the saturated weight method. Briefly, 0.2 g of fresh leaves were weighed and immediately immersed in water for 12 h. Subsequently, they were dried in an oven at 105 °C for 30 min, further dried at 85 °C until reaching a constant weight, cooled to room temperature, and then the dry weight was recorded. This process was repeated three times. The RWC was calculated by the following formula: , where Wf was the fresh weight of the leaves, Wt was the saturated weight, and Wd was the dry weight of the leaves.
2.6. Determination of Relative Electrolyte Conductivity (REL)
Relative electrolyte conductivity (REL) was determined by measuring the electrical conductivity of bermudagrass leaf extracts and comparing it to that of a known standard electrolyte solution. Briefly, leaf samples were collected, thoroughly rinsed with distilled water to eliminate surface contaminants, cut into small segments, and then immersed in deionized water. Vacuum infiltration was applied to ensure the complete extraction of electrolytes. The initial conductivity of the solution (C1) was measured using a conductivity meter. Thereafter, the samples were boiled for 30 min to release all electrolytes, cooled to room temperature, and the final conductivity (C2) was measured. REL was calculated as the ratio of C1 to C2 and expressed as a percentage.
2.7. Determination of Malondialdehyde Content
The measurement of malondialdehyde (MDA) was conducted using the thiobarbituric acid (TBA) method. Specifically, 0.5 g of plant leaf tissue were weighed and placed into a mortar. Subsequently, 0.1 g of PVP, a small amount of quartz sand, and 5 mL of phosphate-buffer solution with a pH of 7.8 were added to the mortar. The mixture was ground on ice, and the homogenate was transferred into a centrifuge tube, frozen, and centrifuged for 10 min at 10,000× g. The supernatant (hereinafter referred to as the enzyme extract) was then transferred to a test tube and stored at 0–4 °C.
To prepare the MDA reaction solution, 0.6 g of TBA were dissolved in a small amount of 1 M NaOH and diluted to 100 mL with 10% TCA. For MDA determination, 1 mL of enzyme extract was mixed with 2 mL of 0.6% TBA solution, sealed in a boiling water bath for 15 min, cooled rapidly, and centrifuged. The supernatant was collected, and its color was measured at three wavelengths: 600, 532, and 450 nm. For each treatment, the procedure was repeated three times. The results were calculated using the following formula: . In the formula, VZ represented the total volume of reaction solution (mL), V1 represented the volume of extraction solution (mL), V2 represented the volume of extraction solution for determination (mL), W represented the fresh weight of sampled leaves (g), and 0.155 represented the extinction coefficient of MDA (nmol/L).
2.8. Determination of Proline Content
The concentration of proline (Pro) in bermudagrass leaves was determined using the ninhydrin colorimetric method. For each treatment, the procedure was repeated four times. Specifically, 0.3 g of leaf tissue were weighed and placed into a 15 mL test tube. Subsequently, 5 mL of 3% sulfosalicylic acid solution were added, and the mixture was covered and boiled for 20 min. After cooling, 2 mL of the extraction supernatant were transferred to another 15 mL test tube. As a reference, 2 mL of distilled water were added. Then, 2 mL of glacial acetic acid and 3 mL of acidic ninhydrin solution (2.5% concentration) were added, and the mixture was heated for 40 min before adding 5 mL of toluene. The solution was shaken for 30 s and allowed to stand for layer separation. The absorbance was measured at a spectrophotometric wavelength of 520 nm using the reference solution as a blank. A standard curve was plotted to determine the Pro concentration of the test solution. The Pro content of the sample was calculated using the following formula: , where C1 represents the Pro concentration in the extraction solution (μg/mL) obtained from the standard curve, V0 represents the total volume of the extraction solution (mL), V1 represents the volume of the test solution added during measurement (mL), W represents the fresh sample weight (g).
2.9. Determination of Soluble Protein Content
A standard curve for protein concentration was prepared using bovine serum albumin as a standard sample and the Coomassie Brilliant Blue G-250 method. For soluble protein content determination, 1 mL of the extract was mixed with 5.0 mL of Coomassie Brilliant Blue G-250 solution. After thorough mixing, the absorbance was measured at 595 nm. The protein content (mg/g) was calculated using the standard curve according to the following formula: , where C is the value obtained from the standard curve (μg), and Vt is the total volume. WF represents the fresh weight of the sample (g). VS represents the sample volume added during measurement (mL).
2.10. Data Statistical Analysis
Microsoft Excel 2021 was used for data collection, and SPSS 26.0 (SPSS Inc., Chicago, IL, USA) was used for the statistical analysis of relevant data. The total difference between each index for control plants and plants exposed to the stress was analyzed using the independent sample t-test. Differences between the genotypes were analyzed using ANOVA, and the salt–alkali tolerance coefficient (D) was calculated according to the following formula: . Correlation analysis was performed on the SAC value of each indicator, and subsequently, the factor weight coefficient (Vi), the membership function value of each comprehensive indicator [μ(xi)], the salt–alkali resistance comprehensive evaluation value (D), and the comprehensive salt–alkali resistance coefficient value (CSAC) were calculated. The value of the positive index membership function is given by , and the negative index membership function value is given by: . Formula: , , . Here, Pi represents the contribution rate of the i composite indicator, indicating the importance of the i relative to all parameters. xi, ximax and ximin represent the value of the i composite indicator and the maximum and minimum values, respectively. The D value and CSAC value were used to perform hierarchical clustering analysis using the Euclidean method, which enabled the classification of salt–alkali resistance levels [18].
3. Results
3.1. Physiological Responses of Bermudagrass under Saline–Alkali Stress
Compared with the control group, bermudagrass seedlings under salt stress exhibited significantly higher average REL, protein, Pro, and MDA content (p < 0.01), but lower average leaf RWC, Chla, Chlb, and Pn (Figure 1). Under saline–alkali stress, the average values of Pn, RWC, Chla, Chlb, Pro, Protein, MDA, and REL were observed to be 73%, 70%, 91%, 77%, 325%, 109%, 102%, and 238%, respectively, relative to the levels measured in non-stressed conditions.
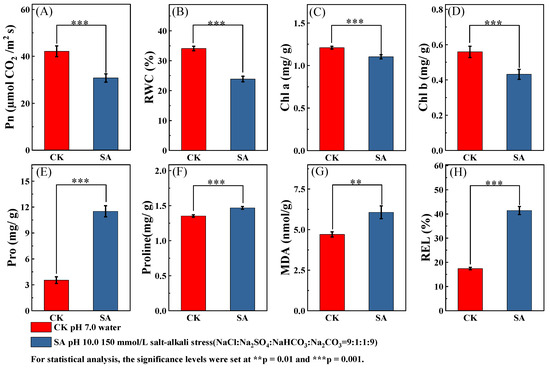
Figure 1.
Effects of salt–alkali stress on the average values of eight indicators of test genotypes. (A) photosynthetic rate (Pn); (B) relative water content (RWC); (C) chlorophyll a (Chla); (D) chlorophyll b (Chlb); (E) proline (Pro); (F) soluble protein (Protein); (G) malondialdehyde (MDA); (H) relative electrical conductivity (REL). significance levels were set at ** p ≤ 0.01 and *** p ≤ 0.001, indicating highly significant and extremely significant differences, respectively.
3.2. Analysis of Eight Indicators of Salt–Alkali Tolerance Coefficient of Bermudagrass
The salt–alkali tolerance coefficient (relative value) was used to eliminate background differences among genotypes and characterize genotype tolerance (Table 3). The results showed that under saline–alkali stress, the number of bermudagrass genotypes with salt–alkali tolerance coefficients ≥100% for Pn, Chla, Chlb, RWC, REL, MDA, Pro, and Protein were, respectively, 7, 1, 8, 11, 36, 20, 38, and 33, which means that salt–alkali tolerance coefficients varied among genotypes, and significant differences existed among genotypes for each indicator (p ≤ 0.05).

Table 3.
The saline–alkali tolerance coefficient (SAC) of eight indicators of test genotypes.
The salt–alkali tolerance coefficients for Pn, Chla and Chlb ranged, respectively, from 0.26 to 2.45, from 0.57 to 1.36, and from 0.24 to 2.79, indicating significant differences in tolerance among the genotypes (Figure 2). Under saline–alkali stress, genotypes such as C111 and C147 showed significant increases in Pn, Chla, or Chlb content, whereas the three indicators of most genotypes significantly decreased. C112 and C14 could maintain higher Chla (salt–alkali tolerance coefficients are 1.36 and 1.24, respectively) and Chlb (salt–alkali tolerance coefficients are 2.25 and 2.79, respectively) contents under salt–alkali stress. The chlorophyll degradation of genotypes such as C135, C40, C50, C1 was more severe compared with other genotypes under saline–alkali stress.
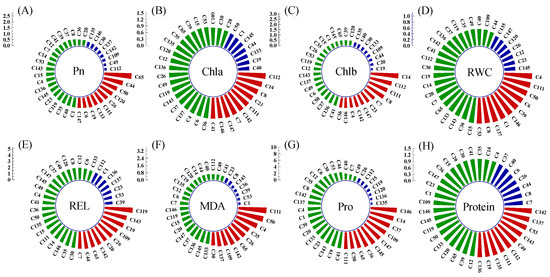
Figure 2.
Ranking of salt–alkali tolerance coefficients of 8 test indicators of 38 bermudagrass genotypes. The red column is the top 20% genotypes, the blue column is the bottom 20% genotypes, and the green column is the middle 60%. (A) photosynthetic rate (Pn); (B) chlorophyll a (Chla); (C) chlorophyll b (Chlb); (D) relative water content (RWC); (H) relative electrical conductivity (REL); (F) malondialdehyde (MDA); (E) proline (Pro); (G) soluble protein (Protein).
The salt–alkali resistance coefficients of REL and MDA ranged from 1.40 to 4.85 and 0.320 to 3.33, respectively (Figure 2). The top 20% of genotypes with the highest REL and MDA salt–alkali resistance coefficients consisted of ten genotypes, including C119 and C143, etc., and ten genotypes, including C111 and C50, etc., respectively, as indicated higher cell membrane damage in these genotypes under salt–alkali stress, with their MDA and REL salt–alkali tolerance coefficients above 1.97 and 2.95, respectively. However, C53, C39, and C1 suffered less damage, with MDA and REL salt–alkali tolerance coefficients below 0.368 and 1.843 respectively. Under saline–alkali stress, some genotypes, including C4 and C111, etc., maintained RWC to support seedling growth, while others, including C145 and C23, had RWC values below 47.10%. Overall, the saline–alkali treatment led to an increase in Pro and protein content, accompanied by a high Pro salt–alkali tolerance coefficient of 8.54.
Under salt–alkali stress, the variation coefficient for eight measured indicators among genotypes spanned from 9.64% to 61.99% (Table 2), highlighting significant variations in salt–alkali tolerance ability across these indicators. The rankings of salt–alkali tolerance coefficients of the eight tested indicators for each genotype have not been consistent (Figure 2), suggesting the difficulty in assessing salt–alkali tolerance based on a single indicator.
3.3. Pearson Correlation Analysis of Salt–Alkali Tolerance of Bermudagrass
Correlation analysis revealed that the eight indicators had correlation coefficients from −0.34 to −0.83 for salt–alkali tolerance, suggesting different sensitivities to salt–alkali stress (Figure 3). Chla and Chlb showed a significant positive correlation (0.83 **), while Chla and Pn had a significant negative correlation (−0.34 *). Other indicators did not display a significant correlation and had minimal information overlap. Therefore, these indicators could be directly used as criteria for comprehensively assessing the salt–alkali tolerance of the test genotypes.
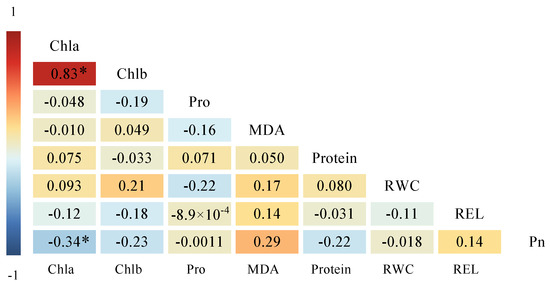
Figure 3.
Correlation analysis of salt–alkali tolerance coefficients among various indicators for test bermudagrass genotypes. * Indicated significance at the 0.05 levels. The indicators included photosynthetic rate (Pn), chlorophyll a content (Chla), chlorophyll b content (Chlb), relative water content (RWC), relative electrical conductivity (REL), malondialdehyde content (MDA), proline content (Pro), and soluble protein content (Protein).
3.4. Comprehensive Evaluation of Salt–Alkali Tolerance of Bermudagrass
Using SAC values, comprehensive evaluation values for 38 bermudagrass genotypes were calculated utilizing the membership function method (Table 4). Normalized inverse membership functions were employed for salt–alkali tolerance REL and MDA content, while standardized membership functions were used for other indicators. D and CSAC values were calculated for each genotype. The D and CSAC values of 38 genotypes ranged from 0.234 to 0.654 and 0.265 to 0.668, with average values of 0.418 and 0.466, respectively. The ranking results were consistent, especially for salt–alkali-resistant and salt–alkali-sensitive genotypes, with stable ranking positions.

Table 4.
D and CSAC value for test genotypes by membership functions.
3.5. Cluster Analysis of Salt–Alkali Tolerance of Bermudagrass
Based on the D and CSAC values, the Euclidean hierarchical clustering method was used to perform cluster analysis on the salt–alkali tolerance of test bermudagrass genotypes (Figure 4). The 38 test genotypes were divided into three clusters: Cluster I (10 genotypes, including C14, C53, C39, C137, C36, C111, C65, C112, C146 and C8), Cluster II (10 genotypes, including C147, C37, C23, C41, C6, C50, C142, C4, C143 and C1), and Cluster III (18 genotypes). Cluster I, Cluster II, and Cluster III accounted for 26.32%, 26.32%, and 47.37% of total genotypes, respectively.
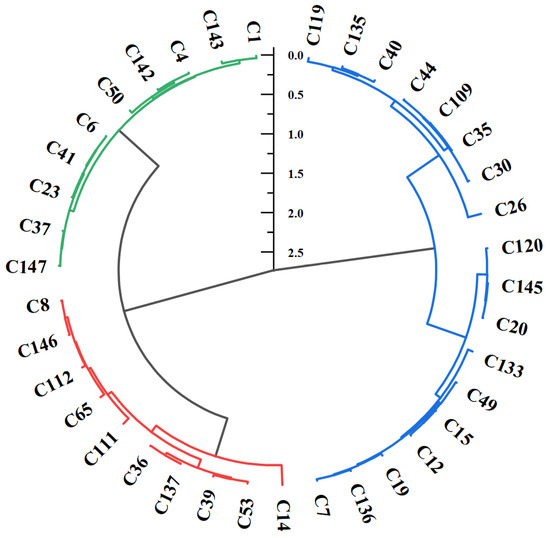
Figure 4.
Cluster analysis of saline–alkali stress resistance of test genotypes based on D value. Cluster I included 10 salt–alkali-tolerant bermudagrass genotypes (red), Cluster II included 10 intermediate-tolerant bermudagrass genotypes (green), and Cluster III consists of 18 salt–alkali-sensitive bermudagrass genotypes (blue).
Based on D value, CSAC value, and the cluster analysis results of various genotypes, C14, C53, C39, C137, and C36 exhibited higher salt–alkali resistance, while C119, C135, C40, C44, and C109 had weaker resistance. A comparison between saline–alkali-tolerant (Cluster I) and salt–alkali-sensitive (Cluster III) genotypes for different indicators (Table 5) revealed that Cluster I genotypes had significantly higher Chlb, RWC, Chla, and Pro levels than Cluster III genotypes by 130.94%, 41.33%, 25.51%, and 68.39%, respectively, under saline–alkali stress.

Table 5.
The difference in the salt–alkali tolerance coefficient for indicators between Cluster I and Cluster III.
3.6. Stepwise Regression Analysis to Screen Key Indicators for Salt–Alkali Tolerance Evaluation
The critical indicators for identifying bermudagrass saline–alkali tolerant genotypes were obtained by stepwise regression analysis (Table 6). Based on the membership function results, the optimal regression model was established using the D value and CSAC value of each indicator. The fitting curve R2 reached 0.930 and 0.874, respectively, with p < 0.01. The critical indicators for D value analysis were Chlb, RWC, Pro, Protein, and Pn, and the critical indicators for CSAC value analysis were Chlb, RWC, REL, Pro, and Protein. The original SAC of 38 genotypes were input into the following regression model:

Table 6.
Model prediction of saline–alkali stress resistance in bermudagrass genotypes.
(I) D = −0.238 + 0.106 × SACChlb + 0.209 × SACRWC + 0.015 × SACPro + 0.284 × SACProtein + 0.051 × SACPn
(II) CSAC = 0.045 + 0.078 × SACChlb + 0.180 × SACRWC − 0.039 × SACREL + 0.013 × SACPro + 0.241 × SACProtein.
The discriminant analysis assessed the agreement between the predicted model values and hierarchical clustering results. The agreement between models (I) and (II) with membership functions was 84.20%. Model (I) exhibited a higher R2 value of 0.930 compared to model (II),which had an R2 value of 0.874. Therefore, model (I) is the best model to determine the salt–alkali tolerance of bermudagrass seedlings.
4. Discussion
4.1. Key Indicators for Evaluating Salt–Alkali Tolerance of Bermudagrass Genotypes
Reliable screening indicators are essential for effectively determining the salt–alkali tolerance of bermudagrass. Under saline–alkali stress, tolerant plants exhibit significantly lower REL values [19]. Pro content acts as a critical molecular switch for salt stress resistance and a crucial indicator for enhancing salt–alkali tolerance [17]. In high-salt environments, Pro accumulation enhances salt tolerance and protects plants from salt stress damage, consistent with the screening of salt–alkali-resistant bermudagrass genotypes in this study. Bai et al. propose that chlorophyll and photosynthesis could be used as essential indicators for assessing oat’s (Avena sativa L.) salt–alkali tolerance [6]. Chlorophyll is sensitive to external pH and is a sensitive indicator of change. This study found that chlorophyll b (Chlb) is particularly essential for selecting salt–alkali-tolerant bermudagrass genotypes, which is in agreement with the findings reported by Ashraf et al. [20]. Because chlorophyll b is less susceptible to degradation than chlorophyll a, plants can remain green and maintain photosynthesis for a while after stress, even though their relative water content (RWC) decreases [21]. Salt–alkali stress reduces Pn and stomatal conductance. Chlb absorbs the blue-violet light band more efficiently than the red light band of Chla, enabling bermudagrass to maintain photosynthesis in adverse environments [22]. Salt–alkali-tolerant bermudagrass may adjust the chlorophyll a/b ratio to inhibit chlorophyll reductase production, maintain Chlb level and improve photosynthetic utilization efficiency under salt–alkali stress, according to Sikder et al. [23]. RWC is suitable for determining the salt tolerance of rice [24], and these findings support the importance of RWC in screening salt-tolerant genotypes, which aligns with the results of this study on bermudagrass salt-tolerant genotypes. Therefore, Chlb, RWC, and Pro content can be critical indicators for evaluating salt–alkali tolerance in bermudagrass.
4.2. Establishment of a Salt–Alkali Tolerance Evaluation Model for Bermudagrass Genotypes
The trends in a single indicator cannot accurately and intuitively reflect the growth ability of different genotypes under salt–alkali stress [25]. Wu et al. (2019) used the membership function fuzzy comprehensive evaluation method to evaluate the salt tolerance of 549 Brassica napus varieties. Similarly, in this study, based on the SAC values of the Pn, Chla, Chlb, RWC, REL, MDA, Pro, and Protein of 38 bermudagrass genotypes, the fuzzy evaluation method was used to evaluate their salt–alkali tolerance. The Euclidean hierarchical clustering method was employed to classify genotypes into salt–alkali-tolerant, intermediate, and sensitive types. A fuzzy comprehensive evaluation can enhance the accuracy of salt–alkali resistance, but the testing indicators are numerous and time-consuming. Therefore, simplifying the evaluation model and screening key indicators can improve accuracy and reduce workload. The stepwise regression analysis model is essential for screening resistance indicators of different genotypes [26]. It is widely used to screen for the critical indicators of drought [10], cold [9], heat [27], and salt–alkali resistance genotypes of different plant genotypes. To conveniently and reliably evaluate the salt–alkali tolerance of bermudagrass genotypes, we established a model using stepwise regression analysis. We performed stepwise regression analysis on the membership function results of the two evaluation methods and the SAC value, respectively. The model’s consistency with clustering results and monitoring R2 value confirm that (D) = −0.238 + 0.106 × SACChlb + 0.209 × SACRWC + 0.015 × SACPro + 0.284 × SACProtein + 0.05 × SACPn can be a predictor for the salt–alkali tolerance evaluation of bermudagrass seedlings.
4.3. Differences in Salt–Alkali Tolerance among Bermudagrass Genotypes
Through research, 10 saline–alkali tolerant genotypes were screened out of the 38 tested genotypes, including C14, C53, C39, C137 and C36. These genotypes exhibited high comprehensive scores (D and CSAC value range from 0.484 to 0.654 and 0.540–0.668) under salt–alkali stress. They exhibit high Chlb, Chla, RWC, and Pro levels, indicating they can maintain high Pn and RWC levels during saline–alkali stress. In contrast, 18 salt–alkali-sensitive genotypes, including C119, C135, C40, C44 and C109 (D and CSAC value are 0.234–0.404 and 0.265–0.455, respectively), exhibited lower Chla, Chlb, Pro and protein content while having higher MDA and REL levels under saline–alkali stress. This study demonstrates significant variations in salt–alkali tolerance among the test genotypes and lays a material and theoretical foundation for future research on salt–alkali tolerance breeding and its mechanisms.
5. Conclusions
This study evaluated the salt–alkali tolerance of 38 bermudagrass genotypes native to Xinjiang by exposing them to 150 mmol/L mixed saline–alkali solution with pH 10.0. Eight indicators were comprehensively analyzed with membership functions, and genotypes were categorized using the Euclidean hierarchical clustering method. The materials under stress exhibited a decrease in photosynthetic rates, leaf water content, and chlorophyll levels compared to the control group. Conversely, there was an elevation in soluble protein content, proline, malondialdehyde, and conductivity. The results classified the genotypes into three distinct clusters: salt–alkali tolerant (Cluster I, ten genotypes, including C14, C53, C39, C137 and C36), intermediate (Cluster II, ten genotypes), and sensitive (Cluster III, 18 genotypes, including C26, C40, C135, C119 and C30). Furthermore, stepwise regression analysis identified Chlb, RWC, and Pro as key indicators for assessing salt–alkali tolerance in bermudagrass genotypes. Additionally, a prediction model for evaluating the salt–alkali tolerance of bermudagrass seedlings was developed: (D) = −0.238 + 0.106 × SACChlb + 0.209 × SACRWC + 0.015 × SACPro + 0.284 × SACProtein + 0.051 × SACPn.
Author Contributions
Conceptualization, L.T., Z.S. and P.L.; Data curation, L.T., Q.Y. and W.L.; Formal analysis, L.T. and P.L.; Investigation, L.T. and W.L.; Methodology, L.T. and P.L.; Project administration, P.L.; Resources, L.T., Q.Y. and W.L.; Software, L.T.; Supervision, Z.S. and P.L.; Validation, L.T. and Q.Y.; Writing—original draft, L.T. and P.L.; Writing—review and editing, Z.S. and P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of China (32260341); the Xinjiang Agricultural University Postgraduate Scientific Research Innovation Program (XJAUGRI2022023).
Data Availability Statement
The research data presented in this article are fully embedded within the text, serving as a vital component supporting our findings. To preserve data integrity and readability, we have not separately archived the dataset. Readers can access and utilize these data through the detailed analyses and charts provided. The absence of an external data link is due to maintaining consistency between data and narrative, not privacy or ethical concerns. We adhere to MDPI’s data policy, promoting sharing and reuse while ensuring research accuracy and verifiability.
Acknowledgments
I sincerely express my gratitude to all individuals and institutions that have provided support and assistance throughout the course of this project. In particular, I am deeply grateful for the financial support from the National Natural Science Foundation of China, as well as the advanced equipment and environment provided by the experimental platform of Xinjiang Agricultural University, which have laid a solid foundation for my research work. At the same time, I am deeply appreciative of the valuable feedback and professional suggestions from the reviewers and editors, which have been invaluable in enhancing the quality of my paper, enabling me to articulate my research viewpoints more accurately and present the experimental data more rigorously. Your diligent work and selfless dedication have collectively driven the progress and development of academic research.
Conflicts of Interest
The authors declare no known conflicts of interest related to this publication.
References
- Li, J.L.; Ma, M.S.; Sun, Y.M.; Lu, P.; Shi, H.F.; Guo, Z.F.; Zhu, H.F. Comparative physiological and transcriptome profiles uncover salt tolerance mechanisms in alfalfa. Front. Plant Sci. 2022, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Qi, A.G.; Wang, B.Q.; Zhang, X.J.; Dong, Q.D.; Liu, J.X. Integrated analyses of transcriptome and chlorophyll fluorescence characteristics reveal the mechanism underlying saline-alkali stress tolerance in Kosteletzkya pentacarpos. Front. Plant Sci. 2022, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.G.; Jin, Y.L.; Guo, W.; Xue, Y.W.; Yu, L.H. Metabolic and physiological changes in the roots of two oat cultivars in response to complex saline-alkali stress. Front. Plant Sci. 2022, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.J.; Wei, M.Y.; Wu, G.H.; Ren, A.Z. Epichloe endophytes improved Leymus chinensis tolerance to both neutral and alkali salt stresses. Front. Plant Sci. 2022, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdallah, H.; Mai, H.J.; Alvarez-Fernandez, A.; Abadia, J.; Bauer, P. Natural variation reveals contrasting abilities to cope with alkaline and saline soil among different Medicago truncatula genotypes. Plant Soil 2017, 418, 45–60. [Google Scholar] [CrossRef]
- Bai, J.H.; Yan, W.K.; Wang, Y.Q.; Yin, Q.; Liu, J.H.; Wight, C.; Ma, B.L. Screening oat genotypes for tolerance to salinity and alkalinity. Front. Plant Sci. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Takano, T.; Liu, S.K. Screening and evaluation of saline-alkaline tolerant germplasm of rice (Oryza sativa L.) in soda saline-alkali soil. Agronomy 2018, 8, 205. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Zhang, Z.H.; Huang, J.F.; Feng, L.L.; Cao, L.Y.; Li, X.; Liu, T.M.; Zong, Q.J.; Wang, H.L. Salt-promoted growth of monolayer tungsten disulfide on hexagonal boron nitride using all chemical vapor deposition approach. Appl. Surf. Sci. 2022, 605, 11. [Google Scholar] [CrossRef]
- Yang, H.L.; Yu, X.H.; Wang, C.F.; Yang, Y.; Wang, X.H.; Yang, Q.H. Evaluation of the cold tolerance of Saccharum spontaneum L. clones with different ploidy levels on the basis of morphological and physiological indices. Plant Biol. 2020, 22, 623–633. [Google Scholar] [CrossRef]
- Katuwal, K.B.; Jespersen, D.; Bhattarai, U.; Chandra, A.; Kenworthy, K.E.; Milla-Lewis, S.R.; Schwartz, B.M.; Wu, Y.Q.; Raymer, P. Multilocational screening identifies new drought-tolerant, warm-season turfgrasses. Crop Sci. 2022, 62, 1614–1630. [Google Scholar] [CrossRef]
- Ye, T.T.; Wang, Y.P.; Feng, Y.Q.; Chan, Z.L. Physiological and metabolomic responses of bermudagrass (Cynodon dactylon) to alkali stress. Physiol. Plant. 2021, 171, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.D.; He, Y.Z.; Wang, H.H.; Zhu, Y.Z. Leaf physiological responses of three psammophytes to combined effects of warming and precipitation reduction in horqin sandy land, northeast China. Front. Plant Sci. 2022, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Fu, Y.; Liu, B. SiFBA5, a cold-responsive factor from Saussurea involucrata promotes cold resilience and biomass increase in transgenic tomato plants under cold stress. BMC Plant Biol. 2021, 21, 75. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, Y.Q.; Geng, G.D.; Yang, R.; Yang, Z.F.; Yang, C.M.; Xu, R.H.; Zhang, Q.Q.; Kakar, K.U.; Li, Z.H.; et al. Comparative analysis of physiological, enzymatic, and transcriptomic responses revealed mechanisms of salt tolerance and recovery in Tritipyrum. Front. Plant Sci. 2022, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xue, H.; Zhang, F. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Iqbal, M.Z.; Jia, T.; Tang, T.; Anwar, M.; Ali, A.; Hassan, M.J.; Zhang, Y.Z.; Tang, Q.L.; Peng, Y. A heat shock transcription factor TRHSFB2A of white clover negatively regulates drought, heat and salt stress tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 12769. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.G.; Zhang, Y.Z.; Wang, N.; He, H.J.; Tan, Q.P.; Wen, B.B.; Zhang, R.; Sun, M.Y.; Zhao, X.H.; Fu, X.L.; et al. Prunus persica terpene synthase PPTPS1 interacts with PPABI5 to enhance salt resistance in transgenic tomatoes. Front. Plant Sci. 2022, 13, 12. [Google Scholar] [CrossRef]
- Lu, Z.K.; Li, J.Y.; Yuan, C.; Xi, B.; Yang, B.H.; Meng, X.Y.; Guo, T.T.; Yue, Y.J.; Gao, Y.Q.; Liu, J.B.; et al. Evaluation of mutton quality characteristics of dongxiang tribute sheep based on membership function and gas chromatography and ion mobility spectrometry. Front. Nutr. 2022, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Ali, Q.; Ayaz, M.; Mu, G.Y.; Hussain, A.; Yuanyuan, Q.; Yu, C.J.; Xu, Y.J.; Manghwar, H.; Gu, Q.; Wu, H.J.; et al. Revealing plant growth-promoting mechanisms of Bacillus strains in elevating rice growth and its interaction with salt stress. Front. Plant Sci. 2022, 13, 1–17. [Google Scholar] [CrossRef]
- Ashraf, M.; Al-Qurainy, F.; Ahmad, M.S.A.; Iqbal, M.Y.; Mehmood, A.; Riffat, A.; Alvi, A.K. Morpho-physiological diversity of barley (Hordeum vulgare L.) germplasm for heat tolerance. Turk. J. Bot. 2022, 46, 37. [Google Scholar]
- Zhao, X.; Jia, T.; Hu, X.Y. Hcar is a limitation factor for chlorophyll cycle and chlorophyll b degradation in chlorophyll-b-overproducing plants. Biomolecules 2020, 10, 1639. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Bai, X.; Jiang, C.F.; Li, Z. Phosphoproteomic analysis of two contrasting maize inbred lines provides insights into the mechanism of salt-stress tolerance. Int. J. Mol. Sci. 2019, 20, 1886. [Google Scholar] [CrossRef] [PubMed]
- Sikder, R.K.; Wang, X.R.; Jin, D.S.; Zhang, H.H.; Gui, H.P.; Dong, Q.; Pang, N.C.; Zhang, X.L.; Song, M.Z. Screening and evaluation of reliable traits of upland cotton (Gossypium hirsutum L.) genotypes for salt tolerance at the seedling growth stage. J. Cotton Res. 2020, 3, 13. [Google Scholar] [CrossRef]
- Kojonna, T.; Suttiyut, T.; Khunpolwattana, N.; Pongpanich, M.; Suriya-Arunroj, D.; Comai, L.; Buaboocha, T.; Chadchawan, S. Identification of a negative regulator for salt tolerance at seedling stage via a genome-wide association study of Thai rice populations. Int. J. Mol. Sci. 2022, 23, 1842. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Guo, J.R.; Wang, C.F.; Li, K.L.; Zhang, X.W.; Yang, Z.; Li, M.T.; Wang, B.S. An effective screening method and a reliable screening trait for salt tolerance of (Brassica napus L) at the germination stage. Front. Plant Sci. 2019, 10, 12. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, M.F.; Ma, H.Y.; Jiang, L.; Zhao, Z.Y.; Ma, J.B.; Wang, L. Differential responses of dimorphic seeds and seedlings to abiotic stresses in the halophyte Suaeda salsa. Front. Plant Sci. 2021, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Al-Ashkar, I.; Sallam, M.; Ghazy, A.; Ibrahim, A.; Alotaibi, M.; Ullah, N.; Al-Doss, A. Agro-physiological indices and multidimensional analyses for detecting heat tolerance in wheat genotypes. Agronomy 2023, 13, 154. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).