Abstract
Simple sequence repeats (SSRs), also known as microsatellites, stand out as the most crucial molecular markers in both animals and plants owing to their high polymorphism, extensive information content, ease of detection through polymerase chain reaction (PCR) assays, and widespread distribution across the genome. In this study, a total of 125,443 SSR loci were identified from the whole-genome sequence of B. oleracea, and 82,948 primer pairs for SSR have been designed. Furthermore, each primer pair is designated with a unique identifier (ranging from BolSSR00001 to BolSSR82984). Our findings indicated that certain markers within them could be transferred to other cruciferous crops. In addition, a total of 336 pairs of SSR primers have been used to screen the polymorphism between the bolting-resistant and bolting-easy gene pools. After the test of verification with F2 generation individual plants, we obtained an SSR dominant marker, BolSSR040196, linked with bolting-resistant locus in cabbage, and the genetic distance between this SSR marker and the bolting-resistant locus was 10.69 cM. Moreover, BolSSR040196 is located on the C05 chromosome with a CT motif, characterized by a repeat of 9 in bolting-easy plants and 11 in bolting-resistant plants. Haplotype analysis showed that the correct prediction rate reached 82.35%. The BolSSR040196 marker can be used in marker-assisted selection (MAS) breeding, offering a straightforward and efficient approach for bolting-resistant cabbage breeding in the future.
1. Introduction
The detection of DNA sequence variation plays a crucial role in the study of Brassica crop genomes. In the last two decades, numerous types of molecular markers, such as restriction fragment length polymorphisms (RFLPs), random amplified polymorphic DNA (RAPD), amplified fragment length polymorphisms (AFLPs), simple sequence repeats (SSRs), sequence-related amplified polymorphisms (SRAPs), sequence-characterized amplified regions (SCARs), and single nucleotide polymorphisms (SNPs), have been used in genetic breeding studies on the Brassica species [1,2,3,4,5,6,7,8]. Among the various types of molecular markers, SSRs, also called microsatellites, possess the characteristics of high polymorphism, high reproducibility, easy detection by PCR, codominance, amenability, high transferability, and genomic abundance. SSRs have been widely employed in a variety of research areas, including the study of genetic diversity, the analysis of quantitative trait loci (QTLs) and genetic maps, gene localization, the classification and evolution of germplasm, and comparative genomics.
The traditional procedure for developing SSRs involves probe hybridization (containing repeated motifs) against genomic and cDNA libraries, DNA sequencing [9] or in silico analysis of publicly available bacterial artificial chromosome (BAC) sequences [10,11], genome survey sequences, and whole-genome shotgun sequences [12,13]. With the release of more and more high-quality genome data, the development of SSRs has become faster and more accurate [14,15,16]. At present, the development of SSR based on the whole genome is widely used in cacao [17], grape [18], maize [19], Chinese bayberry [20], and cabbage [8]. Among other crops, this approach has also been proven useful in the development of SSRs in the expressed sequence tags of many crops, such as rice [21], wheat [22], cotton [23], peanuts [24], cowpea [25], and radish [7].
B. oleracea comprises many important vegetable crops, including cabbage, cauliflower, broccoli, Brussels sprout, kohlrabi, Chinese kale, and kale. B. oleracea has been recognized as a vital vegetable species valued for its substantial food reserves. With the rapid development of genome sequencing technology, the genome sequence of cabbage has been released and is currently available online (http://brassicadb.cn/#/, accessed on 10 May 2023) [26]. Genome sequences provide a powerful tool for genome-wide microsatellite characterization. However, studies on the development of SSR based on the whole genome of B. oleracea are limited [27].
In recent decades, significant progress has been made in research on the inheritance of bolting and the utilization of molecular markers, driven by the continuous advancement of modern breeding techniques. Boudry et al. successfully mapped the B gene associated with bolting between two markers, pkP591 and pkP826, utilizing RFLP markers [28]. Ei-Mezawy identified four markers intricately linked to the sugar beet B gene through the bulked segregant analysis (BSA) method. Among these markers, two were positioned at 0.14 cM and 0.23 cM, respectively, while the other two were located at 0.5 cM [29]. Teutonico and Osborn identified genes that regulate late bolting in Arabidopsis and detected an RFLP locus closely linked to one of the QTLs for flowering time in B. rapa [30]. Rosental et al. discovered significant bolting and flowering QTL located on chromosome 7 (qFLT7.2) using a lettuce mapping population. Through fine mapping, homology analysis, and gene expression analysis, they identified two candidate genes associated with this QTL [31]. Through the mapping and resequencing of two early bolting mutants, Fu et al. discovered that BrSDG8 was related to the bolting of Chinese cabbage, and the mutation of BrSDG8 led to the early bolting of Chinese cabbage [32]. Wang et al. used site-specific amplified fragment BSA (SLAF-BSA) technology to map the bolting time QTLs of cabbage, and the result showed two QTLs located on chromosome C02 at 2.31~3.09 Mb and 33.57~34.40 Mb, respectively, with a total length of 1.61 Mb [33]. Wei et al. revealed that the transition from late bolting to early bolting in Chinese cabbage was controlled by an incomplete dominant gene of BrLb-1 and identified three putative candidate genes for the late bolting trait. These genetic variations represent valuable resources for the development of molecular markers for MAS in Chinese cabbage breeding programs [34]. To date, there have been relatively few reports regarding molecular markers associated with the bolting trait in cabbage [35].
In this study, we performed an analysis of SSR information distribution and developed SSR primers based on the whole genome of B. oleracea. Utilizing the B. oleracea genome, the SSR primers developed in this study cover the entire genome with precise positions. These primers hold significant potential for widespread utilization in the genetic analysis of Brassica crops. Subsequently, the BSA approach was employed to screen the SSR marker, which is closely linked to the bolting-resistance locus, to obtain molecular markers that could be used in the research of molecular marker-assisted breeding in Brassica crops.
2. Materials and Methods
2.1. Source of Whole-Genome Sequence of B. oleracea
A total of 539,673,885 bp genome sequences of B. oleracea (C01~C09) were downloaded from the Brassicaceae Database (BRAD) (http://brassicadb.cn/#/, accessed on 10 May 2023). The genome version of B. oleracea is Braol_JZS_V2.0, and the size of each chromosome is 52,469,534 bp, 66,012,635 bp, 74,510,869 bp, 65,432,303 bp, 57,884,551 bp, 47,844,321 bp, 55,965,641 bp, 52,084,256 bp, and 67,469,775 bp, respectively.
2.2. SSR Identification
The genome sequences were searched for the presence of SSR motifs using the Krait identification tool (v1.3.3) [36]. The parameters were set as follows: the minimum repeats for each perfect SSR type were set to 12 for mononucleotides, 7 for dinucleotides, 5 for trinucleotides, 4 for tetranucleotides, 4 for pentanucleotides, and 4 for hexanucleotides. At the same time, SSRs with incomplete repeats interrupted by a small number of intermediate bases (with intervals less than or equal to 10 bp) were also screened.
2.3. Genomic SSR Primer Design
Primer pairs flanking the SSRs were designed using the SSR Locator I software [37]. The major parameters were set as follows: The primer length was between 18 bp and 27 bp, with an optimum size of 20 bp; the melting temperatures ranged from 58 °C to 65 °C, with an optimum temperature of 60 °C; the optimum GC content was 50%, with a minimum of 30% and a maximum of 70%; and the predicted PCR products ranged from 100 bp to 300 bp. Other parameters were set as defaults.
2.4. Detection and Transferability of SSR Markers
We randomly selected 54 primer pairs, 6 SSR loci per chromosome, from the designed SSR primers based on the B. oleracea whole genome. The transferability of these 54 pairs of primers was validated by 7 species (B. rapa (AA), B. nigra (BB), B. oleracea (CC), B. juncea (AABB), B. napus (AACC), B. carinata (BBCC), and R. sativus). The seeds of these related species came from the Institute of Vegetable Science, Zhejiang University, and total genomic DNA was extracted using the CTAB method [13]. PCR amplification was conducted in a 25 µL reaction mixture that contained 50 ng of genomic DNA, 1 µL (10 µmol/L) of each primer, and 12.5 µL of 2 × T5 Super PCR Mix (PAGE) (TsingKe, Beijing, China). Afterward, ddH2O was added until the total volume of the reaction mixture was 25 µL. The amplification system of PCR was initially denatured at 98 °C for 2 min, followed by 35 cycles of amplification at 98 °C for 10 s, 56 °C annealing for 10 s, 72 °C extension for 10 s, and a final extension at 72 °C for 2 min in a BIO-RAD S1000TM Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). The samples were then stored at 4 °C. The PCR products were detected by 12% PAGE following a method from Molecular Cloning: A Laboratory Manual [38].
2.5. Construction of Bolting-Resistant/Bolting-Easy Segregated Population and Agronomic Traits Investigation
The bolting-resistant male parent (P1 Y9805) and the bolting-easy female parent (P2 Y5314) were crossed by artificial emasculation. Then, the F2 population was developed from a single heterozygous F1 individual plant. Both cabbage accessions originated from the Institute of Vegetable Science, Jiangsu Academy of Agricultural Sciences. The agronomic traits of 224 F2 plants were investigated, with the following specific criteria utilized:
Plant height: The observed value represents the vertical distance from the plant’s highest point to the ground level. To measure this, we determine the natural vertical distance in centimeters from the base of the plant where it meets the ground up to its highest point. The accuracy of the measurement is 0.1 cm.
Plant size: The observation site refers to the outermost foliage of the plant. The measurement technique entails determining the maximum horizontal distance of the outer leaves in their usual vegetative condition, expressed in centimeters, with an accuracy of 0.1 cm.
Number of outer leaves: The observed part is the plant leaves, and the measuring technique is to calculate the sum of the number of leaf scars on detached lotus leaves and the number of remaining lotus leaves, measured in pieces, with precision to the whole position.
Stem diameter: The spot to observe is the shortened stem at the leaf bulb’s base. The measurement technique involves gauging the diameter of the short stem at the leaf bulb’s bottom in centimeters, with a precision of 0.01 cm.
Winterness level: Level 1: shortened stems elongation (<1 cm); Level 3: stem shortening with notable elongation (1–2 cm), devoid of flower stems; Level 5: stems characterized by both brevity and elongation, accompanied by flower stems (<5 cm); Level 7: internode elongation, with flower stems exceeding 5 cm; Level 9: significant internode elongation, featuring flower stems surpassing 20 cm.
2.6. Identification of SSR Markers Linked with Bolting Loci
Genomic DNA was extracted from 15 individuals with winterness level 1 and 15 individuals with winterness level 9 randomly selected from the F2 population, and the same amount of DNA was utilized to generate resistant and easy-bolting DNA pools for further screening of bolting-related SSR markers. For the experiment, a comprehensive set of 336 SSR markers was chosen, all of which originated from the Brassica oleracea entire genome SSR, meticulously developed within our laboratory (Table S6). All of the primer pairs flanking the SSRs were synthesized by Shanghai Invitrogen. The PCR program and PAGE protocol employed remained consistent with prior methodologies. Then, we analyzed the variations in amplified product length between the bolting-resistant gene pool and the bolting-easy gene pool. Validation of each SSR marker was conducted across F2 plants and both parental genotypes. The Kosambi function was subsequently applied to compute the genetic distances among the screened linkage SSR markers.
2.7. Haplotype Analysis with 34 Cabbage Accessions and Sequencing of BolSSR040196
We screened the BolSSR040196 marker to test 34 cabbage accessions from the Institute of Vegetable Science, Jiangsu Academy of Agricultural Sciences, and then investigating their winterness results to validate. PCR amplification systems, product analysis, and the investigation of winterness levels are the same as the aforementioned description in Section 2.4. Meanwhile, a chi-square goodness-of-fit test was employed to ascertain the concordance between the observed results and the expected results.
PCR products were purified using the MolPure® Gel Extraction Kit (Yeasen, Shanghai, China). The purified product was inserted into the T-vectors using the Hieff Clone® Universal Zero TOPO TA/Blunt Cloning Kit (Yeasen, Shanghai, China). Then, the vectors were transformed into E. coli, and picked a single colony for sequencing. MEGA-X (Mega Limited, Auckland, New Zealand) was used to align the sequences of the BolSSR04016 marker.
3. Results
3.1. SSR Information Analysis of the Whole Genome of B. oleracea
A total of 125,443 SSR loci were searched from the whole-genome sequence of B. oleracea, and the frequency (per Mb average occurrence of SSR) of SSR occurrence was 223.6 loci/Mb. Among the total genomic SSRs identified, the mononucleotide repeat motifs (62,204, 49.59%) are the most abundant repeat types, with a frequency of 110.88 loci/Mb. The dinucleotide repeats (41,560, 33.13%) and trinucleotide repeats (13,652, 10.88%) constituted the second and third most prevalent categories, exhibiting frequencies of 74.08 loci/Mb and 24.33 loci/Mb, respectively. The tetranucleotide repeats (4884, 3.89%) and pentanucleotide repeats (1781, 1.42%) represented the fourth and fifth most prevalent repeat motifs, showcasing frequencies of 8.71 loci/Mb and 3.17 loci/Mb, respectively. The hexanucleotide repeat motifs (1362, 1.09%) exhibited the least frequency, with 2.43 loci/Mb (Figure 1A, Table S1).
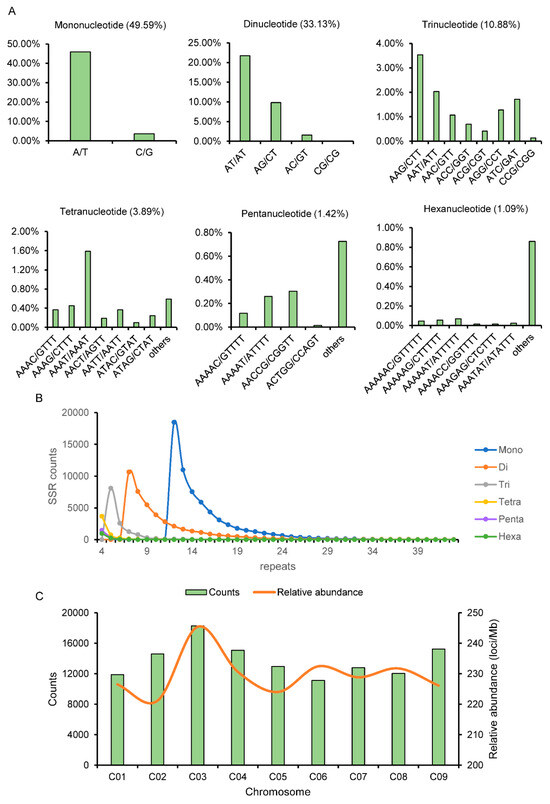
Figure 1.
SSR information analysis of whole genome of B. oleracea. (A) The distribution of major repetitive motifs in B. oleracea. (B) SSR repeats distribution for each type. (C) Distribution of SSR on different chromosomes of B. oleracea.
Among the mononucleotide repeats, the predominant motif was A/T (45.94%), comprising 92.65% of the total. Within the dinucleotide repeats, AT/AT (21.74%) emerged as the most prevalent, followded by AG/CT (9.82%), AC/GT (1.57%), and CG/CG (0.00%). Within the trinucleotide repeats, AAG/CTT (3.54%) emerged as the most prevalent, followed by AAT/ATT (2.03%), AAC/GTT (1.07%), and ACC/GGT (0.70%). Among the tetranucleotide, pentanucleotide, and hexanucleotide repeats in B. oleracea, AAAC/GTTT (0.36%), AAAAC/GTTTT (0.12%), and AAAAAC/GTTTTT (0.05%) notably exhibited higher frequencies compared to other combinations (Figure 1A, Table S1). Among all of the genomic SSRs, 20,697 SSRs with twelve repeats had the highest frequency (16.50%), followed by thirteen repetitions (12,698, 10.12%) and seven repetitions (12,036, 9.59%) (Figure 1B, Table S2).
Variations in distribution counts and relative abundance of SSRs across individual chromosomes were also observed (Table S3). Among these, the C03 chromosome exhibits the highest SSR distribution, totaling 18,286, with a relative abundance of 245.43 loci/Mb. Conversely, the C06 chromosome harbors the lowest SSR distribution, with a relative abundance of 232.44 loci/Mb (Figure 1C).
3.2. SSR Primer Design Based on the Whole Genome of B. oleracea
A total of 125,443 SSR loci were detected in the whole-genome sequence of B. oleracea, sequentially named BolSSR000001 to BolSSR125443. The primer pairs were effectively designed for 82,984 SSR markers using the SSR Locator I software, achieving a commendable success rate of 66.12% in primer design. The primer sequence, Tm value, SSR motif, and length of the predicted product for each SSR marker are listed in Table S4.
Subsequently, we randomly selected 10 primer pairs from the SSR primer set. These primer pairs were selected for PCR simulation using the genomic DNA of B. oleracea (Figure S1). The result indicates that 10 SSR primers effectively amplified the predicted fragment products, suggesting the availability of our previously designed SSR primers.
3.3. Transferability Evaluation of the Genomic SSRs
In order to assess the broad applicability of the primers we have developed, a total of 54 primer pairs (Table S5) were randomly chosen from the pool of SSR primers designed using the entire genome sequence of Brassica oleracea. The primer pairs selected for PCR simulation used the genomic DNA of seven species in Table 1. Results demonstrated that 52 markers (96.30%) were transferable to the CC genome, and 33 SSR markers (61.11%) were transferable to R. sativus. The highest ratio of transfer is cabbage, and the lowest is radish within the previous cruciferous crops. Meanwhile, the ratio of transfer to AA, BB, AABB, AACC, and BBCC is 85.19%, 79.63%, 87.04%, 64.81%, and 87.04%, respectively (Table 1). Most of the SSR markers produced length and quantity variation among the seven accessions (Figure 2). Except for the BolSSR076246 primer pair, the other four primer pairs, BolSSR071665, BolSSR081968, BolSSR021757, and BolSSR053957, showed the polymorphism among the six Brassica species of the U triangle (Figure 2). These findings suggest that the primer pairs, derived from the comprehensive genome of cabbage, hold potential for future utilization in genetic diversity analysis and molecular marker-assisted selection within cruciferous crops.

Table 1.
Transferability detection of the 54 SSR markers in related crucifer species.
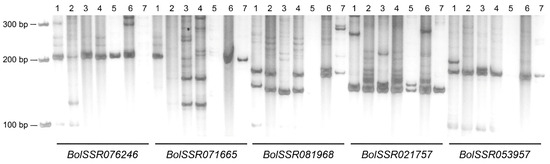
Figure 2.
Validation of B. oleracea genomic SSR primer pairs for the seven crucifer crops. M: 100 bp DNA ladder marker. For one primer pair, the order from left to right, 1~7 lanes, are the DNA of B. rapa (AA), B. nigra (BB), B. oleracea (CC), B. juncea (AABB), B. napus (AACC), B. carinata (BBCC), and R. sativus.
3.4. Construction of BSA Segregated Population and Investigation of Related Traits
In this study, a bolting-related F2 generation segregation population was constructed. Due to the transient segregation of the F2 population and the existence of heterozygous genotypes, a total of 224 individuals were employed to carry out the subsequent trait investigation. We measured the plant height, number of outer leaves, plant size, stem diameter, and winterness level of the F2 generation population. All these traits are in agreement with the normal distribution (Figure 3 and Figure S2). The F2 population obtained in this study is a population with suitable trait segregation, and traits such as plant height, number of outer leaves, stem diameter, and winterness level are quantitative traits controlled by multiple genes. This population holds potential for subsequent marker screening, genetic map construction, and gene mapping investigations.
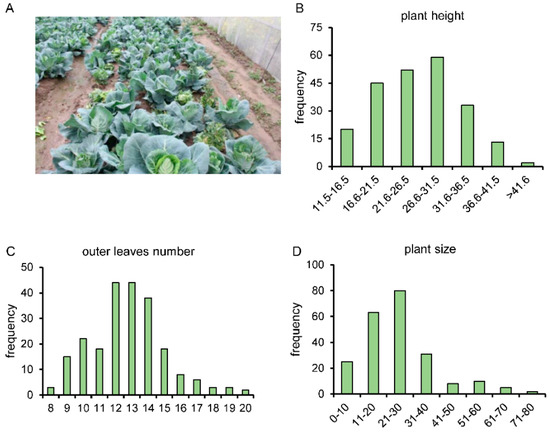
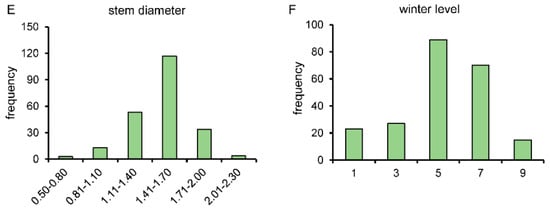
Figure 3.
Investigation on agronomic traits of F2 segregated population. (A) Field-grown condition of F2 generation. (B–F), frequency distribution of plant height (B), number of outer leaves (C), plant size (D), stem diameter (E), and winterness level (F) in F2 population.
3.5. Screening of Bolting Linkage Markers
In order to screen SSR markers related to the bolting locus in cabbage, 336 pairs of primers were randomly selected from the primers previously designed (Table S6). We selected as many varieties of SSR markers as possible, and these markers were evenly dispersed on each chromosome. The BSA method was used to screen the markers associated with the bolting locus. We screened SSR markers showing significant variations between bolting-resistant and bolting-easy DNA pools. Finally, we found that the BolSSR040196 marker has significant differences within the two gene pools. The findings from the PCR amplification of bolting-resistant and bolting-easy gene pools using the BolSSR040196 marker revealed evident polymorphism at 150 bp (Figure 4A). The amplified bands observed in bolting-resistant plants exhibit slight increments in size compared to those detected in bolting-easy plants. Subsequently, a polymorphism validation experiment was conducted utilizing 10 plants each from the bolting-resistant and bolting-easy parental lines (Figure 4B,C), revealing the repeatability of bands exhibiting distinct polymorphisms (Figure S3). The validation remained congruent with the screening results obtained from the gene pool. All findings consistently indicate a close linkage between the BolSSR040196 marker and the locus associated with bolting resistance in cabbage.
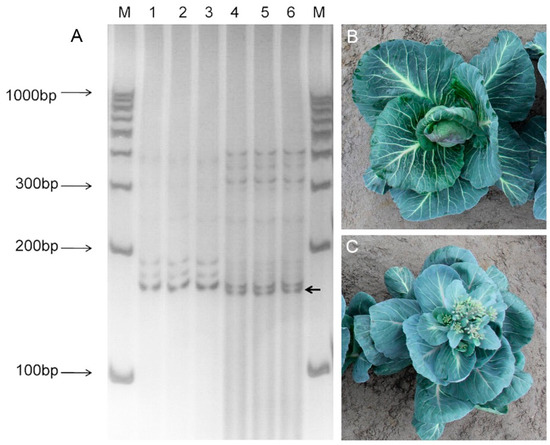
Figure 4.
Amplification results of BolSSR040196 in bolting-resistant and bolting-easy gene pools by 12% PAGE test. (A) Lanes 1–3 were three repeated PCR results of the bolting-resistant gene pool, while lanes 4–6 were three repeated PCR results of the bolting-easy gene pool. Differential fragments between bolting-resistant and easy-bolting gene pools were marked with an arrow. (B) Bolting-resistant male parent (P1 Y9805). (C) Bolting-easy female parent (P2 Y5314).
3.6. Identification of SSR Markers and Linkage Analysis of BolSSR040196
We conducted further verification to assess potential variations in the BolSSR040196 markers across additional F2 progeny individuals. The results revealed that 20 individuals prone to easy bolting exhibited the same band pattern as depicted in Figure 4A, while 3 individuals did not (Figure 5A). Within 15 bolting-resistant individuals, it was observed that one individual, indicated by an arrowhead, amplified a smaller fragment (Figure 5B). The bulk of the F2 progeny conformed to our anticipated outcomes; however, a few recombinants were identified. These findings underscore the close linkage between BolSSR040196 and the bolting-resistance locus. In other words, there were four instances of genetic recombination among the 38 plants tested in the F2 generation. Calculating based on the definition of genetic distance, the recombination rate was determined to be 10.53%. Employing the Kosambi mapping function, the analysis revealed a genetic linkage distance of 10.69 cM between BolSSR040196 and the bolting-resistant locus in B. oleracea.
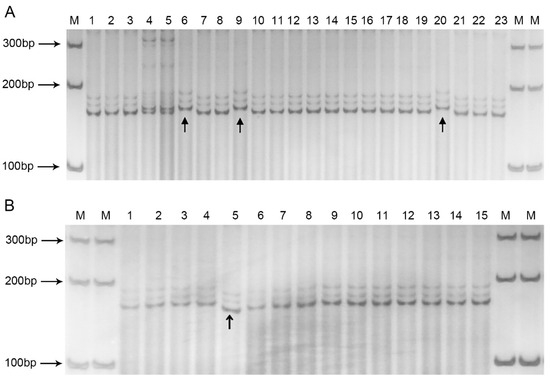
Figure 5.
Validation results of bolting-resistant and bolting-easy individuals in F2 generation with the primers BolSSR040196 tested by 12% PAGE. There were 3 recombinants among the 23 bolting-easy individuals (A) and 1 recombinant among the 15 bolting-resistant individuals (B).
3.7. Haplotype Analysis with 34 Cabbage Accessions
The previous study, spanning parental and F2 generations, amplified the target fragment using the BolSSR040196 primer, consistently demonstrating stable polymorphism of BolSSR040196. This observation suggests a close linkage between BolSSR040196 and the bolting-resistant locus, indicating its potential for further development into stable SSR markers. Subsequently, the haplotype analysis experiment was carried out with 34 cabbage accessions. Only 6 out of the 34 cabbage accessions examined using BolSSR040196 showed discrepancies with the anticipated outcomes, while the remaining 28 varieties exhibited conformity with the expected results. The haplotype analysis results indicated a predictive accuracy rate of 82.35% (Table 2). The result of the chi-square goodness-of-fit test showed that the χ2 value was 1.12, and the χ2(1)0.05 = 3.84. Therefore, with a p-value > 0.05, the ratio of bolting-easy plants to bolting-resistant plants was consistent with the expected value.

Table 2.
Haplotype analysis results with 34 varieties tested by the BolSSR040196 marker.
3.8. Sequencing of BolSSR040196
The sequencing results revealed that BolSSR040196 had a length of 151 bp, comprising a (CT)n dinucleotide repeat type, with (CT)9 repeats in bolting-easy plants and (CT)11 repeats in bolting-resistant plants (Figure 6). Following this, a BLAST analysis was conducted utilizing the Brassicaceae Database [31], revealing that the molecular marker was located on the C05 chromosome.

Figure 6.
Sequence alignment result between the sequences amplified from the bolting-resistant and bolting-easy individuals with the primers BolSSR040196. Part of shading was (CT)9 in the bolting-easy plant and (CT)11 in the bolting-resistant plant.
4. Discussion
4.1. Distribution Feature of the SSR on the Whole Genome of B. oleracea
A comprehensive search conducted by the Krait software (v1.3.3) revealed a total of 125,443 SSRs within the most recent whole-genome sequence of B. oleracea. The frequency of SSR occurrence averaged 223.6 loci/Mb. Contrarily, a prior report utilizing the PERL5 script MIcroSAtellite (MISA: http://pgrc.ipkgatersleben.de/misa/, accessed on 17 June 2023) identified a larger set of SSR markers, totaling 185,662, within B. oleracea [27]. Among mononucleotide repeats, the A/T repeat motif stands out with 57,629 occurrences, constituting 45.94% of the total. This motif holds significance as it encompasses 92.65% of all mononucleotide repeats. In contrast, the G/C repeat motif is notably less prevalent, with only 4575 occurrences, representing 7.35% of mononucleotide repeats. This finding aligns with a prior study that employed the entire genome of Vitis vinifera for SSR analysis [18]. Among dinucleotide repeats, the AT/AT repeat motif has 27,267 (21.74%). Therefore, it is the most important for this type of repeat as it accounts for 65.61% of the dinucleotide repeats. Among trinucleotide repeats, the AAG/CTT repeat motif has 4436 (3.54%). These findings deviate slightly from earlier investigations that leveraged the entire B73 maize genome for SSR analysis. Prior research identified AGC/GCT as the predominant motif among trinucleotide repeats [19]. Among all types of repeat motifs, the mononucleotide repeats exhibit the highest frequency, with a proportion of 110.88 loci/Mb, encompassing 62,204 motifs, which accounts for 49.59% of the total. In contrast to this result, Xu et al. reported a dominance of dinucleotide repeats (40.9%), followed by mononucleotide repeats (29.2%) [8]. The disparities observed could be attributed to variations in the algorithm employed by the SSR software, differences in parameter settings, variations in database versions, and discrepancies in the SSR information available for different species.
Shi et al. suggested that the microsatellite distribution pattern might exhibit conservation within Brassica species. This hypothesis was based on their comprehensive genome-wide characterization of microsatellites and subsequent marker development across three sequenced Brassica crops: B. rapa, B. oleracea, and B. napus. It is noteworthy that only 3974 tested SSR markers from B. oleracea have been published to date [27]. However, 82,948 pairs of SSR primers were designed and released in this study based on the whole-genome sequence of B. oleracea. Xu identified a total of 64,546 perfect SSR motifs and 93,724 imperfect SSR motifs within the genome of the cabbage cultivar “TO1000” [8]. These SSR markers provide a powerful tool for studying the genomics and genetics of B. oleracea in the future.
4.2. Application of the Newly Developed Genome-Wide B. oleracea SSR Markers
High transferability has been previously reported for SSRs across various plant species. For instance, in “Chiifu” of B. rapa, approximately 95% of its SSRs demonstrated the ability to amplify fragments in other species [39]. Moreover, it was found that 47% of the EST-SSR markers developed from B. rapa, B. oleracea, and B. napus exhibited transferability to six different Brassica species [40]. Gupta and Gopalakrishna conducted a random selection of 41 SSR markers from thistle and alfalfa. Their findings revealed transferability rates ranging from 53% to 71% in the leguminous plant (alfalfa) and from 33% to 44% in the non-leguminous plant (thistle) [25]. About 57% of cereal EST-SSRs could also be amplified in ryegrass [41]. Roughly 60% of EST-SSR markers derived from barley demonstrated amplification in wheat and rye as well [42]. Cui et al. employed 69 pairs of SSR primers designed for non-heading Chinese cabbage across eight varieties of Brassica crops. Their investigation revealed a transferability amplification rate ranging from 49.3% to 85.5%. Additionally, they observed that 33% of the SSR primers exhibited abundant diversity in inter-specific hybrids of Brassica [43]. Xu et al. randomly picked 24 stable and reliable SSR primers for amplification on 10 different species in the Brassicaceae family and found that 87.5% of the SSR primers exhibited transferability and applicability to one or more of the 10 related Brassica species [8].
Based on the 1176 SSR-containing ESTs in cabbage, a total of 978 primer pairs have been successfully designed and assessed for amplification efficiency using two inbred lines [44]. Subsequently, results indicated that the developed SSRs from ESTs of B. oleracea were valid and valuable in marker-assisted selection and QTL analysis in cabbage [45,46]. In addition, some useful information about SSRs and sequence analysis in Brassica crops can also be obtained on the website of the Brassicaceae Database. In this study, approximately 70% exhibited transferability to all six Brassica U triangle species, while around 60% demonstrated transferability to R. sativus (cruciferous) crops. This finding suggests that genomic SSR markers with high transferability are valuable tools that can be effectively utilized across various Brassica species and even extended to non-Brassica species. Therefore, these genomic SSR markers with definite locations and a uniform naming systems could be more widely applied to a variety of areas, such as gene location, genetic mapping, evolutionary analysis, molecular marker-assisted breeding, and supply marker material for genetic analysis and comparative genomics analysis. These markers are a significant contribution to the B. oleracea research community.
4.3. The Genetic Linkage Distance between BolSSR040196 and Bolting-Resistant Locus Is 10.69 cM
Cao and Wang employed the BSA approach to explore the molecular markers linked to the late bolting gene of B. oleracea. A RAPD marker N1-750 with a linkage distance of 7.9 cM was obtained, and one of the late bolting major genes is similar to the quality trait [47]. Mao et al. applied the BSA method to identify molecular markers linked to bolting genes in B. juncea. Through amplification mapping of parent and gene pools, they identified differences in the amplification patterns of four primers (E13M4, ESM7, UBC834, and UBC876). These differences were observed primarily in the early bolting cultivar CT07 and the early bolting pool, suggesting their potential association with bolting-related traits. The genetic distance of marker E13M4 was 16.8 cM [48]. In this study, SSR-BSA was applied to screen 336 pairs of SSR primers, and then different SSR markers were found between the bolting-resistant gene pool and the easy-bolting gene pool. Following that, a validation program was executed against parental and F2 generation single plants. Ultimately, a linkage SSR dominant marker, BolSSR040196, was identified, exhibiting a genetic distance of 10.69 cM to the bolting-resistant locus. Due to the independent development of markers in each laboratory, comparisons with reported maps become challenging. Additionally, determining the linkage group to which the gene belongs also poses difficulty. However, despite these challenges, the marker holds significant potential for employment in molecular marker-assisted selection breeding. It offers a straightforward and rapid approach to breeding B. oleracea varieties resistant to bolting.
4.4. Application of the BolSSR040196 Marker in Genetic Improvement of Cabbage
It is widely acknowledged that plants must flower at the appropriate time to ensure successful reproduction, and early flowering can significantly reduce crop yield during agricultural production. Breeding a bolting-resistant cultivars is specifically important for biennials, especially for crops originated in temperate regions, such as sugar beet [49], spinach [50], onion [51], radish [52], Brassica crops [53], and so on. In recent years, substantial advancements have been achieved in the development and utilization of molecular markers in the aforementioned crops [35,53,54].
According to findings from Gao et al., microsatellites in plants are universal and highly mutable. They also coexist and coevolve with transposable elements and are under selective pressure, and the generation, loss, functionality, and evolution of microsatellites can be related to plant gene expression and functional alterations [55]. According to the alignment result, the BolSSR040196 SSR marker is located on the 5′ UTR of the uncharacterized gene BolC05g008270.2J. Therefore, following the fine mapping process, the key candidate gene will be identified, and subsequent functional analysis will be conducted in the near future. This endeavor will enhance our comprehension of the biological function of the BolSSR040196 SSR marker. Presently, the BolSSR040196 SSR marker can be effectively utilized in molecular marker-assisted selection breeding. This approach will provide a more expedient and streamlined method for selecting bolting-resistant cabbage varieties.
5. Conclusions
A total of 125,443 SSR loci were identified within the whole-genome sequence of B. oleracea, and 82,948 primer pairs for SSR were designed. Moreover, each primer pair was assigned a unique name, ranging from BolSSR00001 to BolSSR82984, and a percentage of these primers has been confirmed to exhibit transferability to cruciferous crops. Additionally, we identified an SSR marker, BolSSR040196, located at a genetic distance of 10.69 cM from the bolting-resistant locus in B. oleracea. This SSR marker presents cabbage breeders with a valuable tool for selecting bolting-resistant cabbage varieties through MAS.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10050443/s1, Table S1: Distribution of the major SSR types identified from the genome sequence of B. oleracea; Table S2: Major SSR types and their repetition number as identified from the genome sequence of B. oleracea; Table S3: Distribution of SSR on different chromosomes of B. oleracea; Table S4: Characteristics of SSR loci and primer sequences in B. oleracea; Table S5: The sequence of the 54 pairs of SSR primers for transferability evaluation; Table S6: The sequence and Tm value of the 336 pairs of SSR primers used to screen the markers associated with the bolting gene; Figure S1: PCR amplification of B. oleracea genomic SSR markers, M: marker; 1: BolSSR000026; 2: BolSSR007539; 3: BolSSR014499; 4: BolSSR057533; 5: BolSSR027265; 6: BolSSR040726; 7: BolSSR056868; 8: BolSSR066830; 9: BolSSR064485; 10: BolSSR082001; Figure S2: Identification of the winterness level in cabbage. Figure S3: Amplification results of parental individuals with the primers BolSSR040196 tested by 12% PAGE; all of the female individuals could amplify the target fragment that appeared in bolting-resistant individuals in F2 generation while none of the male individuals could amplify.
Author Contributions
T.Z.: Methodology, Investigation, Writing—Original Draft, and Visualization; L.M.: Writing—Review and Editing; M.Z.: Methodology, Validation, and Formal Analysis; I.H.: Writing—Review and Editing; H.Y.: Investigation; J.L. (Jia Li): Visualization; N.S.: Investigation and Methodology; L.K.: Investigation and Visualization; S.W.: Resources and Supervision; J.L. (Jianbin Li): Investigation; X.Y.: Conceptualization, Methodology, Formal Analysis, Writing—Review and Editing, Resources, and Funding Acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Project of Sci-tech Foundation of Zhejiang Province (2022C02030 and 2022C02032), the 948 Program of Ministry of Agriculture, China (Grant No. 2014-Z28), the Breeding Project of the Sci-tech Foundation of Zhejiang Province (2021C02065), the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS[2021]067), the Basic Public Welfare Research Plan of Zhejiang Province (LTGN23C150008), and the Zhejiang Province SanNongJiuFang Science and Technology Cooperation Project (2023SNJF009).
Data Availability Statement
All data in this study can be found in the manuscript or the Supplementary Materials.
Acknowledgments
The authors express gratitude to Rimsha Azhar for proofreading the manuscript and acknowledge the programs that provided financial support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ananga, A.O.; Cebert, E.; Soliman, K.; Kantety, R.; Pacumbaba, R.P.; Konan, K. RAPD markers associated with resistance to blackleg disease in Brassica species. Afr. J. Biotechnol. 2006, 5, 2041–2048. [Google Scholar] [CrossRef]
- Christensen, S.; von Bothmer, R.; Poulsen, G.; Maggioni, L.; Phillip, M.; Andersen, B.A.; Jorgensen, R.B. AFLP analysis of genetic diversity in leafy kale (Brassica oleracea L. convar. acephala (DC.) Alef.) landraces, cultivars and wild populations in Europe. Genet. Resour. Crop Evol. 2010, 58, 657–666. [Google Scholar] [CrossRef]
- Rahman, M.; Li, G.; Schroeder, D.; McVetty, P.B.E. Inheritance of seed coat color genes in Brassica napus (L.) and tagging the genes using SRAP, SCAR and SNP molecular markers. Mol. Breed. 2010, 26, 439–453. [Google Scholar] [CrossRef]
- Zeng, X.H.; Wen, J.; Wan, Z.J.; Yi, B.; Shen, J.X.; Ma, C.Z.; Tu, J.X.; Fu, T.D. Effects of Bleomycin on microspore embryogenesis in Brassica napus and detection of somaclonal variation using AFLP molecular markers. Plant Cell Tiss. Org. 2009, 101, 23–29. [Google Scholar] [CrossRef]
- Panigrahi, J.; Kole, P.; Kole, C. RFLP mapping of loci controlling self-incompatibility in Brassica campestris and their comparative mapping with B. napus and B. oleracea. Biol. Plant. 2011, 55, 54–60. [Google Scholar] [CrossRef]
- Rezaeizad, A.; Wittkop, B.; Snowdon, R.; Hasan, M.; Mohammadi, V.; Zali, A.; Friedt, W. Identification of QTLs for phenolic compounds in oilseed rape (Brassica napus L.) by association mapping using SSR markers. Euphytica 2010, 177, 335–342. [Google Scholar] [CrossRef]
- Shirasawa, K.; Oyama, M.; Hirakawa, H.; Sato, S.; Tabata, S.; Fujioka, T.; Kimizuka-Takagi, C.; Sasamoto, S.; Watanabe, A.; Kato, M.; et al. An EST-SSR linkage map of Raphanus sativus and comparative genomics of the Brassicaceae. DNA Res. 2011, 18, 221–232. [Google Scholar] [CrossRef]
- Xu, Y.; Xing, M.; Song, L.; Yan, J.; Lu, W.; Zeng, A. Genome-wide analysis of simple sequence repeats in cabbage (Brassica oleracea L.). Front. Plant Sci. 2021, 12, 726084. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.J.; Moule, C.; Trick, M.; Edwards, K.J. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 2004, 108, 1103–1112. [Google Scholar] [CrossRef]
- Burgess, B.; Mountford, H.; Hopkins, C.J.; Love, C.; Ling, A.E.; Spangenberg, G.C.; Edwards, D.; Batley, J. Identification and characterization of simple sequence repeat (SSR) markers derived in silico from Brassica oleracea genome shotgun sequences. Mol. Ecol. Notes 2006, 6, 1191–1194. [Google Scholar] [CrossRef]
- Xu, J.S.; Qian, X.J.; Wang, X.F.; Li, R.Y.; Cheng, X.M.; Yang, Y.A.; Fu, J.; Zhang, S.C.; King, G.J.; Wu, J.S.; et al. Construction of an integrated genetic linkage map for the a genome of Brassica napus using SSR markers derived from sequenced BACs in B. rapa. BMC Genom. 2010, 11, 594. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.M.; Xu, J.S.; Xia, S.; Gu, J.X.; Yang, Y.; Fu, J.; Qian, X.J.; Zhang, S.C.; Wu, J.S.; Liu, K.D. Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor. Appl. Genet. 2009, 118, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Li, H.T.; Chen, X.; Yang, Y.; Xu, J.S.; Gu, J.X.; Fu, J.; Qian, X.J.; Zhang, S.C.; Wu, J.S.; Liu, K.D. Development and genetic mapping of microsatellite markers from whole genome shotgun sequences in Brassica oleracea. Mol. Breed. 2011, 28, 585–596. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.B.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.Y.; Maghirang, R.; Li, Z.K.; Xing, Y.Z.; et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.J.; Shi, J.R.; Singh, S.; Fickus, E.W.; Costa, J.M.; Lewis, J.; Gill, B.S.; Ward, R.; Cregan, P.B. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 2005, 110, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.C.; Grant, D.; Olson, T.; Warren, W.C.; Wing, R.; Yu, Y.; Kim, H.; Cregan, P.; Joseph, B.; Futrell-Griggs, M.; et al. Microsatellite discovery from BAC end sequences and genetic mapping to anchor the soybean physical and genetic maps. Genome 2008, 51, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.C.; Wang, Y.; Huang, L.S.; Wang, Y.J.; Yu, Y.J.; Yang, L. Large-scale development of SSR markers in the genome of cacao. J. Shandong Agric. Univ. Nat. Sci. 2013, 44, 340–344. [Google Scholar]
- Cai, B.; Li, C.H.; Yao, Q.H.; Zhou, J.; Tao, J.M.; Zhang, Z. Analysis of SSRs in grape genome and development of SSRs database. J. Nanjing Agric. Univ. 2009, 32, 28–32. [Google Scholar]
- Qu, J.T.; Liu, J. A genome-wide analysis of simple sequence repeats in maize and the development of polymorphism markers from next-generation sequence data. BMC Res. Notes 2013, 6, 403. [Google Scholar] [CrossRef]
- Jia, H.M.; Shen, Y.T.; Jiao, Y.; Wang, G.Y.; Dong, X.; Jia, H.J.; Du, F.; Liang, S.M.; Zhou, C.C.; Mao, W.H.; et al. Development of 107 SSR markers from whole genome shotgun sequences of Chinese bayberry (Myrica rubra) and their application in seedling identification. J. Zhejiang Univ. Sci. B 2014, 15, 997–1005. [Google Scholar] [CrossRef]
- La Rota, M.; Kantety, R.V.; Yu, J.K.; Sorrells, M.E. Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genom. 2005, 6, 23. [Google Scholar] [CrossRef]
- Cardle, L.; Ramsay, L.; Milbourne, D.; Macaulay, M.; Marshall, D.; Waugh, R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 2000, 156, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.H.; Alabady, M.S.; Ulloa, M.; Sickler, B.; Wilkins, T.A.; Yu, J.; Stelly, D.M.; Kohel, R.J.; El-Shihy, O.M.; Cantrell, R.G. Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Mol. Genet. Genom. 2005, 274, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.Q.; Chen, X.P.; Hong, Y.B.; Liu, H.Y.; Zhou, G.Y.; Li, S.X.; Guo, B.Z. Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol. 2009, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Gopalakrishna, T. Development of unigene-derived SSR markers in cowpea (Vigna unguiculata) and their transferability to other Vigna species. Genome 2010, 53, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Huang, S.; Zhan, J.; Yu, J.; Wang, X.; Hua, W.; Liu, S.; Liu, G.; Wang, H. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res. 2014, 21, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Boudry, P.; Wieber, R.; Saumitou-Laprade, P.; Pillen, K.; Dijk, H.V.; Jung, C.J.T.; Genetics, A. Identification of RFLP markers closely linked to the bolting gene B and their significance for the study of the annual habit in beets (Beta vulgaris L.). Theor. Appl. Genet. 1994, 88, 852–858. [Google Scholar] [CrossRef]
- El-Mezawy, A.; Dreyer, F.; Jacobs, G.; Jung, C. High-resolution mapping of the bolting gene B of sugar beet. Theor. Appl. Genet. 2002, 105, 100–105. [Google Scholar] [CrossRef]
- Teutonico, R.A.; Osborn, T.C. Mapping loci controlling vernalization requirement in Brassica rapa. Theor. Appl. Genet. 1995, 91, 1279–1283. [Google Scholar] [CrossRef]
- Rosental, L.; Still, D.W.; You, Y.; Hayes, R.J.; Simko, I. Mapping and identification of genetic loci affecting earliness of bolting and flowering in lettuce. Theor. Appl. Genet. 2021, 134, 3319–3337. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Huang, S.N.; Gao, Y.; Zhang, M.D.; Qu, G.Y.; Wang, N.; Liu, Z.Y.; Feng, H. Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 2020, 133, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Wang, J.L.; Li, B.Y.; Wei, Q.Z.; Hu, T.H.; Hu, H.J.; Bao, C.L. Genetic and QTL Mapping Analysis of Bolting Time in Cabbage(Brassica oleracea). Acta Hortic. Sin. 2020, 47, 974–982. [Google Scholar] [CrossRef]
- Wei, X.C.; Rahim, M.A.; Zhao, Y.Y.; Yang, S.J.; Su, H.N.; Wang, Z.Y.; Shahriar, S.A.; Li, J.D.; Yang, Z.Y.; Yuan, Y.X.; et al. Inheritance and Genetic Mapping of Late-Bolting to Early-Bolting Gene, BrEb-1, in Chinese Cabbage (Brassica rapa L.). Agronomy 2022, 12, 1048. [Google Scholar] [CrossRef]
- Zhu, X.W.; Bo, T.Y.; Chen, J.X.; Tai, X.; Ren, Y.Y. Research Progress on Mapping of Genes Associated with Main Agronomic Traits of Cabbage. Acta Hortic. Sin. 2017, 44, 1729–1737. [Google Scholar] [CrossRef]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B.; Hancock, J. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- da Maia, L.C.; Palmieri, D.A.; de Souza, V.Q.; Kopp, M.M.; de Carvalho, F.I.; Costa de Oliveira, A. SSR Locator: Tool for Simple Sequence Repeat Discovery Integrated with Primer Design and PCR Simulation. Int. J. Plant Genom. 2008, 2008, 412696. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W.; Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- An, Z.S.; Gao, C.H.; Li, J.N.; Fu, D.H.; Tang, Z.L.; Ortegon, O. Large-scale development of functional markers in Brassica species. Genome 2011, 54, 763–770. [Google Scholar] [CrossRef]
- Sim, S.C.; Yu, J.K.; Jo, Y.K.; Sorrells, M.E.; Jung, G. Transferability of cereal EST-SSR markers to ryegrass. Genome 2009, 52, 431–437. [Google Scholar] [CrossRef]
- Castillo, A.; Budak, H.; Varshney, R.K.; Dorado, G.; Graner, A.; Hernandez, P. Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol. 2008, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.M.; Hou, X.L.; Dong, Y.X. Development of SSR Primers of Non-heading Chinese Cabbage and Transferability among Closely Related Species. Sci. Technol. Rev. 2005, 23, 20–25. [Google Scholar]
- Chen, C.; Zhuang, M.; Li, K.N.; Liu, Y.M.; Yang, L.M.; Zhang, Y.Y.; Cheng, F.; Sun, P.T.; Fang, Z.Y. Development and Utility of EST-SSR Marker in Cabbage. Acta Hortic. Sin. 2010, 37, 221–228. [Google Scholar] [CrossRef]
- Chen, C.; Zhuang, M.; Fang, Z.Y.; Wang, Q.B.; Zhang, Y.Y.; Liu, Y.M.; Yang, L.M.; Cheng, F. A Co-Dominant Marker BoE332 Applied to Marker-Assisted Selection of Homozygous Male-Sterile Plants in Cabbage (Brassica oleracea var. capitata L.). J. Integr. Agr. 2013, 12, 596–602. [Google Scholar] [CrossRef]
- Su, Y.; Liu, Y.; Li, Z.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y. QTL Analysis of Head Splitting Resistance in Cabbage (Brassica oleracea L. var. capitata) Using SSR and InDel Makers Based on Whole-Genome Re-Sequencing. PLoS ONE 2015, 10, e0138073. [Google Scholar] [CrossRef]
- Cao, W.R.; Wang, C. RAPD Marker of Later Bolting Gene on Cabbage. Biotechnol. Bull. 2007, 5, 167–169. [Google Scholar] [CrossRef]
- Mao, W.W.; Gao, H.S.; Bo, T.Y.; Ma, J.J.; Xu, D.J.; Chen, X.H.; Jia, Z.M.; Wang, Y.L. Analysis of Bolting Trait of Brassica campestris L. ssp. Chinensis var. utilis Tsen et Lee By ISSR and SRAP Markers. Jiangsu J. Agric. Sci. 2009, 25, 829–833. [Google Scholar]
- Tränkner, C.; Pfeiffer, N.; Kirchhoff, M.; Kopisch-Obuch, F.J.; van Dijk, H.; Schilhabel, M.; Hasler, M.; Emrani, N. Deciphering the complex nature of bolting time regulation in Beta vulgaris. Theor. Appl. Genet. 2017, 130, 1649–1667. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Liu, Z.Y.; Feng, C.D.; Zhang, H.L.; Xu, Z.S.; Wang, X.W.; Wu, J.; She, H.B.; Qian, W. Quantitative trait locus mapping and identification of candidate genes controlling bolting in spinach (Spinacia oleracea L.). Front. Plant Sci. 2022, 13, 850810. [Google Scholar] [CrossRef] [PubMed]
- Wako, T.; Tsukazaki, H.; Yaguchi, S.; Yamashita, K.; Ito, S.; Shigyo, M. Mapping of quantitative trait loci for bolting time in bunching onion (Allium fistulosum L.). Euphytica 2016, 209, 537–546. [Google Scholar] [CrossRef]
- Wang, Q.B.; Wang, Y.P.; Zhang, L. Inheritance and Molecular Marker for Flowering Time in Radish (Raphanus sativus L.). Plant Mol. Biol. Rep. 2018, 36, 878–887. [Google Scholar] [CrossRef]
- Dai, Y.; Sun, X.; Wang, C.G.; Li, F.; Zhang, S.F.; Zhang, H.; Li, G.L.; Yuan, L.Y.; Chen, G.H.; Sun, R.F.; et al. Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization. BMC Genom. 2021, 22, 236. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A century of gibberellin research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.H.; Ren, X.D.; Mason, A.S.; Li, J.N.; Wang, W.; Xiao, M.L.; Fu, D.H. Revisiting an important component of plant genomes: Microsatellites. Funct. Plant Biol. 2013, 40, 645–661. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).