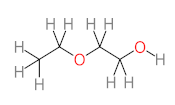
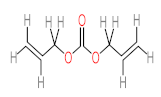
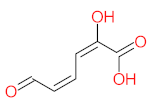
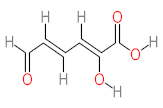
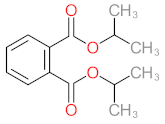
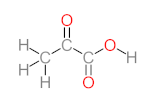
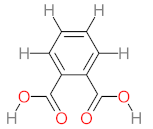
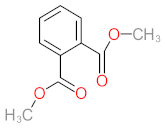
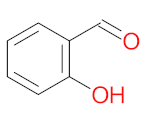
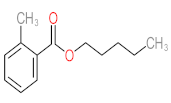
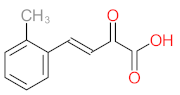
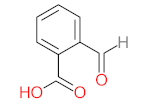
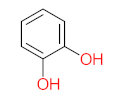
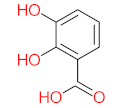
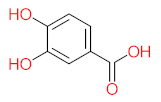
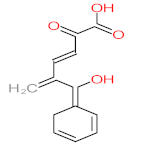
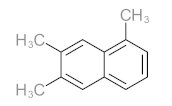
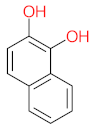
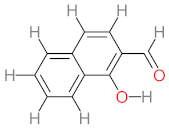
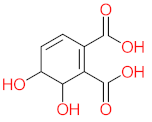
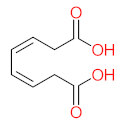
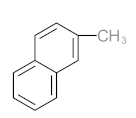
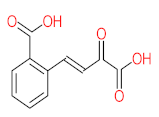
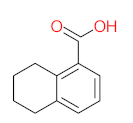
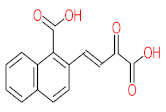
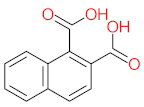
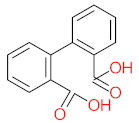
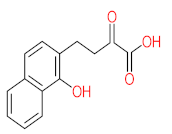
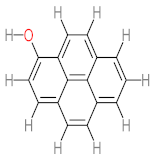
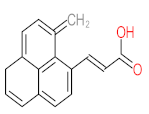
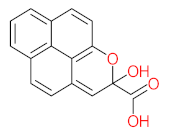
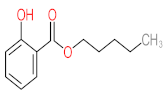
Abstract
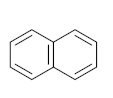
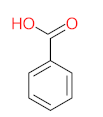
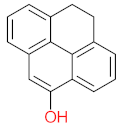
Polycyclic aromatic hydrocarbons are compounds with 2 or more benzene rings, and 16 of them have been classified as priority pollutants. Among them, pyrene has been found in higher concentrations than recommended, posing a threat to the ecosystem. Many bacterial strains have been identified as pyrene degraders. Most of them belong to Gram-positive strains such as Mycobacterium sp. and Rhodococcus sp. These strains were enriched and isolated from several sites contaminated with petroleum products, such as fuel stations. The bioremediation of pyrene via Mycobacterium strains is the main objective of this review. The scattered data on the degradation efficiency, formation of pyrene metabolites, bio-toxicity of pyrene and its metabolites, and proposed degradation pathways were collected in this work. The study revealed that most of the Mycobacterium strains were capable of degrading pyrene efficiently. The main metabolites of pyrene were 4,5-dihydroxy pyrene, phenanthrene-4,5-dicarboxylate, phthalic acid, and pyrene-4,5-dihydrodiol. Some metabolites showed positive results for the Ames mutagenicity prediction test, such as 1,2-phenanthrenedicarboxylic acid, 1-hydroxypyrene, 4,5-dihydropyrene, 4-phenanthrene-carboxylic acid, 3,4-dihydroxyphenanthrene, monohydroxy pyrene, and 9,10-phenanthrenequinone. However, 4-phenanthrol showed positive results for experimental and prediction tests. This study may contribute to enhancing the bioremediation of pyrene in a different matrix.
1. Introduction
The demand for water and energy is increasing, putting additional strain on water and environmental resources. Water scarcity has been identified as a socioeconomic and environmental problem that challenges the world in the twenty-first century, affecting approximately four billion people worldwide at least one month per year [1,2,3]. The continuous release of harmful chemicals such as persistent organic pollutants (POPs) is considered one of the most threatening environmental problems, as mentioned in the Stockholm convention, 2004, Basel convention, 1989, the Rotterdam convention, 1998, Barcelona resolution, 1995, 8 Aarhus Protocol, 1998, and the Arctic environmental protection strategy, 1991 [4]. According to the listed protocols and conventions, harmful chemicals should be eliminated, or their production decreased. POPs are a group of toxic chemicals that stay in the environment long-term and resist natural degradation. There are two major sources of POPs, (i) natural sources, including forest fires, volcanic eruptions, and biogenic sources (microbial metabolites, plants, and algae), and (ii) anthropogenic sources, including incomplete combustion (oil, wood, petroleum, and coal), synthetic fertilizers, pesticides formulations, and industrial process [5]. POPs groups include personal care products, polychlorinated compounds, dibenzo-p-dioxins and dibenzofurans, and polycyclic aromatic hydrocarbons (PAHs) [6]. The increasing discharge of POPs into the environment leads to them bioaccumulating and becoming biomagnified until they reach a specific concentration leading to bio-toxicity. POPs can cross boundaries, move freely away from their original sources, and be absorbed by soil particles; they are volatile in the atmosphere, can run off into water bodies, enter into the food chain, be uptaken by plants, or leach into groundwater. PAHs are compounds that contain two or more benzene rings. The United States Environmental Protection Agency (USEPA) classified 16 PAHs as priority pollutants due to their low solubility, non-polarity, hydrophobicity, high boiling point, high melting point, corrosion resistance, conductivity, heat resistance, light-sensitive, bioaccumulation, biomagnification, and bio-toxicity [7,8]. Many adverse effects of PAHs have been reported on human health, aquatic organisms, and wildlife, such as genetic mutation, endocrine disruption, cardiovascular disorders, hypertension, immune system suppression, and birth defects. In this work, pyrene was the main target pollutant and was detected at higher concentrations than the standard values or maximum contaminant levels [9]. According to the United States Environmental Protection Agency (USEPA), the standard values of pyrene for human health for the consumption of water and organism is 20 µg/L, while for human health for the consumption of water is only 30 µg/L [10]. Many biological, physical, and chemical treatment techniques have been used to eliminate pyrene from different mediums. The advanced oxidation process is one of the most efficient treatment methods capable of oxidizing pyrene [11]. However, advanced oxidation processes need to inject a large number of chemicals to complete mineralization, which increases the treatment costs and the discharge of chemicals into the environment [12]. Bioremediation approaches have gained attention due to their advantages, such as being cost-effective and environmentally friendly. Biological treatment is a biological process that uses target pollutants as a source of energy and carbon to degrade, mineralize, transform, and detoxify the target pollutant in a specific medium. Bioremediation can use indigenous biological agents (biostimulation) or external biological agents (bioaugmentation) and can be applied either in situ or ex situ based on many factors such as, (i) type of pollutants, (ii) cost of treatment, (iii) geological site, (iv) pollutants’ concentration, and (v) depth of pollution. The most popular treatment techniques use in situ natural attenuation, bio-slurping, bioventing, disparaging, and phytoremediation. Many ex situ techniques are used to treat many pollutants, including POPs, such as landfarming, bioreactor, windrow, and biopile windrow [13]. Quintella et al. [14] applied a strengths, weaknesses, opportunities, and threats (SWOT) analysis for the study of bioremediation technologies. They revealed that most of the studies have been conducted in the United States of America and China, and the most common biological agents used were bacteria, enzymes, fungi, algae, plants, and protozoa, with percentages of 57%, 19%, 13%, 6%, 4%, and 1%, respectively. Water, soil, and sludge were the most common degradation matrixes that were treated, with percentages of 53%, 36%, and 11%, respectively. They reported that the target pollutants that were degraded via biological agents the most were oil, metals, organic waste, polymers, food, and cellulose, and their percentages were 38%, 21%, 21%, 10%, and 5%, respectively. Recently, the degradation of pyrene via isolation of bacterial strains has increased. Mycobacterium strains and Rhodococcus strains were the most dominant bacterial species used for PAHs degradation; these strains were enriched and isolated from different sites contaminated with petroleum products, such as fuel stations [15,16]. Figure 1 represents the number of documents by year and the percentage of each type of document found using keywords (oxidation of pyrene by bacteria) through the Scopus database. In 1995, the number of studies related to the biodegradation of pyrene for each type of document was 5, while in 2021, the number of studies was 55, which means that this topic has been gaining researchers’ attention. More than 94% of these studies were articles, 2.3%, 2.0%, 0.6%, 0.5%, 0.5, and 2.0% were reviews, conference papers, notes, book chapters, short surveys, and errata, respectively. In this review, the main objective is to collect and organize the scattered information related to the studies that investigated the degradation of pyrene by Mycobacterium strains. The major topics that are investigated in this review are degradation efficiency, pyrene metabolites, bio-toxicity, and the proposed degradation pathways.
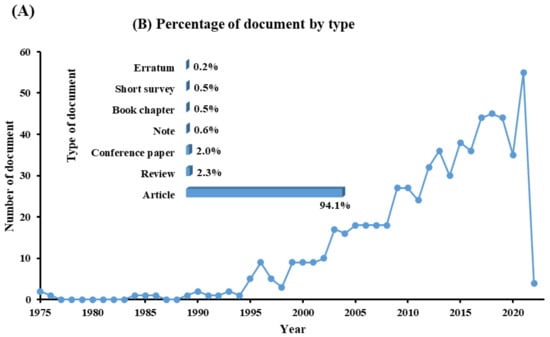
Figure 1.
(A) represents the number of documents by year, and (B) the percentage for each type of document when using keywords (oxidation of pyrene by bacteria) on the Scopus database.
2. Degradation of Pyrene by Mycobacterium sp.
Several bacterial strains have been isolated to use pyrene as a sole carbon and energy source; most of them are Gram-positive, such as Mycobacterium and Rhodococcus [17,18]. Mycobacteria are catalase-positive, non-motile, non-spore-forming, rod-shaped bacteria (0.2–0.6 mm wide and 1.0–10 mm long). The colony morphology of Mycobacteria varies, with some species growing as rough or smooth colonies. Colony color ranges from white to orange or pink [19]. It has been reported that the first isolation of a bacterial strain to mineralize pyrene was in 1988 [20]. Mycobacterium was the most dominant strain to mineralize pyrene [21]. The successful mineralization of pyrene by Mycobacterium strains refers to their ability to produce several functional enzymes capable of metabolizing high molecular weight polycyclic aromatic hydrocarbons, such as pyrene. Dioxygenase is a complex, multi-component enzymatic system containing iron sulfur-containing terminal oxygenase, reductase, and ferredoxin [22]. It has been reported that hydroxylation is the initial biochemical step in the pyrene degradation process. It introduces a couple of oxygen atoms into aromatic pyrene rings [23]. The complete mineralization of pyrene occurs through different enzymatic reactions such as dioxygenase, dihyrogendiol, dehydrogenase, ring cleavage dioxygenase, epoxide hydrolase, alcohol dehydrogenase, acetaldehyde dehydrogenation, and decarboxylation [24]. Figure 2 illustrates the biodegradation of pyrene by Mycobacterium.
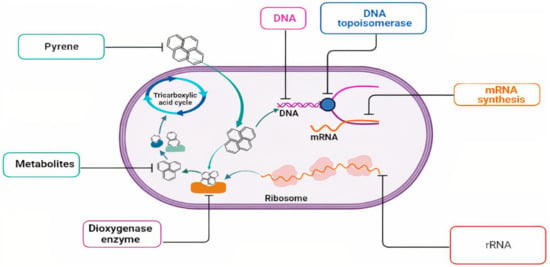
Figure 2.
Illustrated the biodegradation of pyrene by Mycobacterium.
Many functional genes have been identified in the Mycobacterium strains, such as NidA, NidB, NidAB, NidA3B3, PhdA, PhdB, PdoA, PdoB, and PdoAB. Among Mycobacterium strains, the vanbaalenii PYR-1 strain has many functional genes capable of degrading pyrene and its metabolites. Table 1 includes some of the enzymes produced by different Mycobacterium strains during pyrene degradation. Miller et al. [25] identified NidB and NidA genes that are responsible for producing dioxygenase enzyme when Mycobacterium sp. JLS is used to catabolize pyrene. Zeng et al. [26] reported that the PdoAB gene is responsible for encoding a dioxygenase capable of oxidizing pyrene. Costa et al. [27] observed that PhdA and PhdB are the main genes of the dioxygenase enzyme in the Mycobacterium fortuitum strain.

Table 1.
Summary of the functional genes for each Mycobacterium strain.
In this review article, more than 40 studies related to pyrene degradation via Mycobacterium strains or consortium culture were collected. In general, Mycobacterium strains showed high degradation efficiency, most of them 80–100%. There are numerous Mycobacterium strains that can degrade pyrene. The phylogenetic tree of Mycobacterium strains is depicted in Figure 3. The Mycobacterium sequences were collected using the NCBI gene bank (Home Nucleotide—www.ncbi.nlm.nih.gov (accessed on 30 December 2021)). The sequences were assembled, aligned, and analyzed with MEGA software version 11.0
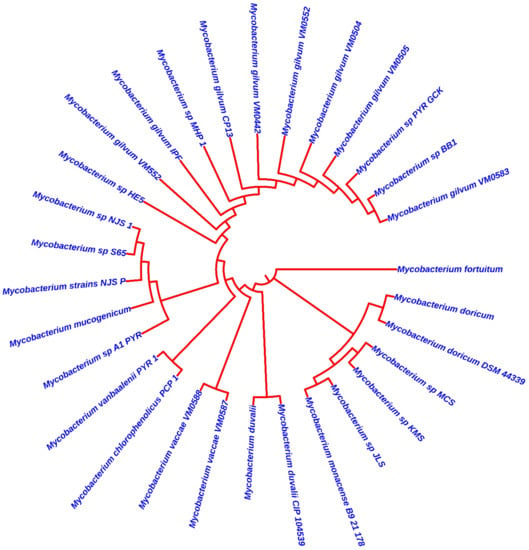
Figure 3.
Phylogenetic tree for Mycobacterium strains that have been used for pyrene degradation.
Wanapaisan et al. [28] used a consortium culture containing five bacterial strains (Mycobacterium sp. PO1, PO2, Bacillus sp. FW1, Ochrobactrum sp. PW1, and Novosphingobium pentaromativorans PY1). The result showed that 100 mg/L of pyrene was completely eliminated within 6 days of incubation. In addition, the Mycobacterium sp. NJS-1 strain was used to mineralize pyrene on metal-modified montmorillonite. This study revealed that around 93.6% of 15 mg/L of pyrene was degraded within 3 days at neutral pH conditions, and the degradation rate was first-order kinetics 0.62 k/d [34]. Additionally, Zhang et al. [35] applied a consortium of bacterial strains (Micrococcus sp. PHE9 and Mycobacterium sp. NJS-P) to decompose pyrene. About 58% of 100 mg/L of pyrene was removed after 18 days of incubation, and the degradation rate was 3.24 mg/L × day. Sun et al. [36] isolated the Mycobacterium sp. WY10 strain to oxidize 50 mg/L of pyrene in a mineral salt medium. Around CFU/mL was inoculated, and the degradation was 83% after 72 h of treatment. Xiaoning Li et al. [37] examined the Mycobacterium sp. NJS-1 strain to treat and remove high molecular weight polycyclic aromatic hydrocarbons, such as pyrene. The author used a mineral medium, and around CFU/mL was inoculated to degrade 200 mg/L of pyrene; the degradation was 90% of pyrene in the presence of humic acid, while about 10.5% was in the absence of humic acid within 7 days of incubation. The Mycobacterium gilvum CP13 strain was isolated for oxidizing pyrene in a mineral salt medium at alkaline conditions. The bacteria were inoculated at an optical density of 600 nm = 0.5, and 95% of 50 mg/L of pyrene was oxidized after 7 days of degradation treatment [38]. Furthermore, Chen et al. [39] applied biotechnology to treat agricultural and industrial soils contaminated with 16 priority polycyclic aromatic hydrocarbons, including pyrene. The Mycobacterium strain was capable of removing 85% of 100 mg/kg of pyrene during 35 days of treatment in both soils. Also, Terzaghi et al. [40] examined the Mycobacterium gilvum VM552 strain to degrade pyrene suspended on the leaf surface of holm oak (Quercus ilex). The results indicated that after 2 weeks of treatment, the removal was only 17%. Chen et al. [15] attempted to stimulate a microbial degradation approach for soil-containing pyrene. In this study, the active bacterial strains were identified; among them, Mycobacterium strains were the most dominant, and the degradation was 80% of 60 mg/kg within 35 days; the experiment was conducted at pH 8. Sarma and Pakshirajan [41] isolated the Mycobacterium frederiksbergense strain to mineralize pyrene using a batch shake flask reactor. After 200 h of incubation, the pyrene was totally eliminated at neutral pH conditions. Moreover, Peng et al. [17] reported that approximately 81% of 50 mg/kg of soil-containing pyrene was oxidized after 60 days of bioremediation under acidic conditions using the Mycobacterium strain. They pointed out that the NidA gene in Mycobacterium was responsible for generating the dioxygenase enzyme. In addition, the Mycobacterium vanbaalenii PYR-1strain was used in a phosphate-based mineral medium, and 25 µM of pyrene was completely oxidized after 24 h of treatment [18]. Table 2 provides a summary of the studies that used Mycobacterium strains to degrade pyrene in a different medium, pH, optical density, degradation efficiency, incubation time, and initial concentration.

Table 2.
Summary of the studies that used Mycobacterium strains to treat pyrene in a different medium.
3. Identification of Pyrene Metabolites Degraded by Mycobacterium sp. and Their Biotoxicity
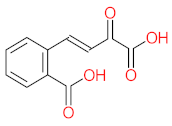
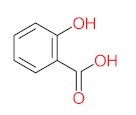
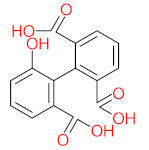
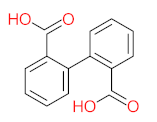
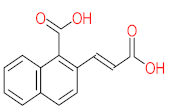
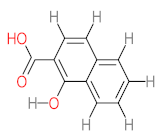
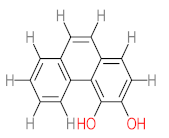
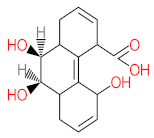
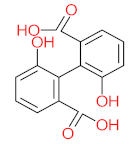
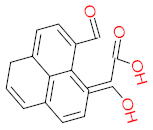
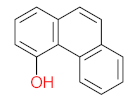
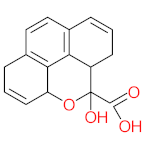
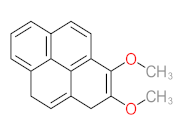
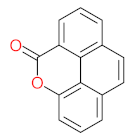
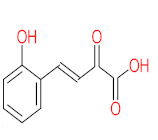
The specific aim of the remediation is to achieve complete mineralization or convert the target pollutant into harmless products. Metabolites (by-products, also called intermediate products) are products that are partially degraded and are generated during and after the treatment process. Some metabolites could be more toxic to public health and the environment than the original pollutant. It has been illustrated that the risk of the pollutant’s metabolites is like an iceberg. The pollutants themselves are just the tip of the iceberg, while the metabolites’ products represent the majority of the iceberg, which is hidden underwater. The researchers monitor the metabolites for many reasons: (i) to examine the effectiveness of the treatment approach, (ii) to detect any ecotoxic by-products after the end of treatment operation, and (iii) to build an oxidation pathway based on detected by-products. Many metabolites have been detected during and after the remediation process. The studies that investigated the degradation of pyrene via the Mycobacterium strains detected many metabolites. For example, Seo et al. [70] pointed out that phenantharene-4,5-dicarboxylic acid and naphthalene-1,2-dicarboxylic acid were the major intermediate products when Mycobacterium aromativorans strain JS19b1 was applied for pyrene degradation. Sun et al. [36] observed many metabolites produced by Mycobacterium sp. WY10 during pyrene degradation, such as cis-pyrene dihydrodiol, cis-pyrene-4,5-dihydrodiol, dihydroxy pyrene, methylated-phenanthrene-4,5-dicarobxylic acid, 4-phenanthrene-4-carboxylic acid, phenantharene-4,5-dicarboxylic acid, and phenanthrene-4-carboxylic acid. In addition, the Mycobacterium sp. flavescens PYR-1 strain was used for pyrene degradation. The major by-products were 4,5-dihydroxy-4,5-4,5-dihydropyrene, 4-phenanthroic, phthalic acid, and 4,5-phenanthrenedioic acid [43]. Mycobacterium sp. AP1 grew with pyrene as a sole carbon and energy source. The identified metabolites were trans- or cis-4,5-dihydroxy-4,5-4,5-dihydropyrene, phenantharene-4,5-dicarboxylic acid, phenanthrene-4-carboxylic acid, and 6,6-dihydroxy-2,2-biphenyl dicarboxylic acid [44]. Additionally, Zhong et al. [45] mentioned that the by-products of pyrene were dihydroxy phenanthrene, monohydroxy pyrene, dihydroxy pyrene, 4-phenanthrene-carboxylic acid, and 4-phenanthroic when Mycobacterium sp. A1-PYR was applied. Rehmann et al. [21] used Mycobacterium sp. KR2 to remove pyrene. After 8 days of incubation, the metabolites were cis-4,5-pyrene dihydrodiol, 4,5-phenanthrene dicarboxylic acid, 1-hydroxy-2-naphthoic acid, 2-carboxybenzaldehyde, phthalic acid, and protocatechuic acid. Also, pyrene cis-4,5-dihydrodiol and dihydroxypyrene were the main metabolites produced after 24 h of incubation of Mycobacterium vanbaalenii PYR-1 [18]. Furthermore, Luo et al. [46] used synergistic microbes (Selenastrum capricornutum and Mycobacterium sp. A1-PYR) to oxidize pyrene, and the metabolites were dihydroxy pyrene, 1-hydroxypyrene, 4-phenanthrol, 4-phenanthrene-carboxylic acid, hydroxyphenyl acetic acid, phenylacetic acid, salicylic acid, and benzoic acid. Kim et al. [71] observed 1,2-dicarboxynaphthalene, phenanthrene and pyrene-diols, and cis-4-(1-hydroxynaphth-2-yl)-2-oxobut-3-enoic acid. Liang et al. [72] detected pyrene-4,5-dione, cis-4,5-pyrene-dihydrodiol, phenanthrene-4,5-dicarboxylic acid, and 4-phenanthroic acid as a metabolite when Mycobacterium sp. strain KMS was applied. Moreover, Zhong et al. [47] examined a bacterial culture (Mycobacterium sp. A1-PYR and Sphingomonas sp. PheB4) for pyrene decomposition. The metabolites in this system were monohydroxy pyrene, pyrene diol, and dihydroxy pyrene. Xiaoning Li et al. [37] used Mycobacterium sp. NJS-1 to oxidize pyrene in the presence of and without humic acid. The by-product in the absence of humic acid was phenanthrene 3,4-diol, while 1,2-dimethoxypyrene was detected in the presence of humic acid. In addition, Wu et al. [38] studied the metabolites of Mycobacterium gilvum CP13 when pyrene was used as a sole carbon and energy source. The major metabolites were 4-phenanthrenecarboxylic acid, 4-phenanthrenol, 1-naphthol, and phthalic acid. Also, phthalic acid, naphthalene-1,8-dicarboxylic acid, diphenic acid, 6,6′-dihydroxy-2,2′-biphenyl dicarboxylic acid, Z-9-carboxymethylenefluorene-1-carboxylic acid, and phenantharene-4,5-dicarboxylic acid were detected by [48]. Many metabolites were detected by [49] when the Mycobacterium sp. strain RJGII-135 was isolated to degrade pyrene. The metabolites were 4,5-phenanthrene dicarboxylic acid, 4-phenanthrene-carboxylic acid, and 4,5-pyrene-dihydrodiol. In conclusion, according to these studies, phenantharene-4,5-dicarboxylic acid, dihydroxy pyrene, phenanthrene-4-carboxylic acid, phthalic acid, and pyrene-4,5-dihydrodiol were the most frequent metabolites that were detected when Mycobacterium sp. strains were used for pyrene degradation. Table 3 represents the metabolites of several organisms, such as plants, algae, earthworms, bacteria, and fungi, that have been utilized to remove pyrene from different mediums.

Table 3.
Summary of the metabolites of pyrene from many species used for pyrene oxidation.
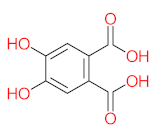
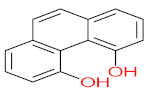
It should be noted that phthalic acid, 1-hydroxypyrene, 1-hydroxy-2-naphthoic acid, 4,5-dihydroxy pyrene, phenanthrene 4,5-dicarboxylate, and pyrene-4,5-dihydrodiol are the most frequent metabolites in the last table. It has been observed that the metabolites of Mycobacterium sp. strains and the species mentioned in Table 3 share 4,5-dihydroxy pyrene, phenanthrene-4,5-dicarboxylate, phthalic acid, and pyrene-4,5-dihydrodiol as the most frequent metabolites. That may be attributed to the enzymes that are shared between them, which in turn, leads to shared degradation pathways of pyrene.
The mass consumption of petroleum products and increase in their demand around the world leads to an increase in the opportunity for pyrene leakage into the environment and increases the opportunity for exposure to pyrene by organisms and humans. Frequent and long-term exposure to pyrene leads to bioaccumulation and biomagnification in the organism cell, which increases the possibility of carcinogenicity and mutagenicity. Many studies have mentioned the negative impacts of pyrene and its metabolites on animals and humans. The toxicity evaluation of pyrene metabolites is important to increase system efficiency. The toxicity assessment of pyrene and its metabolites was carried out using the United States Environmental Protection Agency’s software, called Toxicity Estimation Software Tool (TEST) version 5.1. This software is capable of applying mathematical models to predict pollutant toxicity based on Quantitative Structure-Activity Relationship (QSAR) methodology. The data were introduced by inputting the name of each by-product. The lethal concentration of 50% (LC50) (96 h) in fathead minnow and Ames mutagenicity were the considered toxicity for pyrene metabolites using Mycobacterium strain and other biological agents, represented in Table 4. Some metabolites showed positive results for the Ames mutagenicity prediction test, such as 1,2-phenanthrenedicarboxylic acid, 1-hydroxypyrene, 4,5-dihydropyrene, 4-phenanthrene-carboxylic acid, 3,4-Dihydroxyphenanthrene, Monohydroxy pyrene, and 9,10-phenanthrenequinone. However, 4-phenanthrol showed positive results for experimental and prediction tests.

Table 4.
Summary of the results of LC50 (96 h) fathead minnow and the Ames mutagenicity test for the main pyrene metabolites after treatment by using Mycobacterium strain and other biological agents.
4. Proposed Biodegradation Pathways
Many bacterial strains have been applied to degrade pyrene in a different medium. Some bacterial strains share the same functional enzymes, which leads to the same degradation pathways, as shown in Table 5. Some genes in the Mycobacterium sp. strain produce enzymes capable of oxidizing pyrene. There are numerous advantages to determining the degradation pathway, including the ability to control the effectiveness of remediation systems, eliminating the influence of degradation on analytical results, and knowledge of degradation pathways for specific compounds can facilitate the assessment of environmental pollution with POPs based on the presence of degradation products. In addition, identifying the degradation pathway is useful for the future development of bioremediation [93,94].

Table 5.
Summary of the main genes of Mycobacterium sp. strain that responsible in pyrene degradation and their functions.
The following studies are examples of the degradation of pyrene by using the Mycobacterium sp. strain. Yuan et al. [29] proposed a detailed pyrene degradation pathway via Mycobacterium sp. strain A1-PYR. The first step of pyrene degradation was hydroxylation using NidAB and PodA3B3, leading to forming cis-4,5-dihydroxy-4,5-hydropyrene, then PhdE acting to convert cis-4,5-dihydroxy-4,5-hydropyrene into 4,5-dihydroxypyren, then phenanthrene-4,5-dicarboxylate via PhdF, further degradation leading to form phenanthrene-4-carboxylate. PodA2B2 enzyme works to produce cis-3,4-phenanthrene-dihydrodiol-4-carboxylate, then PhdE acts to generate 3,4-dihydroxy-phenanthrene. More decomposition of 3,4-dihydroxy-phenanthrene via PhdF leading to form 2-hydroxy-2H-benzo[h]chromene-2-carboxylate then cis-4-(1′-hydroxy-naphth-2′-yl)-2 oxobut-3-enoate. PhdG leading to form 1-hydroxy-2-naphthaldehyde 1-hydroxy-2-naphthoate, further degradation of 1-hydroxy-2-naphthoate leading to produce 2-cis-2′-carboxy-benzalpyruvate. Additionally, the PhdJ enzyme converts 2-cis-2′-carboxy-benzalpyruvate into phthalate then the ring cleavage via PhtC results to form carboxylic acids compounds. The final metabolite step was that the small carboxylic acids enter the tricarboxylic acid cycle to produce energy,. In addition, Krivobok et al. [95] proposed the degradation pathway of pyrene by Mycobacterium sp. Strain-6 PY1. They observed that PhdABCD, PhdE, PhdF, PhdG, PhdH, PhdI, and PhdK enzymes were detected in the Mycobacterium sp. Strain-6 PY1. The degradation of pyrene started with the hydroxylation process of C4 and C5 positions to form pyrene cis-4,5-dihydrodiol then 4,5-dihydroxypyrene, further oxidation of 4,5-dihydroxypyrene generates 4,5-phenanthrenedioic → 4-phenanthrene acid → phenanthrene-3,4-diol → phenanthrene → cis-3,4-phenanthrene-dihydrodiol → 3,4-dihydroxy-phenanthrene → 2-hydroxy-2H-1-oxa-pyrene-2-carboxylic acid → 2-Hydroxy-2H-benzo[h]chromene-2-carboxylate → 1-Hydroxy-2-naphthaldehyde → trans-2′-carboxybenzal pyruvic acid → 2-2-Carboxybenzaldehyde → O-phthalic acid → tricarboxylic acid cycle. Wu et al. [38] studied the degradation of pyrene via Mycobacterium gilvum and the proposed the degradation pathway as the following: pyrene → 4-phenanthrenecarboxylic acid → 3,4-dihydroxy-phenanthrene → 2-Hydroxy-2H-benzo[h]chromene-2-carboxylate → 1-naphthol → phthalic acid. A simple degradation pathway of pyrene through Mycobacterium sp. is shown in Figure 4. The most common transformation metabolites that have been proposed to build degradation pathways are shown in Table 6, while Table 7 shows an example of pyrene degradation pathways via different microbial species.
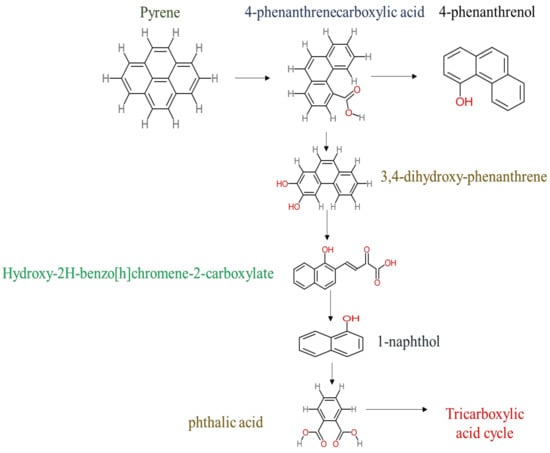
Figure 4.
Proposed degradation pathway of pyrene by Mycobacterium gilvum.

Table 6.
Summary of the most frequent transformation metabolites of pyrene.

Table 7.
Proposed degradation pathways and the active genes during pyrene oxidation.
5. Future Perspectives and Challenges
The current techniques for the biodegradation of pyrene by Mycobacterium strains still need further investigation for future works:
- A knowledge gap between pyrene oxidation at the field site compared to laboratory conditions needs to be addressed for each product seeking commercial success.
- The degradation of pyrene by Mycobacterium strains generates many metabolites. Some of the metabolites and their bio-toxicity have been identified, while most of them need bio-toxicity assessment.
- The main biodegradation drawback is the limitation of the bioavailability of the target pollutant. Therefore, it is highly recommended to add a biosurfactant to increase the bioavailability.
- The literature revealed that the biodegradation of pyrene via consortium microbial gives a better result than a single strain. That is referred to diverse enzymes capable of oxidizing pyrene and its metabolites.
- There are several studies that applied successful synergetic biodegradation systems for pyrene degradation, such as biofuel cells and coupling of the advanced oxidation process and biodegradation system.
6. Conclusions
This article attempted to provide a review of pyrene bioremediation using Mycobacterium strains in various biodegradation mediums. This study’s findings are summarized as follows:
- Mycobacterium strains are efficient biological agents to degrade pyrene, that is, referring to their ability to produce many functional enzymes able to metabolite pyrene and its transformation molecules.
- Phenantharene-4,5-dicarboxylic acid, dihydroxy pyrene, phenanthrene-4-carboxylic acid, phthalic acid, and pyrene-4,5-dihydrodiol were the most frequent metabolites that were detected when Mycobacterium sp. strains were used for pyrene degradation.
- Some metabolites showed positive results for the Ames mutagenicity prediction test, such as 1,2-phenanthrenedicarboxylic acid, 1-hydroxypyrene, 4,5-dihydropyrene, 4-phenanthrene-carboxylic acid, 3,4-Dihydroxyphenanthrene, Monohydroxy pyrene, and 9,10-phenanthrenequinone. However, 4-phenanthrol showed positive results for experimental and prediction tests.
Author Contributions
Conceptualization, M.Q., S.A.M. and M.R.; writing—original draft preparation, M.Q.; writing—review and editing, S.A.M., A.M.A., H.S.A., M.A.H. and M.R.; supervision, S.A.M. and M.R.; funding acquisition, S.A.M. and M.R. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to express their appreciation to the Ministry of Higher Education Malaysia for the Fundamental Research Grant Scheme with Project Code: FRGS/1/2019/STG07/USM/02/12.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mekonnen, M.M.; Hoekstra, A.Y. Sustainability: Four Billion People Facing Severe Water Scarcity. Sci. Adv. 2016, 2, e1500323. [Google Scholar] [CrossRef] [PubMed]
- Qutob, M.; Rafatullah, M.; Qamar, M.; Alor, H.S.; Al Romaizan, A.N.; Hussein, M.A. A Review on Heterogeneous Oxidation of Acetaminophen Based on Micro and Nanoparticles Catalyzed by Di Ff Erent Activators. Nanotechnol. Rev. 2022, 11, 497–525. [Google Scholar] [CrossRef]
- Degefu, D.M.; Weijun, H.; Zaiyi, L.; Liang, Y.; Zhengwei, H.; Min, A. Mapping Monthly Water Scarcity in Global Transboundary Basins at Country-Basin Mesh Based Spatial Resolution. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Miraji, H.; Ripanda, A.; Moto, E. A Review on the Occurrences of Persistent Organic Pollutants in Corals, Sediments, Fish and Waters of the Western Indian Ocean. Egypt. J. Aquat. Res. 2021, 47, 373–379. [Google Scholar] [CrossRef]
- Gutierrez-Urbano, I.; Villen-Guzman, M.; Perez-Recuerda, R.; Rodriguez-Maroto, J.M. Removal of Polycyclic Aromatic Hydrocarbons (PAHs) in Conventional Drinking Water Treatment Processes. J. Contam. Hydrol. 2021, 243, 103888. [Google Scholar] [CrossRef]
- Dinç, B.; Çelebi, A.; Avaz, G.; Canlı, O.; Güzel, B.; Eren, B.; Yetis, U. Spatial Distribution and Source Identification of Persistent Organic Pollutants in the Sediments of the Yeşilırmak River and Coastal Area in the Black Sea. Mar. Pollut. Bull. 2021, 172, 112884. [Google Scholar] [CrossRef]
- IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. IARC Monogr. Eval. Carcinog. Risks Hum. 2010, 93, 9–38. [CrossRef]
- Hussain, K.; Hoque, R.R.; Balachandran, S.; Medhi, S.; Idris, M.G.; Rahman, M.; Hussain, F.L. Monitoring and Risk Analysis of PAHs in the Environment. In Handbook of Environmental Materials Management; Springer: New York, NY, USA, 2019; pp. 973–1007. [Google Scholar] [CrossRef]
- Yang, Y.; Woodward, L.A.; Li, Q.X.; Wang, J. Concentrations, Source and Risk Assessment of Polycyclic Aromatic Hydrocarbons in Soils from Midway Atoll, North Pacific Ocean. PLoS ONE 2014, 9, e86441. [Google Scholar] [CrossRef]
- EPA. National Recommended Water Quality Criteria—Human Health Criteria Table. Available online: https://www.epa.gov/wqc/national-recommended-water-quality-criteria-human-health-criteria-table (accessed on 24 May 2022).
- Nikolić, V.M.; Karić, S.D.; Nikolić, Ž.M.; Tošić, M.S.; Tasić, G.S.; Milovanovic, D.M.; Kaninski, M.P.M. Novel Photochemical Advanced Oxidation Process for the Removal of Polycyclic Aromatic Hydrocarbons from Polluted Concrete. Chem. Eng. J. 2017, 312, 99–105. [Google Scholar] [CrossRef]
- Oller, I.; Malato, S.; Sánchez-Pérez, J.A. Combination of Advanced Oxidation Processes and Biological Treatments for Wastewater Decontamination-A Review. Sci. Total Environ. 2011, 409, 4141–4166. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation Techniques–Classification Based on Site of Application: Principles, Advantages, Limitations and Prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef] [PubMed]
- Quintella, C.M.; Mata, A.M.T.; Lima, L.C.P. Overview of Bioremediation with Technology Assessment and Emphasis on Fungal Bioremediation of Oil Contaminated Soils. J. Environ. Manag. 2019, 241, 156–166. [Google Scholar] [CrossRef]
- Chen, S.; Peng, J.; Duan, G. Enrichment of Functional Microbes and Genes during Pyrene Degradation in Two Different Soils. J. Soils Sediments 2016, 16, 417–426. [Google Scholar] [CrossRef]
- Subashchandrabose, S.R.; Venkateswarlu, K.; Naidu, R.; Megharaj, M. Biodegradation of High-Molecular Weight PAHs by Rhodococcus Wratislaviensis Strain 9: Overexpression of Amidohydrolase Induced by Pyrene and BaP. Sci. Total Environ. 2019, 651, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.J.; Cai, C.; Qiao, M.; Li, H.; Zhu, Y.G. Dynamic Changes in Functional Gene Copy Numbers and Microbial Communities during Degradation of Pyrene in Soils. Environ. Pollut. 2010, 158, 2872–2879. [Google Scholar] [CrossRef]
- Kim, S.J.; Kweon, O.; Jones, R.C.; Freeman, J.P.; Edmondson, R.D.; Cerniglia, C.E. Complete and Integrated Pyrene Degradation Pathway in Mycobacterium Vanbaalenii PYR-1 Based on Systems Biology. J. Bacteriol. 2007, 189, 464–472. [Google Scholar] [CrossRef]
- Percival, S.L.; Williams, D.W. Mycobacterium, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar] [CrossRef]
- Heitkamp, M.A.; Franklin, W.; Cerniglia, C.E. Microbial Metabolism of Polycyclic Aromatic Hydrocarbons: Isolation and Characterization of a Pyrene-Degrading Bacterium. Appl. Environ. Microbiol. 1988, 54, 2549–2555. [Google Scholar] [CrossRef]
- Rehmann, K.; Noll, H.P.; Steinberg, C.E.W.; Kettrup, A.A. Pyrene Degradation by Mycobacterium sp. Strain KR2. Chemosphere 1998, 36, 2977–2992. [Google Scholar] [CrossRef]
- Zada, S.; Zhou, H.; Xie, J.; Hu, Z.; Ali, S.; Sajjad, W.; Wang, H. Bacterial Degradation of Pyrene: Biochemical Reactions and Mechanisms. Int. Biodeterior. Biodegrad. 2021, 162, 105233. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Biodegradation Aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Elyamine, A.M.; Kan, J.; Meng, S.; Tao, P.; Wang, H.; Hu, Z. Aerobic and Anaerobic Bacterial and Fungal Degradation of Pyrene: Mechanism Pathway Including Biochemical Reaction and Catabolic Genes. Int. J. Mol. Sci. 2021, 22, 8202. [Google Scholar] [CrossRef]
- Miller, C.D.; Hall, K.; Liang, Y.N.; Nieman, K.; Sorensen, D.; Issa, B.; Anderson, A.J.; Sims, R.C. Isolation and Characterization of Polycyclic Aromatic Hydrocarbon-Degrading Mycobacterium Isolates from Soil. Microb. Ecol. 2004, 48, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zhu, Q.; Wu, Y.; Chen, H.; Lin, X. Characterization of a Polycyclic Aromatic Ring-Hydroxylation Dioxygenase from Mycobacterium sp. NJS-P. Chemosphere 2017, 185, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Costa, K.C.; Moskatel, L.S.; Meirelles, L.A.; Newman, D.K. PhdA Catalyzes the First Step of Phenazine-1-Carboxylic Acid Degradation in Mycobacterium Fortuitum. J. Bacteriol. 2018, 200, 10. [Google Scholar] [CrossRef] [PubMed]
- Wanapaisan, P.; Laothamteep, N.; Vejarano, F.; Chakraborty, J.; Shintani, M.; Muangchinda, C.; Morita, T.; Suzuki-Minakuchi, C.; Inoue, K.; Nojiri, H.; et al. Synergistic Degradation of Pyrene by Five Culturable Bacteria in a Mangrove Sediment-Derived Bacterial Consortium. J. Hazard. Mater. 2018, 342, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Xie, X.; Wang, X.; Lin, L.; Yang, L.; Luan, T.; Chen, B. Transcriptional Response of Mycobacterium sp. Strain A1-PYR to Multiple Polycyclic Aromatic Hydrocarbon Contaminations. Environ. Pollut. 2018, 243, 824–832. [Google Scholar] [CrossRef]
- Habe, H.; Kanemitsu, M.; Nomura, M.; Takemura, T.; Iwata, K.; Nojiri, H.; Yamane, H.; Omori, T. Isolation and Characterization of an Alkaliphilic Bacterium Utilizing Pyrene as a Carbon Source. J. Biosci. Bioeng. 2004, 98, 306–308. [Google Scholar] [CrossRef]
- McLellan, S.L.; Warshawsky, D.; Shann, J.R. The Effect of Polycyclic Aromatic Hydrocarbons on the Degradation of Benzo[a]Pyrene by Mycobacterium sp. Strain RJGII-Environ. Toxicol. Chem. 2002, 21, 253–259. [Google Scholar] [CrossRef]
- Churchill, P.F.; Morgan, A.C.; Kitchens, E. Characterization of a Pyrene-Degrading Mycobacterium sp. Strain CH-2. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2008, 43, 698–706. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, K.; Lee, D.H.; Cha, C.J. Comparative Genomic Analysis of Pyrene-Degrading Mycobacterium Species: Genomic Islands and Ring-Hydroxylating Dioxygenases Involved in Pyrene Degradation. J. Microbiol. 2018, 56, 798–804. [Google Scholar] [CrossRef]
- Wang, Z.; Sheng, H.; Xiang, L.; Bian, Y.; Herzberger, A.; Cheng, H.; Jiang, Q.; Jiang, X.; Wang, F. Different Performance of Pyrene Biodegradation on Metal-Modified Montmorillonite: Role of Surface Metal Ions from a Bioelectrochemical Perspective. Sci. Total Environ. 2022, 805, 150324. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, F.; Zhu, X.; Zeng, J.; Zhao, Q.; Jiang, X. Extracellular Polymeric Substances Govern the Development of Biofilm and Mass Transfer of Polycyclic Aromatic Hydrocarbons for Improved Biodegradation. Bioresour. Technol. 2015, 193, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, H.; Chen, Y.; Lou, J.; Wu, L.; Xu, J. Salicylate and Phthalate Pathways Contributed Differently on Phenanthrene and Pyrene Degradations in Mycobacterium sp. WY10. J. Hazard. Mater. 2019, 364, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, H.; Yang, W.; Sheng, H.; Wang, F.; Harindintwali, J.D.; Herath, H.M.S.K.; Zhang, Y. Humic Acid Enhanced Pyrene Degradation by Mycobacterium sp. NJS-1. Chemosphere 2021, 288, 132613. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Guo, C.; Liu, S.; Liang, X.; Lu, G.; Dang, Z. Pyrene Degradation by Mycobacterium Gilvum: Metabolites and Proteins Involved. Water Air Soil Pollut. 2019, 230, 67. [Google Scholar] [CrossRef]
- Chen, S.C.; Duan, G.L.; Ding, K.; Huang, F.Y.; Zhu, Y.G. DNA Stable-Isotope Probing Identifies Uncultivated Members of Pseudonocardia Associated with Biodegradation of Pyrene in Agricultural Soil. FEMS Microbiol. Ecol. 2018, 94, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Terzaghi, E.; Posada-Baquero, R.; Di Guardo, A.; Ortega-Calvo, J.J. Microbial Degradation of Pyrene in Holm Oak (Quercus Ilex) Phyllosphere: Role of Particulate Matter in Regulating Bioaccessibility. Sci. Total Environ. 2021, 786, 147431. [Google Scholar] [CrossRef]
- Sarma, S.J.; Pakshirajan, K. Surfactant Aided Biodegradation of Pyrene Using Immobilized Cells of Mycobacterium Frederiksbergense. Int. Biodeterior. Biodegrad. 2011, 65, 73–77. [Google Scholar] [CrossRef]
- Mahanty, B.; Pakshirajan, K.; Dasu, V.V. A Two Liquid Phase Partitioning Bioreactor System for the Biodegradation of Pyrene: Comparative Evaluation and Cost-Benefit Analysis. J. Chem. Technol. Biotechnol. 2010, 85, 349–355. [Google Scholar] [CrossRef]
- Dean-Ross, D.; Cerniglia, C.E. Degradation of Pyrene by Mycobacterium Flavescens. Appl. Microbiol. Biotechnol. 1996, 46, 307–312. [Google Scholar] [CrossRef]
- Vila, J.; López, Z.; Sabaté, J.; Minguillón, C.; Solanas, A.M.; Grifoll, M. Identification of a Novel Metabolite in the Degradation of Pyrene by Mycobacterium sp. Strain AP1: Actions of the Isolate on Two- and Three-Ring Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2001, 67, 5497–5505. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Luan, T.; Zhou, H.; Lan, C.; Tam, N.F.Y. Metabolite Production in Degradation of Pyrene Alone or in a Mixture with Another Polycyclic Aromatic Hydrocarbon by Mycobacterium sp. Environ. Toxicol. Chem. 2006, 25, 2853–2859. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Chen, B.; Lin, L.; Wang, X.; Tam, N.F.Y.; Luan, T. Pyrene Degradation Accelerated by Constructed Consortium of Bacterium and Microalga: Effects of Degradation Products on the Microalgal Growth. Environ. Sci. Technol. 2014, 48, 13917–13924. [Google Scholar] [CrossRef]
- Zhong, Y.; Luan, T.; Lin, L.; Liu, H.; Tam, N.F.Y. Production of Metabolites in the Biodegradation of Phenanthrene, Fluoranthene and Pyrene by the Mixed Culture of Mycobacterium sp. and Sphingomonas sp. Bioresour. Technol. 2011, 102, 2965–2972. [Google Scholar] [CrossRef]
- Lease, C.W.M.; Bentham, R.H.; Gaskin, S.E.; Juhasz, A.L. Isolation and Identification of Pyrene Mineralizing Mycobacterium spp. from Contaminated and Uncontaminated Sources. Appl. Environ. Soil Sci. 2011, 2011, 1–11. [Google Scholar] [CrossRef][Green Version]
- Schneider, J.; Grosser, R.; Jayasimhulu, K.; Xue, W.; Warshawsky, D. Degradation of Pyrene, Benz[a]Anthracene, and Benzo[a]Pyrene by Mycobacterium sp. Strain RJGII-135, Isolated from a Former Coal Gasification Site. Appl. Environ. Microbiol. 1996, 62, 13–19. [Google Scholar] [CrossRef]
- Fan, R.; Tian, H.; Wu, Q.; Yi, Y.; Yan, X.; Liu, B. Mechanism of Bio-Electrokinetic Remediation of Pyrene Contaminated Soil: Effects of an Electric Field on the Degradation Pathway and Microbial Metabolic Processes. J. Hazard. Mater. 2022, 422, 126959. [Google Scholar] [CrossRef]
- Lu, H.; Sun, J.; Zhu, L. The Role of Artificial Root Exudate Components in Facilitating the Degradation of Pyrene in Soil. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Wang, S.; Li, X.; Liu, W.; Li, P.; Kong, L.; Ren, W.; Wu, H.; Tu, Y. Degradation of Pyrene by Immobilized Microorganisms in Saline-Alkaline Soil. J. Environ. Sci. 2012, 24, 1662–1669. [Google Scholar] [CrossRef]
- Deng, F.; Zhang, Z.; Yang, C.; Guo, C.; Lu, G.; Dang, Z. Pyrene Biodegradation with Layer-by-Layer Assembly Bio-Microcapsules. Ecotoxicol. Environ. Saf. 2017, 138, 9–15. [Google Scholar] [CrossRef]
- Toyama, T.; Furukawa, T.; Maeda, N.; Inoue, D.; Sei, K.; Mori, K.; Kikuchi, S.; Ike, M. Accelerated Biodegradation of Pyrene and Benzo[a]Pyrene in the Phragmites Australis Rhizosphere by Bacteria-Root Exudate Interactions. Water Res. 2011, 45, 1629–1638. [Google Scholar] [CrossRef]
- Mahanty, B.; Pakshirajan, K.; Venkata Dasu, V. Biodegradation of Pyrene by Mycobacterium Frederiksbergense in a Two-Phase Partitioning Bioreactor System. Bioresour. Technol. 2008, 99, 2694–2698. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Calvo, J.J.; Gschwend, P.M. Influence of Low Oxygen Tensions and Sorption to Sediment Black Carbon on Biodegradation of Pyrene. Appl. Environ. Microbiol. 2010, 76, 4430–4437. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Liao, C.; Yang, C.; Guo, C.; Dang, Z. Enhanced Biodegradation of Pyrene by Immobilized Bacteria on Modified Biomass Materials. Int. Biodeterior. Biodegrad. 2016, 110, 46–52. [Google Scholar] [CrossRef]
- Chang, B.V.; Chang, I.T.; Yuan, S.Y. Biodegradation of Phenanthrene and Pyrene from Mangrove Sediment in Subtropical Taiwan. J. Environ. Sci. Health Part A Toxic/Hazardous Subst. Environ. Eng. 2008, 43, 233–238. [Google Scholar] [CrossRef]
- Zeng, J.; Lin, X.; Zhang, J.; Li, X. Isolation of Polycyclic Aromatic Hydrocarbons (PAHs)-Degrading Mycobacterium Spp. and the Degradation in Soil. J. Hazard. Mater. 2010, 183, 718–723. [Google Scholar] [CrossRef]
- Jacques, R.J.S.; Okeke, B.C.; Bento, F.M.; Teixeira, A.S.; Peralba, M.C.R.; Camargo, F.A.O. Microbial Consortium Bioaugmentation of a Polycyclic Aromatic Hydrocarbons Contaminated Soil. Bioresour. Technol. 2008, 99, 2637–2643. [Google Scholar] [CrossRef]
- Cerniglia, C.E. Degradation Of A Mixture Of High-Molecular-Weight Polycyclic Aromatic Hydrocarbons By A Mycobacterium Strain Pyr-1. J. Soil Contam. 1995, 4, 77–91. [Google Scholar] [CrossRef]
- Sho, M.; Hamel, C.; Greer, C.W. Two Distinct Gene Clusters Encode Pyrene Degradation in Mycobacterium sp. Strain S65. FEMS Microbiol. Ecol. 2004, 48, 209–220. [Google Scholar] [CrossRef]
- López, Z.; Vila, J.; Ortega-Calvo, J.J.; Grifoll, M. Simultaneous Biodegradation of Creosote-Polycyclic Aromatic Hydrocarbons by a Pyrene-Degrading Mycobacterium. Appl. Microbiol. Biotechnol. 2008, 78, 165–172. [Google Scholar] [CrossRef][Green Version]
- Child, R.; Miller, C.D.; Liang, Y.; Sims, R.C.; Anderson, A.J. Pyrene Mineralization by Mycobacterium sp. Strain KMS in a Barley Rhizosphere. J. Environ. Qual. 2007, 36, 1260–1265. [Google Scholar] [CrossRef] [PubMed]
- Badejo, A.C.; Badejo, A.O.; Shin, K.H.; Chai, Y.G. A Gene Expression Study of the Activities of Aromatic Ring-Cleavage Dioxygenases in Mycobacterium Gilvum PYR-GCK to Changes in Salinity and PH during Pyrene Degradation. PLoS ONE 2013, 8, e58066. [Google Scholar] [CrossRef]
- Ramirez, N.; Cutright, T.; Ju, L.K. Pyrene Biodegradatin in Aqueous Solutions and Soil Slurries by Mycobacteriurn PYR-1 and Enriched Consortium. Chemosphere 2001, 44, 1079–1086. [Google Scholar] [CrossRef]
- Vila, J.; Grifoll, M. Actions of Mycobacterium sp. Strain AP1 on the Saturated- and Aromatic-Hydrocarbon Fractions of Fuel Oil in a Marine Medium. Appl. Environ. Microbiol. 2009, 75, 6232–6239. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, I.Y.; Bartha, R. Solvent-Augmented Mineralization of Pyrene by a Mycobacterium sp. Appl. Environ. Microbiol. 1996, 62, 2311–2316. [Google Scholar] [CrossRef]
- Guo, C.; Dang, Z.; Wong, Y.; Tam, N.F. Biodegradation Ability and Dioxgenase Genes of PAH-Degrading Sphingomonas and Mycobacterium Strains Isolated from Mangrove Sediments. Int. Biodeterior. Biodegrad. 2010, 64, 419–426. [Google Scholar] [CrossRef]
- Seo, J.S.; Keum, Y.S.; Kim, K.; Li, Q.X. Degradation of Pyrene by Mycobacterium Aromativorans Strain JS19b1. J. Appl. Biol. Chem. 2010, 53, 323–329. [Google Scholar] [CrossRef]
- Kim, Y.H.; Freeman, J.P.; Moody, J.D.; Engesser, K.H.; Cerniglia, C.E. Effects of PH on the Degradation of Phenanthrene and Pyrene by Mycobacterium Vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 2005, 67, 275–285. [Google Scholar] [CrossRef]
- Liang, Y.; Gardner, D.R.; Miller, C.D.; Chen, D.; Anderson, A.J.; Weimer, B.C.; Sims, R.C. Study of Biochemical Pathways and Enzymes Involved in Pyrene Degradation by Mycobacterium sp. Strain KMS. Appl. Environ. Microbiol. 2006, 72, 7821–7828. [Google Scholar] [CrossRef]
- Sarma, P.M.; Duraja, P.; Deshpande, S.; Lal, B. Degradation of Pyrene by an Enteric Bacterium, Leclercia Adecarboxylata PS4040. Biodegradation 2010, 21, 59–69. [Google Scholar] [CrossRef]
- Zheng, T.; Liu, R.; Chen, J.; Gu, X.; Wang, J.; Li, L.; Hou, L.; Li, N.; Wang, Y. Fire Phoenix Plant Mediated Microbial Degradation of Pyrene: Increased Expression of Functional Genes and Diminishing of Degraded Products. Chem. Eng. J. 2021, 407, 126343. [Google Scholar] [CrossRef]
- Agrawal, N.; Shahi, S.K. Degradation of Polycyclic Aromatic Hydrocarbon (Pyrene) Using Novel Fungal Strain Coriolopsis Byrsina Strain APC5. Int. Biodeterior. Biodegrad. 2017, 122, 69–81. [Google Scholar] [CrossRef]
- Lu, J.; Guo, C.; Zhang, M.; Lu, G.; Dang, Z. Biodegradation of Single Pyrene and Mixtures of Pyrene by a Fusant Bacterial Strain F14. Int. Biodeterior. Biodegrad. 2014, 87, 75–80. [Google Scholar] [CrossRef]
- Al Farraj, D.A.; Hadibarata, T.; Yuniarto, A.; Syafiuddin, A.; Surtikanti, H.K.; Elshikh, M.S.; Al Khulaifi, M.M.; Al-Kufaidy, R. Characterization of Pyrene and Chrysene Degradation by Halophilic Hortaea sp. B15. Bioprocess Biosyst. Eng. 2019, 42, 963–969. [Google Scholar] [CrossRef]
- Ma, J.; Xu, L.; Jia, L. Characterization of Pyrene Degradation by Pseudomonas sp. Strain Jpyr-1 Isolated from Active Sewage Sludge. Bioresour. Technol. 2013, 140, 15–21. [Google Scholar] [CrossRef]
- Rathour, R.; Gupta, J.; Tyagi, B.; Kumari, T.; Thakur, I.S. Biodegradation of Pyrene in Soil Microcosm by Shewanella sp. ISTPL2, a Psychrophilic, Alkalophilic and Halophilic Bacterium. Bioresour. Technol. Rep. 2018, 4, 129–136. [Google Scholar] [CrossRef]
- Swati; Ghosh, P.; Thakur, I.S. Biodegradation of Pyrene by Pseudomonas sp. ISTPY2 Isolated from Landfill Soil: Process Optimisation Using Box-Behnken Design Model. Bioresour. Technol. Rep. 2019, 8, 100329. [Google Scholar] [CrossRef]
- Swati; Kumari, M.; Ghosh, P.; Thakur, I.S. Evaluation of a Biosurfactant Producing Bacterial Strain Pseudomonas sp. ISTPY2 for Efficient Pyrene Degradation and Landfill Soil Bioremediation through Soil Microcosm and Proteomic Studies. Bioresour. Technol. Rep. 2020, 12, 100607. [Google Scholar] [CrossRef]
- Jin, J.; Yao, J.; Zhang, Q.; Liu, J. Biodegradation of Pyrene by Pseudomonas sp. JPN2 and Its Initial Degrading Mechanism Study by Combining the Catabolic NahAc Gene and Structure-Based Analyses. Chemosphere 2016, 164, 379–386. [Google Scholar] [CrossRef]
- Fernández-López, C.; Posada-Baquero, R.; García, J.L.; Castilla-Alcantara, J.C.; Cantos, M.; Ortega-Calvo, J.J. Root-Mediated Bacterial Accessibility and Cometabolism of Pyrene in Soil. Sci. Total Environ. 2021, 760, 143408. [Google Scholar] [CrossRef]
- Kashyap, N.; Moholkar, V.S. Intensification of Pyrene Degradation by Native Candida Tropicalis MTCC 184 with Sonication: Kinetic and Mechanistic Investigation. Chem. Eng. Process. Process Intensif. 2021, 164, 108415. [Google Scholar] [CrossRef]
- Ghosh, I.; Jasmine, J.; Mukherji, S. Biodegradation of Pyrene by a Pseudomonas Aeruginosa Strain RS1 Isolated from Refinery Sludge. Bioresour. Technol. 2014, 166, 548–558. [Google Scholar] [CrossRef]
- Nzila, A.; Ramirez, C.O.; Musa, M.M.; Sankara, S.; Basheer, C.; Li, Q.X. Pyrene Biodegradation and Proteomic Analysis in Achromobacter Xylosoxidans, PY4 Strain. Int. Biodeterior. Biodegrad. 2018, 130, 40–47. [Google Scholar] [CrossRef]
- Umar, Z.D.; Nor Azwady, A.A.; Zulkifli, S.Z.; Muskhazli, M. Effective Phenanthrene and Pyrene Biodegradation Using Enterobacter sp. MM087 (KT933254) Isolated from Used Engine Oil Contaminated Soil. Egypt. J. Pet. 2018, 27, 349–359. [Google Scholar] [CrossRef]
- Ghosh, I.; Mukherji, S. Substrate Interaction Effects during Pyrene Biodegradation by Pseudomonas Aeruginosa RS1. J. Environ. Chem. Eng. 2017, 5, 1791–1800. [Google Scholar] [CrossRef]
- Gupta, B.; Puri, S.; Thakur, I.S.; Kaur, J. Enhanced Pyrene Degradation by a Biosurfactant Producing Acinetobacter Baumannii BJ5: Growth Kinetics, Toxicity and Substrate Inhibition Studies. Environ. Technol. Innov. 2020, 19, 100804. [Google Scholar] [CrossRef]
- Jiang, B.; Chen, Y.; Xing, Y.; Lian, L.; Shen, Y.; Zhang, B.; Zhang, H.; Sun, G.; Li, J.; Wang, X.; et al. Negative Correlations between Cultivable and Active-yet-Uncultivable Pyrene Degraders Explain the Postponed Bioaugmentation. J. Hazard. Mater. 2022, 423, 127189. [Google Scholar] [CrossRef]
- Yang, W.; Hadibarata, T.; Mahmoud, A.H.; Yuniarto, A. Biotransformation of Pyrene in Soil in the Presence of Earthworm Eisenia Fetida. Environ. Technol. Innov. 2020, 18, 100701. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Li, L.; Lin, C.; Dong, W.; Shen, W.; Yong, X.; Jia, H.; Wu, X.; Zhou, J. Anaerobic Biodegradation of Pyrene by Klebsiella sp. LZ6 and Its Proposed Metabolic Pathway. Environ. Technol. 2020, 41, 2130–2139. [Google Scholar] [CrossRef]
- Giménez, B.N.; Conte, L.O.; Alfano, O.M.; Schenone, A.V. Paracetamol Removal by Photo-Fenton Processes at near-Neutral PH Using a Solar Simulator: Optimization by D-Optimal Experimental Design and Toxicity Evaluation. J. Photochem. Photobiol. A Chem. 2020, 397, 112584. [Google Scholar] [CrossRef]
- Dabrowska, D.; Kot-Wasik, A.; Namieśnik, J. Pathways and Analytical Tools in Degradation Studies of Organic Pollutants. Crit. Rev. Anal. Chem. 2005, 35, 155–176. [Google Scholar] [CrossRef]
- Krivobok, S.; Kuony, S.; Meyer, C.; Louwagie, M.; Willison, J.C.; Jouanneau, Y. Identification of Pyrene-Induced Proteins in Mycobacterium sp. Strain 6PY1: Evidence for Two Ring-Hydroxylating Dioxygenases. J. Bacteriol. 2003, 185, 3828–3841. [Google Scholar] [CrossRef]
- Maruyama, T.; Ishikura, M.; Taki, H.; Shindo, K.; Kasai, H.; Haga, M.; Inomata, Y.; Misawa, N. Isolation and Characterization of O-Xylene Oxygenase Genes from Rhodococcus Opacus TKN14. Appl. Environ. Microbiol. 2005, 71, 7705–7715. [Google Scholar] [CrossRef] [PubMed]
- Pagnout, C.; Frache, G.; Poupin, P.; Maunit, B.; Muller, J.F.; Férard, J.F. Isolation and Characterization of a Gene Cluster Involved in PAH Degradation in Mycobacterium sp. Strain SNP11: Expression in Mycobacterium Smegmatis Mc2155. Res. Microbiol. 2007, 158, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kweon, O.; Freeman, J.P.; Jones, R.C.; Adjei, M.D.; Jhoo, J.W.; Edmondson, R.D.; Cerniglia, C.E. Molecular Cloning and Expression of Genes Encoding a Novel Dioxygenase Involved in Low- and High-Molecular-Weight Polycyclic Aromatic Hydrocarbon Degradation in Mycobacterium Vanbaalenii PYR-1. Appl. Environ. Microbiol. 2006, 72, 1045–1054. [Google Scholar] [CrossRef]
- Adamou, J.E.; Heinrichs, J.H.; Erwin, A.L.; Walsh, W.; Gayle, T.; Dormitzer, M.; Dagan, R.; Brewah, Y.A.; Barren, P.; Lathigra, R.; et al. Identification and Characterization of a Novel Family of Pneumococcal Proteins That Are Protective against Sepsis. Infect. Immun. 2001, 69, 949–958. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, Y.; Zhou, N. A Newly Defined Dioxygenase System from Mycobacterium Vanbaalenii PYR-1 Endowed with an Enhanced Activity of Dihydroxylation of High-Molecular-Weight Polyaromatic Hydrocarbons. Front. Environ. Sci. Eng. 2020, 14, 1–11. [Google Scholar] [CrossRef]
- Levieux, J.A.; Medellin, B.; Johnson, W.H.; Erwin, K.; Li, W.; Johnson, I.A.; Zhang, Y.J.; Whitman, C.P. Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily. Biochemistry 2018, 57, 3524–3536. [Google Scholar] [CrossRef]
- Kumari, S.; Regar, R.K.; Bajaj, A.; Ch, R.; Satyanarayana, G.N.V.; Mudiam, M.K.R.; Manickam, N. Simultaneous Biodegradation of Polyaromatic Hydrocarbons by a Stenotrophomonas Sp: Characterization of Nid Genes and Effect of Surfactants on Degradation. Indian J. Microbiol. 2017, 57, 60–67. [Google Scholar] [CrossRef]
- Saito, A.; Iwabuchi, T.; Harayama, S. A Novel Phenanthrene Dioxygenase from Nocardioides sp. Strain KP7: Expression in Escherichia Coli. J. Bacteriol. 2000, 182, 2134–2141. [Google Scholar] [CrossRef]
- Walter, U.; Beyer, M.; Klein, J.; Rehm, H. Appeal Microbiology Biote Nology Degradation of Pyrene by Rhodococcus sp. UW1. Power 1991, 671–676. [Google Scholar]
- Sakshi; Singh, S.K.; Haritash, A.K. Catabolic Enzyme Activity and Kinetics of Pyrene Degradation by Novel Bacterial Strains Isolated from Contaminated Soil. Environ. Technol. Innov. 2021, 23, 101744. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, S.; Xie, J.; Wei, H.; Hu, Z.; Wang, H. Pyrene Biodegradation and Its Potential Pathway Involving Roseobacter Clade Bacteria. Int. Biodeterior. Biodegrad. 2020, 150, 104961. [Google Scholar] [CrossRef]
- Khanna, G.P.; Goyal, D.; Khanna, S. Pyrene Biodegradation by Bacillus Spp. Isolated from Coal Tar-Contaminated Soil. Bioremediat. J. 2011, 15, 12–25. [Google Scholar] [CrossRef]
- Vasconcelos, M.R.S.; Vieira, G.A.L.; Otero, I.V.R.; Bonugli-Santos, R.C.; Rodrigues, M.V.N.; Rehder, V.L.G.; Ferro, M.; Boaventura, S.; Bacci, M.; Sette, L.D. Pyrene Degradation by Marine-Derived Ascomycete: Process Optimization, Toxicity, and Metabolic Analyses. Environ. Sci. Pollut. Res. 2019, 26, 12412–12424. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).