Abstract
Background. Currently, most of the research on breast cancer has been carried out in conventional two-dimensional (2D) cell cultures due to its practical benefits, however, the three-dimensional (3D) cell culture is becoming the model of choice in cancer research because it allows cell–cell and cell–extracellular matrix (ECM) interactions, mimicking the native microenvironment of tumors in vivo. Methods. In this work, we evaluated the effect of 3D cell organization on the expression pattern of miRNAs (by Small-RNAseq) and mRNAs (by microarrays) in the breast cancer SKBR3 cell line and analyzed the biological processes and signaling pathways regulated by the differentially expressed protein-coding genes (DE-mRNAs) and miRNAs (DE-microRNAs) found in the organoids. Results. We obtained well-defined cell-aggregated organoids with a grape cluster-like morphology with a size up to 9.2 × 105 μm3. The transcriptomic assays showed that cell growth in organoids significantly affected (all p < 0.01) the gene expression patterns of both miRNAs, and mRNAs, finding 20 upregulated and 19 downregulated DE-microRNAs, as well as 49 upregulated and 123 downregulated DE-mRNAs. In silico analysis showed that a subset of 11 upregulated DE-microRNAs target 70 downregulated DE-mRNAs. These genes are involved in 150 gene ontology (GO) biological processes such as regulation of cell morphogenesis, regulation of cell shape, regulation of canonical Wnt signaling pathway, morphogenesis of epithelium, regulation of cytoskeleton organization, as well as in the MAPK and AGE–RAGE signaling KEGG-pathways. Interestingly, hsa-mir-122-5p (Fold Change (FC) = 15.4), hsa-mir-369-3p (FC = 11.4), and hsa-mir-10b-5p (FC = 20.1) regulated up to 81% of the 70 downregulated DE-mRNAs. Conclusion. The organotypic 3D cell-organization architecture of breast cancer SKBR3 cells impacts the expression pattern of the miRNAs–mRNAs network mainly through overexpression of hsa-mir-122-5p, hsa-mir-369-3p, and hsa-mir-10b-5p. All these findings suggest that the interaction between cell–cell and cell–ECM as well as the change in the culture architecture impacts gene expression, and, therefore, support the pertinence of migrating breast cancer research from conventional cultures to 3D models.
1. Introduction
Breast cancer is the most common cancer around the world, affecting 2.26 million individuals and placing it in fifth place worldwide in 2020 [1]. In addition, Siegel et al. estimate that in the United States alone the number of new cases of breast cancer will be 300,590 and the estimated number of deaths will be 43,700 in 2023 [2]. Breast cancer exhibits a high degree of heterogeneity, encompassing distinct genotypic, phenotypic, and anatomical characteristics that exert profound influences on the clinical outcomes of patients [3]. Although the histological and molecular classifications of breast cancer may appear simplistic, they can predict biological behavior and facilitate the development of targeted therapeutic approaches [4]. Approximately 84 breast cancer cell lines have been categorized based on the presence of four major molecular subtypes: human epidermal growth factor receptor 2 (HER2)-enriched, basal-like, luminal A, and luminal B [5]. HER2-positive (HER2+) and triple-negative breast cancers (TNBC) exhibit the most unfavorable prognosis, primarily attributable to the intrinsic resistance or acquired insensitivity of certain HER2+ tumors to anti-HER2 therapies, as well as the unresponsiveness of TNBC to hormonal therapies or agents targeting HER2 receptors [6]. Consequently, extensive research efforts have been dedicated to the development of innovative therapeutic strategies. These approaches focus on either employing diverse combinations of drugs and treatment regimens that target multiple receptors or exploiting compensatory and downstream crosstalk signaling pathways associated with HER2 [7]. Interestingly, a large part of the available research on the HER2+ subtype has been performed using the SKRB3 cell line mostly cultured in monolayer with a conventional two-dimensional (2D) cell culture, this is due to its practical benefits and a huge amount of substantial information has been obtained until now; however, 2D culture has been gradually replaced, since cells grow up in a monolayer on a flat solid surface, lacking cell–cell and cell–extracellular matrix (ECM) interactions that can be found in native tumors, and acquiring artificial polarity, which may cause aberrant gene expression [8,9,10,11].
By contrast, 3D cell culture allows cancer cells to interact among themselves and with the ECM, closely mimicking the native microenvironment of tumors in vivo [12,13]. This results in the acquisition of morphological and cellular characteristics similar to the tumors in vivo [14], as well as the activation of cell signaling pathways leading to changes in expression of protein-coding genes (mRNAs) and non-coding genes such as microRNAs (miRNAs) [15,16]. For instance, lower cell proliferation, higher resistance to docetaxel and paclitaxel as well as changes in gene expression were shown in 3D cell culture compared with 2D in PC3, LNCaP, and DU145 prostate tumor cell lines [17]; moreover, in a report using these same cell lines, it was shown that integrin-mediated cell–ECM interactions can modulate tumor cell morphology as well as the expression of chemokine receptors which are associated with the invasive phenotype and progression of prostate cancer [18]. In a report, spheroids from the laryngeal carcinoma HLaC78 cell line showed upregulation of genes involved in cell adhesion and cell junctions, and downregulation of genes controlling cell cycle, DNA-replication, and DNA mismatch repair [19].
MicroRNAs are small RNA molecules of approximately 22 nucleotides that participate in the post-transcriptional downregulation of gene expression through interaction with the 3′ untranslated regions (3′UTR) of their target mRNA [20], resulting in mRNA degradation or translational repression into cytoplasmic P-bodies [21]. Nowadays, miRNAs have an important role in cancer since have been implicated in the initiation and progression of cancers as well as in chemotherapy resistance [21,22,23,24]. However, most of the studies reported on the expression of RNAs that code or do not code for proteins in cancer research have been derived from studies carried out in 2D cell cultures [25]. There are few studies of 3D cell culture-based global miRNA expression analysis of different types of cancer such as colorectal cancer [26], and basal and luminal breast cancer cells [27], as such, it is necessary to study the change of gene and miRNAs expression patterns in a 3D context to a better understanding of the cell signaling pathways of the HER2+ breast cancer. Therefore, in this study, a comparative analysis was conducted between the 3D culture and the conventional two-dimensional (2D) culture of HER2+ breast cancer SKRB3 cells to comprehensively assess the consequences of pattern changes in miRNA and mRNA expression on the putative cellular signaling pathways and associated biological processes. In our study, we obtained well-defined cell-aggregated organoids with a grape cluster-like morphology. In this discovery phase, the transcriptomic assays showed that cell growth in organoids significantly affected the expression patterns of miRNAs and mRNAs. In silico analysis showed that a subset of 11 upregulated DE-microRNAs target 70 downregulated DE-mRNAs involved in the regulation of cell morphogenesis, the regulation of cell shape, the regulation of the canonical Wnt signaling pathway, morphogenesis of the epithelium, and the regulation of cytoskeleton organization, as well as in the MAPK and AGE–RAGE signaling KEGG-pathways. The miRNA–mRNA interaction analysis showed that hsa-mir-122-5p, hsa-mir-369-3p, and hsa-mir-10b-5p are responsible for downregulating 81% of the 70 downregulated DE-mRNAs.
2. Results
2.1. Organotypic 3D On-Top Cultures of SKBR3 form Grape Cluster-like Organoids
We tested the Geltrex On-Top and Embedded cultures to obtain SKRB3 organoids with cell–cell and cell–ECM interactions and used ultra-low attachment (ULA) as aggrupation control (Figure 1A). We found that, at 120 h of incubation, the organoids of the Embedded culture showed no significant differences in size, but were more numerous (p = 0.010), in comparison with those of ULA (Figure 1B,C). In contrast, the On-Top culture generated well-defined organoids with the biggest size and numbers (p < 0.001), and, therefore, was selected for the transcriptome analysis.
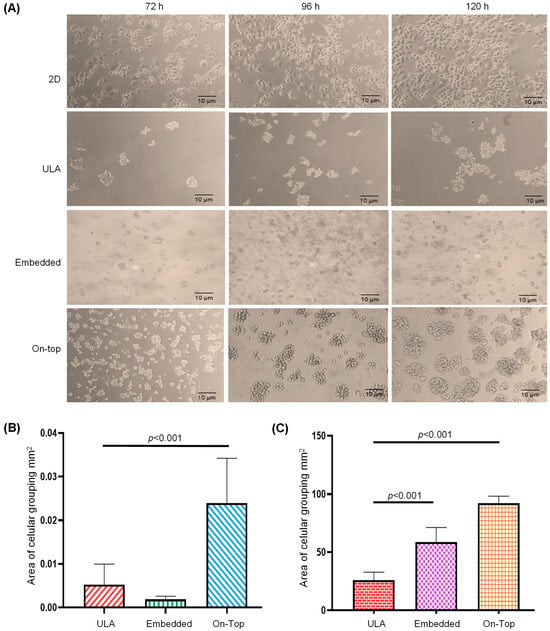
Figure 1.
Formation and analysis of SKBR3 breast cancer cell organoids. Cell line was incubated for 72, 96, and 120 h under conventional 2D culture, ultra-low attachment (ULA), or with Geltrex in a 3D Embedded and On-Top culture (A). The area (mm2) (B) and number (C) of cell-aggregates were determined in 2D, ultra-low attachment (ULA), 3D Embedded, and On-Top cultures.
The morphology of SKBR3 cells in conventional 2D culture has the typical epithelial shape, with a wide cytoplasm and well-organized actin fibers, observing lamellipodia and filopodia structures (Figure 2A,B). By contrast, in the 3D On-Top culture, cells had a rounded morphology with little cytoplasm, forming grape cluster-like organoids evidencing weak cell–cell contact (Figure 2C,D), and were up to 150 μm in size. Interestingly, discrete extensions of the cytoplasmic membrane in SKRB3 cells were observed, establishing direct connections with neighboring spheroid cells reminiscent of the tunneled nanotubes documented in prior studies involving 3D-cultured cancer cells [28]. These observations imply the existence of intricate cellular intercommunication mechanisms in 3D structures. In addition, the reconstruction allowed us to determine its size, reaching about 9.2 × 105 μm3 (Figure 3A, Supplementary Video File S1). Based on previously established classifications, SKBR3 organoids fall into the category of loosely aggregated cell clusters due to their low level of compaction [29]. The 3D reconstruction of the organoid shows similar zones to those identified by Kelly et al. [30], which consist of a core, a quiescent zone, and a proliferation zone (as shown in Figure 3B). However, further investigation is necessary to confirm whether these zones are indeed necrotic, quiescent, or proliferative.
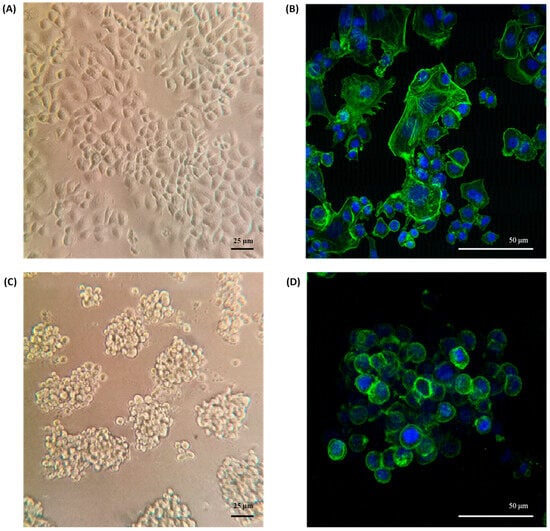
Figure 2.
Morphology of SKBR3 organoids. (A) Conventional 2D culture on the plastic adherent surface observed by phase contrast microscopy at 200× magnification and (B) Confocal with maximum projection at 400× magnification. (C) Morphology of organoids grown on Geltrex by phase contrast and (D) confocal, labeling f-actin green and DAPI blue.
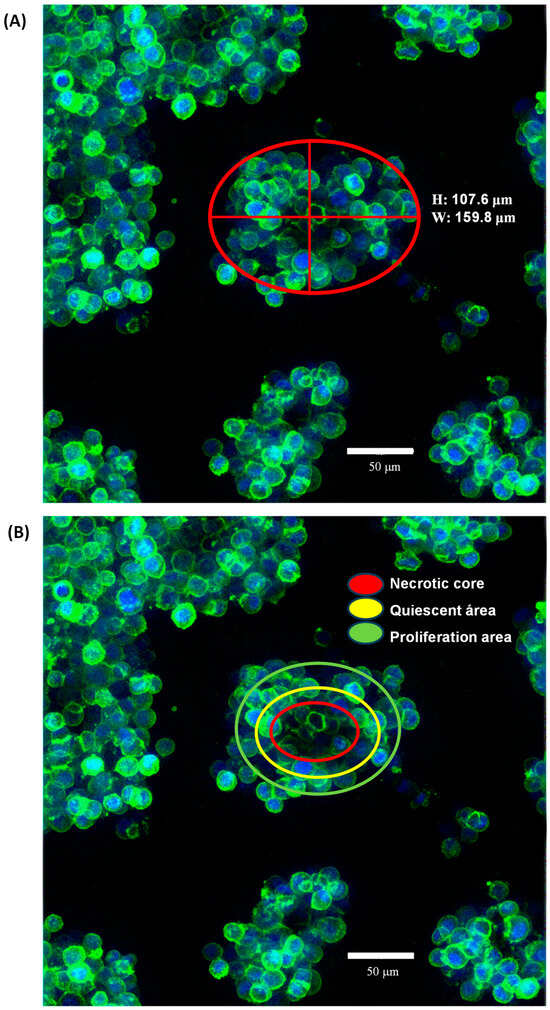
Figure 3.
The morphology of SK-BR-3 organoids cultured using the On-Top method was analyzed at 120 h of cultivation through staining with dyes which enabled visualization of the actin cytoskeleton (phalloidin) and nuclei present in each organoid (DAPI) using a confocal microscope at a 40× objective. Based on previously established classifications by various authors, SK-BR-3 organoids fall into the category of loosely aggregated cell clusters due to their low level of compaction, with a morphology resembling clusters of grapes due to their poor cell–cell interaction (A). Nevertheless, the 3D reconstruction resembled the very well-established zones of the organoid that could be a necrotic core (red), a quiescent area (yellow), and a proliferation area (green). However, further investigation is necessary to confirm whether these zones are indeed necrotic, quiescent, or proliferative (B).
2.2. 3D Culture Induces Important Changes in Small RNAs Expression in SKBR3 Cells
RNAseq analysis found different types of small RNAs present in the samples, 79.4% were identified as known miRNAs, 2.16% as novel miRNAs, 17.6% as piRNAs, 1.49% as snoRNAs, and 2.36% as tRNAs; therefore, we selected only the known miRNAs for further analysis. There were about 1320 known miRNAs in the samples (1359–1367 for 2D culture and 1322–1396 for 3D culture), of which 39 were differentially expressed (DE-microRNAs) with the established criteria (p < 0.01), 20 upregulated, and 19 downregulated (Table 1 and Figure 4A), which demonstrates that the cellular organization present in organoids, together with cell–ECM interactions, have a remarkable effect on gene expression.

Table 1.
Differentially expressed up- and downregulated miRNAs (DE-microRNAs) found in SKBR3 organoids, in comparison to 2D culture.
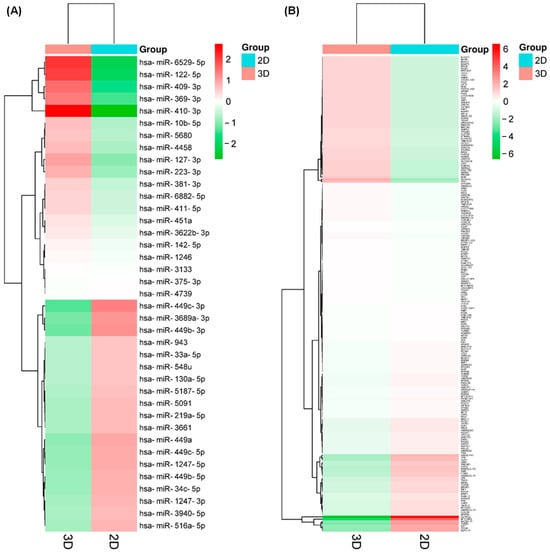
Figure 4.
Differentially expressed protein-coding genes (DE-mRNAs) and miRNAs (DE-microRNAs) found in the organoids. Unsupervised hierarchical clustering analysis displaying the differential expression of DE-microRNAs (A) and DE-mRNAs (B) in both 2D and 3D cultures (Euclidean distance). Each column represents an individual sample, and each row represents a different miRNA. Red represents upregulation levels meanwhile green represents downregulation levels.
Among the DE-microRNAs, those which were most upregulated are hsa-miR-410-3p (FC = 20.1), hsa-miR-6529-5p (FC = 15.9), hsa-miR-122-5p (FC = 15.4), hsa-miR-409-3p (FC = 12.2), and hsa-miR-369-3p (FC = 11.4). Among the DE-microRNAs, those which were most downregulated are hsa-miR-449c-3p (FC = −6.9), hsa-miR-449b-3p (FC = −5.7), hsa-miR-3689a-3p (FC = −5.3), hsa-miR-449a (FC = −4.2), and hsa-miR-1247-5p (FC = −3.9). We found that a set of 14 upregulated DE-microRNAs matched with 4370 experimentally validated microRNA-target interactions (Figure 5), while a set of 15 downregulated DE-microRNAs matched with 2547 gene targets (Figure 6).
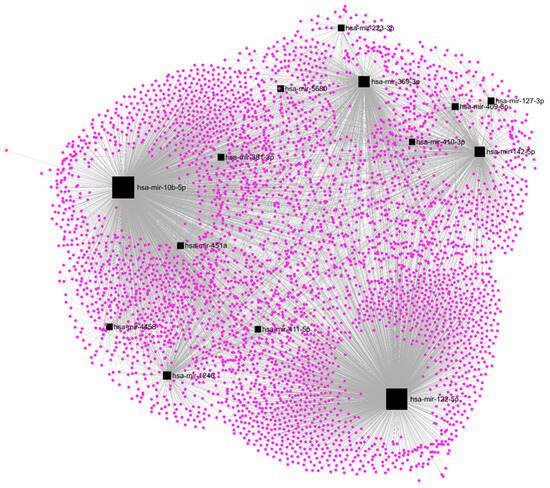
Figure 5.
Regulatory networks among the set of upregulated DE-microRNAs and their experimentally validated target genes in SKBR3 organoids.
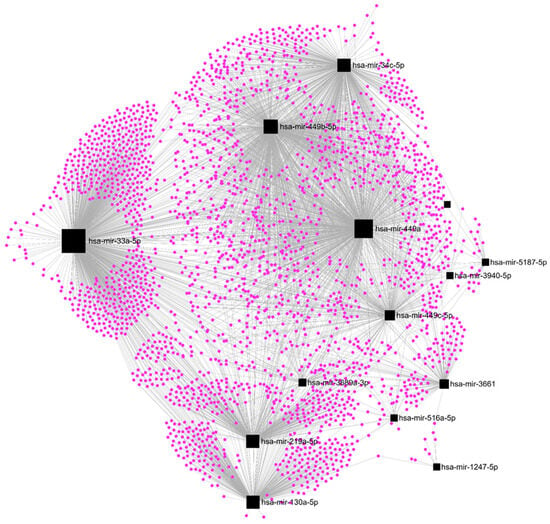
Figure 6.
Regulatory networks among the set of downregulated DE-microRNAs and their experimentally validated target genes in SKBR3 organoids.
2.3. The Expression Profile of mRNAs Is Downregulated under 3D Culture Conditions
On the other hand, the analysis of transcript expression, using Clariom D Assay human microarrays, showed 819 transcripts downregulated and 92 upregulated in the organoids compared to the conventional 2D culture of SKBR3 cells; of which, 172 have a gene annotation corresponding to known protein-coding genes or mRNAs (DE-mRNAs) and had the established criteria (adjusted p < 0.01), where 123 were downregulated and 49 were upregulated (Table 2 and Figure 4B), demonstrating, as in the case of the DE-microRNAs, the effect of the cellular organization present in the organoids, together with the cell–ECM interactions, on gene expression.

Table 2.
Differentially expressed up- and downregulated protein-coding genes (DE-mRNAs) found in SKBR3 organoids, in comparison to 2D culture.
Among the DE-mRNAs, those which were most upregulated are SLC44A4 (FC = 6.7), TFF1 (FC = 5.5), BGN (FC = 3.9), PRODH (3.1), and SLC22A18 (2.9). Among the DE-mRNAs, those which were most downregulated are GLYATL2 (FC = −36.1), TGFB2 (FC = −16.1), DST (FC = −14.4), OLR1 (FC = −12.2), and TPR (FC = −12.0). Notably, signaling pathways such as FOXO and HIPPO are affected, and a decreased level of TGF-β and SMAD3 expression in the 3D-cultured cell line is present.
2.4. Upregulation of hsa-mir-122-5p, hsa-mir-369-3p, and hsa-mir-10b-5p Affects Most of the DE-mRNAs
The cross-matching between DE-microRNAs and DE-mRNAs found in the organoids derived from the SKBR3 cells showed that the magnitude of FC values was well correlated, since FC of upregulated DE-microRNAs were higher than the observed in the downregulated ones, and FC values of downregulated DE-mRNAs were higher than the upregulated ones. These observations were in alignment with the number of targeted genes from the sets of miRNAs, where 14 upregulated DE-microRNAs matched 4370 target genes, while 15 downregulated DE-microRNAs matched with 2547 targets.
A subset of 11 upregulated DE-microRNAs targets 70 of the 123 downregulated DE-mRNAs, giving a correlation of 57% (Table 3). These selected genes are involved in 150 biological processes, such as the regulation of morphogenesis of an epithelium, cell morphogenesis, cell shape, cytoskeleton organization, MAPK cascade, Ras protein signal transduction, and Wnt signaling, among others, as well as in the MAPK and AGE–RAGE signaling KEGG-pathways, with CCL2, WNT5A, NRP1, WNK1, and TGFB2 being involved in most of the processes (Table 4). Of note, hsa-mir-122-5p (FC = 15.4), hsa-mir-369-3p (FC = 11.4), and hsa-mir-10b-5p (FC = 20.1) regulated up to 81% of the 70 DE-mRNAs, highlighting their pivotal roles in downregulating genes, thus affecting the 3D architecture of organoids. Moreover, five downregulated DE-microRNAs regulated only five of the forty-nine upregulated DE-mRNAs, giving a correlation of 10% (Table 5), and for that reason, we cannot further study the grouped function of these genes.

Table 3.
Correlation between differentially expressed upregulated miRNAs (DE-microRNAs) and downregulated protein-coding genes (DE-mRNAs) found in SKBR3 organoids culture.

Table 4.
Biological Processes and pathways in which correlated downregulated protein coding genes (DE-mRNAs) found in SKBR3 organoids participates.

Table 5.
Correlation between differentially expressed downregulated miRNAs (DE-microRNAs) and upregulated protein coding genes (DE-mRNAs) found in SKBR3 organoids culture.
3. Materials and Methods
3.1. Cell Culture
The human cell line SKBR3 (HTB-30. ATCC, Manassas, VA, USA), derived from a patient with breast carcinoma HER2+, was grown in Dulbecco’s modification of Eagle’s minimal medium (DMEM/F-12) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (Invitrogen, Waltham, MA, USA) at 37 °C, in a 5% CO2 atmosphere. Cells between passages 2 and 6 were used for all experiments.
3.2. 3D Cell Culture System
To obtain SKBR3 organoids, a standard On-Top protocol was used [4,27]; briefly, 24-well flat-bottom plates were coated with 150 μL of LDEV-free growth factor-reduced Geltrex (Gibco, USA) and incubated at 37 °C for 30 min to promote its gelation. Then, 4.2 × 104 cells were seeded, and DMEM/F-12 supplemented medium was added to incubate for up to 5 days. The only modification was that the medium had no additional 5% Geltrex as reported. Wells without Geltrex were used as a control (2D), maintaining the rest of the conditions. Similarly, a standard Embedded (In-Gel) protocol was used; briefly, the same number of cells were mixed with Geltrex pregel and then placed in 24-well flat-bottom plates before gelation [31]. Additionally, single cells were diluted to a density of 5 × 103 cells and placed into ultra-low attachment (ULA) 24-well plates and incubated under the same conditions to facilitate the formation of cell spheroids. Cells were monitored at 72, 96, and 120 h to measure the number and size of spheres using ImageJ software v 1.53t in which regions of interest were surrounded manually around the entire organoid from a bright-field microscopy image, and then the surface area was calculated in the ImageJ module (“Area Measurements of a Complex Object”).
3.3. Confocal Immunofluorescence Microscopy
Cells were fixed in 4% formaldehyde in 1 × PBS for 10 min at room temperature, and then were washed three times with 1 × PBS and permeabilized with 0.1% Triton X-100 for 10 min. Samples were washed and blocked with 22.5 mg/mL glycine and 1% bovine serum albumin, and then actin filaments were stained with 1:2000 Phalloidin-iFluor 488 (ab176753, Abcam, Waltham, MA, USA) for 5 min, washed, and mounted with a Fluoroshield with DAPI (Sigma-Aldrich, St. Louis, MO, USA). Visualization was performed at a 40× magnification using a Leica LSCM (TCS SP8, Leica Microsystems, Wetzlar, Germany).
3.4. RNA Isolation
RNA was extracted using Trizol reagent (Invitrogen) following the manufacturer’s instructions. The quantity and quality of total RNA were evaluated using Nanodrop (ThermoFisher Scientific, Waltham, MA, USA) and RNA integrity was assessed first using agarose gel electrophoresis at 60 V, stained with GelRed 0.5×, (Biotium, San Francisco, CA, USA) and visualized on a UV transilluminator. Additionally, it was evaluated by capillary gel electrophoresis using an RNA 6000 Nano Chip (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA). Samples with RNA integrity numbers ≥ 9.0 were used for these studies [32].
3.5. Small RNA Sequencing (RNAseq)
MicroRNA expression analysis of SKBR3 cells cultured in 2D and 3D culture was performed by whole Small RNAseq. The sequencing data obtained from this study has been deposited in the Gene Expression Omnibus (GEO) under Accession No. GSE239998 provided by NCBI, (NIH, Bethesda, MD, USA). The TruSeq Small RNA Library Prep kit (Illumina, San Diego, CA, USA) was used for the preparation of the library. The sequencing was performed with a total of 125 M reads for each sample. rRNAs were removed. Reads with Phred Quality Score values greater than 30 across all sequencing cycles were conserved. The filtered reads had a distribution of 6 to 36 bp in length, with a mean of 22 bp, which corresponds to the common size of miRNAs. Reads were aligned to the reference genome (GRCh38), miRBase v22.1, and non-coding RNA database (RNAcentral release 14.0) to classify the different types of small RNAs present in the samples. For the quantification of the miRNAs in the samples, Bowtie alignment mapping was performed in miRBase v21 and the counting of mature miRNAs was performed with the miRDeep2 Quantifier.
To find the differentially expressed miRNAs (DE-microRNAs) between the experimental groups (3D vs. 2D), miRNAs were normalized using TMM (Trimmed mean of M-values), and Principal Component Analysis (PCA) was performed to assess the homogeneity of the experimental replicates. The detection of the DE-miRNAs was performed using Fold Change, and an exact test using edgeR per comparison pair. From the general list, miRNAs with a fold-change (FC) greater than 2 or less than -2 were filtered with significance values of p < 0.01.
3.6. Microarrays Hybridization and Analysis
Transcript expression analysis of SKBR3 cells cultured in both 2D and 3D conditions was conducted using Clariom D Assay human microarrays (GeneChip, Affymetrix, Singapore, CA, USA). The Microarray data obtained from this study has been deposited in the Gene Expression Omnibus (GEO) under Accession No. GSE239813 provided by NCBI. The hybridization for whole-genome transcriptome analysis was performed following the instructions provided by the manufacturer. Briefly, cDNA preparation and biotin labeling were carried out using the Affymetrix GeneChip WT Pico Kit. Subsequently, cRNA purification was performed using the Affymetrix magnetic bead protocol. Array processing was performed using the Affymetrix GeneChip™ Hybridization, Wash, and Stain Kit. The arrays were incubated for 16 h in an Affymetrix GeneChip 645 hybridization oven at 45 °C with rotation at 60 rpm. Fluorescence amplification was achieved by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin–phycoerythrin stain. A confocal scanner (Affymetrix GeneChip Scanner 3000 7G plus) was utilized to capture the fluorescence signal at a resolution of 3 μm after excitation at 570 nm. The average signal from two sequential scans was calculated for each microarray.
For the subsequent analysis, Partek Genomic Suite v8.0 software was employed. All samples were normalized using the Robust Multiarray Average (RMA) method, which encompasses background correction, normalization, and calculation of expression values. Differential expression analysis was performed using one-way ANOVA. Differentially expressed protein-coding genes (DE-mRNAs) were selected between the groups based on an absolute fold-change of 2, and the Benjamini and Hochberg false discovery rate [23] was applied to account for multiple hypothesis testing. Genes with an adjusted p-value < 0.01 were considered significant.
3.7. In Silico Analysis
The TarBase v8.0 database was used to find experimentally validated target genes regulated by the DE-microRNAs. miRNet software v. 4.2 was used to define how many validated genes are regulated by each set of DE-microRNAs and to graph the regulation networks formed by them. The web-based portal Metascape v3.5 was used for pathway enrichment analysis of selected genes using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and GO Biological Processes ontology sources.
3.8. Statistical Analysis
Experiments were performed in triplicate and results were represented as mean ± standard deviation (SD). Independent sample’s t-test was used to compare the means of both groups, considering p < 0.05 as statistically significant. Statistical analyses were performed using the software GraphPad v8.0.
4. Discussion
In the present study, we aimed to elucidate the effect of 3D organization cells and the presence of an ECM on the pattern of expression of miRNAs and mRNAs using a breast cancer HER2+ SKBR3 cell line. The most common 3D culture systems are based on the use of scaffolds, scaffold-free, and derived from tissues [31]. There are no studies evaluating differential expression patterns between On-Top, Embedded, and ULA cultures, nevertheless, it is expected that the presence of a scaffold could modify the gene expression of organoids. For instance, the first evidence that the ECM component, laminin regulates the gene expression and differentiation of the primary mammary cells through direct interaction with the cell integrins was seen in the 90s [33]. It is believed that modifications in the position of cells in the matrix can affect gene expression. Embedded cells have a strong interaction with the ECM but limited interaction with other cells, while On-Top cells have less interaction with the ECM they have better interactions with neighboring cells, which allows for organoid formation. Our research indicates that the On-Top culture resulted in well-defined organoids with larger sizes and greater numbers compared to Embedded cells (as shown in Figure 1A). Although ULA culture is typically used for cell aggregation, we used it as a 3D control culture because it lacks scaffolding and cell–ECM interactions, unlike the Geltrex matrix [34].
In the conventional 2D cell culture of SKBR3, we obtained the common cell morphology in clusters with a great amount of free-floating cells or very loosely attached round cells (Figure 2A,B), which have been previously reported [35,36]. However, this changed when the cells were grown in the 3D On-Top cell culture (Figure 2C,D), where the cells turned to a cell morphology frankly rounded, smaller in size with less cytoplasm and a grape-like appearance distinguished by their poor cell–cell contacts. It has been seen that grape-like cells formed less-closely associated colonies with reduced cell–cell adhesion which could be a reflection of the acquisition of the ability to escape from their neighbors in the primary tumor over their evolution as they acquired the ability to metastasize [30]. These changes in cell morphology influenced by 3D cell culture have been previously reported in SKBR3 cells on Matrigel [30,37]. Our On-Top generated organoids, derived from cultures using Geltrex, present a larger volume of up to 150 µm, and a better definition than the organoids derived from the SKBR3 cultivated in Matrigel, maintaining grape-like appearance [30] and also presenting discrete extensions of the cytoplasmic membrane, establishing direct connections with neighboring spheroid cells reminiscent of the tunneled nanotubes documented in previous studies involving 3D-cultured cancer cells [29].
We next evaluated the miRNA expression in the organoids, finding 39 differentially expressed miRNAs (DE-microRNAs): 20 miRNAs upregulated and 19 downregulated (Table 1). Interestingly, none of these sets of dysregulated miRNAs have been associated previously with the 3D cell culture system in breast cancer [27,38]. However, some of these downregulated DE-microRNAs, such as hsa-miR-449a, hsa-miR-4739, hsa-miR-449a, hsa-miR-34c-5p, hsa-miR-219a-5p, hsa-miR-34c-5p, hsa-miR-34c-5p, hsa-miR-219a-5p, has-miR-5091, and has-miR-943 have been previously associated with poor breast cancer prognosis [39], among other processes [40,41,42,43,44,45]. In addition, the upregulated miRNAs hsa-miR-127-3p, hsa-miR-223-3p, has-miR-4458, hsa-miR-10b-5p, hsa-miR-381-3p, has-miR-451a, hsa-miR-142-5p, has-miR-1246, hsa-miR-375-3p, and hsa-miR-4739, have been associated with several breast cancer processes, such as malignancy [40,46], tumor progression [41], and poor prognosis [47], as well as, cancer hallmarks’ activation as proliferation [48,49], apoptosis [49] and metastasis [49,50,51], among others [51,52,53,54,55,56].
In two previous reports from our group, using the triple-negative breast cancer (TNBC) cell line Hs578T and the luminal B breast cancer cell line BT-474 cultivated under the same cell culture conditions used here, we also obtained compact and large organoids, but with different morphology [4,27]. In the case of the BT-474 cell line, it has been shown that a 3D microenvironment can reprogram the oncogenic lncRNAs/mRNAs coexpression networks [4]. Similarly, in our case, SKBR3 cells reprogram the oncogenic miRNA/mRNA coexpresion network, which makes it obvious that lncRNA expression is also modified in our HER2+ subtype.
In the case of both basal Hs578T and luminal T47D breast cancer cell lines, the adoption of 3D organotypic cultures resulted in notable morphological alterations, including perturbations in cell–cell and cell–ECM interactions, the loss of cellular polarity, the reorganization of bulk structures, and the downregulation of specific miRNAs in Hs578T 3D cultures when compared to the 2D condition. This contributes to the positive modulation of crucial biological processes, such as the cellular response to hypoxia and focal adhesion, whereas the upregulation of miRNAs is associated with negative regulation of the cell cycle [20]. Meanwhile, in SKBR3 cells, in silico analysis showed that a subset of 11 upregulated DE-microRNAs target 70 downregulated DE-mRNAs involved in the regulation of cell morphogenesis, the regulation of cell shape, the regulation of canonical Wnt signaling pathway, the morphogenesis of epithelium, and the regulation of cytoskeleton organization, as well as in the MAPK and AGE–RAGE signaling KEGG-pathways. These data showed the impact of the cell culture system in each breast cancer subtype, in fact, in these reports, we did not find any deregulated DE-microRNAs that were observed in this work.
We also found, through the expression profile of mRNAs, that most of the genes are downregulated under 3D-cultured conditions. Interestingly, within the differentially expressed protein-coding genes (DE-mRNAs), we found genes involved in the tumoral microenvironment, such as CCL2, a chemokine involved in the tumoral progression of various cancers by modulating the tumor microenvironment [57], promoting cellular growth, migration, angiogenesis, and the recruitment of immunosuppressive cells through its activation at different stages of tumorigenesis. Additionally, its involvement as an autocrine or paracrine growth factor have been described [58,59]. WNT5A belongs to the Wnt signaling pathway family, which modulates different crucial processes for normal cellular development, including cell proliferation, adhesion, migration, and differentiation [60]. Loss of Wnt5 is associated with cancer relapse and poor survival, as it plays an important role as a tumor suppressor and inhibits cell migration by decreasing the production of matrix metalloproteinases (MMPs), contributing to cellular dispersion, reduced cell–collagen interaction, increased motility, and decreased adhesion [61,62]. Borcherding et al. reported that WNT5A is expressed in early breast cancer tumors, but as the tumor progresses to later stages and migrates to other tissues, its expression decreases [63]. NRP1 encodes neuropilins (NRPs), which are transmembrane glycoprotein receptors that act as co-receptors for vascular endothelial growth factor (VEGF) [64]. Overexpression of NRP1 has been reported in lung, colorectal, and breast cancer [65,66]. In breast cancer, its upregulation is identified as a tumor promoter, and its downregulation results in apoptosis promotion and inhibition of tumor growth [64]. PMEPA1 encodes the androgen-induced prostate transmembrane protein 1 and is highly regulated in prostate cancer [67], lung cancer [68], and breast cancer [69]. PMEPA1 negatively regulates TGF-β/SMAD signaling, thereby suppressing its tumor-suppressive capacity [70]. Previous work showed that the elimination of PMEPA1 in breast cancer cells significantly decreased their ability to form spheroids in Hs578t and BT-549 cells. Additionally, HCC1359 breast cancer cells lack phosphorylation levels of SMAD3, which correlate with decreased TMEPA1 expression [71]. This information is consistent with the decreased expression of TGF- β, SMAD3, and TMEPA1 in our 3D cultures. The dysregulation of all these genes constitutes a relevant pathway for the understanding and investigation of cancer biology.
One of the significant pathways implicated in the gene deregulation uncovered in our study is the Forkhead Box O (FOXO) pathway. The FOXO family of transcription factors exerts notable effects on cellular fate and tumor suppression across a broad spectrum of cancers, governed by stress and growth factors [72]. Reports have indicated that FOXO is downregulated in various types of cancers, including breast, colon, gastric, lung, and leukemias [73,74]. A prior study highlighted that the deletion of FOXO1 in adult mice heightens tumor incidence [75], while its activation halts the cell cycle and induces apoptosis in tumor cells [74]. This aligns with our findings, as evidenced by its downregulation in 3D cultures, reflecting closer concordance with in vivo observations. Another relevant gene in this pathway is HOMER1, which encodes a scaffold protein contributing to intracellular signal transduction [76], and is crucially engaged in Ca2+-dependent signaling [77]. As such, it has been reported to promote the proliferation, migration, and invasion in colorectal cancer cells via the G3BP1 pathway [78]. However, no reports of HOMER1’s involvement in breast cancer were found. In addition, an important finding is the HIPPO signaling pathway which is regarded as a tumor-suppressive pathway due to its pivotal role in regulating organ size, cell number, and tissue homeostasis [79]. Therefore, aberrant expression of some of its genes leads to increased cellular proliferation, tumorigenesis, and cancer metastasis [80,81]. This pathway encompasses mammalian sterile 20-like kinases 1/2 (MST1/2), large tumor suppressor kinases 1/2 (LATS1/2), Salvador 1 protein (SAV1), MOB kinases 1A/B (MOB1A/B), Yes-associated protein (YAP), and transcriptional coactivator with PDZ-binding motif (TAZ) [82]. Furthermore, the components of this pathway are reported to act as transducers, conferring cellular structural characteristics, polarity, shape, and cytoskeletal organization. These properties are closely associated with cells’ capacity to adhere to other cells and the extracellular matrix, and they are also influenced by the cellular microenvironment [81,82]. In our study, we observed that YAP1 expression is positively regulated in 2D-cultured conditions and decreased in 3D-cultured, consistent with findings from various research groups [83,84,85,86]. YAP activation occurs when cells grow on a rigid matrix and spread extensively, while YAP is inactivated when cells are seeded on a soft matrix and aggregate in a small area (round and compact geometry) [83,84,85]. Additionally, cell–cell contact, cellular geometry, matrix stiffness, and cell–matrix interactions [87,88] generate crucial signals that regulate the Hippo pathway. In low-density cell cultures, YAP is predominantly located in the nucleus, promoting proliferation. Conversely, when cell density is high, YAP is expressed in the cytoplasm, suppressing proliferation [84,86].
On the other hand, when the cross-matching between upregulated DE-miRNAs and downregulated DE-mRNA was performed, our results showed that 11 upregulated DE-microRNAs correlate with 70 downregulated DE-mRNAs (Table 3), which are involved in several biological processes and pathways (Table 4) that contribute to the development of breast cancer, such as Wnt [89,90] and MAPK [91] signaling. Additionally, the downregulated DE-mRNAs, such as TGFB2, DST, OLR1, GEN1, RASA2, FOXO1, JMJD1C, ROCK1, and WNT5A, have been previously associated with breast cancer as biomarkers [92,93,94,95] and therapeutic targets [96], or by participating in several processes such as metastasis [62,97,98], apoptosis [99], chemosensitivity [99], and epigenetic changes [100]. A highlighted downregulated DE-gene is WNT5A which in normal tissue belongs to Wnt/β-catenin-independent signaling, binding to different receptors to promote normal development in breast tissue [101,102,103]; its loss plays an important role in breast cancer progression [104]. This downregulated gene is a target for the upregulated hsa-mir-381-3p; its downregulation could be related to the overexpression of these miRNAs, which promote mammary carcinogenesis acting as oncomiRs.
Notably, hsa-mir-122-5p (FC = 15.4), hsa-mir-369-3p (FC = 11.4), and hsa-mir-10b-5p (FC = 20.1) collectively controlled approximately 81% of the 70 DE-mRNAs. This underscores their crucial function in suppressing genes and consequently impacting the 3D structure of organoids. In the case of hsa-mir-369-3p it has been found in patients according to their lymph node status and associated with biological processes such as regulation of the epithelial-mesenchymal transition, cell proliferation, and transcriptional regulation [105]; it is downregulated in triple-negative breast cancer [106] and has been found in male breast cancer samples [107].
Hsa-mir-10b-5p has been extensively investigated in the context of breast cancer, particularly in comparison to other cancer types. Its upregulated expression has been consistently associated with various outcomes, including enhanced metastasis [108,109], increased invasive potential in both in vitro [110,111] and in vivo settings, augmented migration [112], elevated epithelial–mesenchymal transition [113], angiogenesis [114], and enhanced proliferation [112]. Collectively, these alterations contribute to unfavorable clinical outcomes, such as larger tumor size, advanced clinical stage, and shorter relapse-free survival periods [115,116,117,118]. Furthermore, hsa-mir-10b-5p exhibits associations with the expression of established biomarkers in breast cancer. Irrespective of metastatic status, hsa-mir-10b-5p expression positively correlates with HER2 positivity [117,119], while it negatively correlates with estrogen and progesterone receptor positivity [116,117]. This association further reinforces the connection between hsa-mir-10b-5p and the metastatic potential of breast cancer, as HER2 positivity and hormone receptor negativity serve as known predictors of tumor aggressiveness. Additional evidence supporting the pleiotropic effects of hsa-mir-10b-5p as a driving force in breast cancer invasiveness and metastasis comes from the observed positive correlation between hsa-mir-10b-5p expression and stemness, or self-renewal, in breast cancer stem cells [113]. Specifically, researchers discovered that stable overexpression of hsa-mir-10b-5p in MCF-7 cells led to heightened self-renewal capabilities and upregulation of genes associated with stemness and epithelial–mesenchymal transition. Conversely, the use of synthetic antagomirs against hsa-mir-10b-5p resulted in decreased self-renewal of stem cells [113].
Regarding hsa-mir-122-5p, it has a very important role in breast cancer. For instance, it has been shown that the downregulation of SDC1 mediated by hsa-mir-122-5p or liver-cell-derived exosomes would significantly augment the migratory capacity of breast cancer cells; furthermore, the metastatic potential or mobility of breast cancer cells is probably influenced by the presence of circulating hsa-mir-122-5p, which may not be directly correlated with the progression of breast cancer [120]. Fong et al. showed that elevated levels of miR-122 were observed in the blood of breast cancer patients with metastasis, indicating that cancer cells produce hsa-mir-122-5p, thereby facilitating metastasis through increased nutrient availability in the premetastatic niche. In vitro and in vivo experiments demonstrated that hsa-mir-122-5p derived from cancer cells inhibited glucose uptake by niche cells by suppressing pyruvate kinase, a key glycolytic enzyme. Notably, inhibiting hsa-mir-122-5p in vivo restored glucose absorption in distant organs such as the brain and lungs, thereby reducing the risk of metastasis. These findings suggest that extracellular hsa-mir-122-5p released by cancer cells can modulate systemic energy metabolism to drive disease progression by altering glucose utilization in recipient premetastatic niche cells [121]. It has also has been observed that, in TNBC cells, a substantial increase in hsa-mir-122-5p expression and a marked downregulation of the CHMP3 gene concluding that hsa-mir-122-5p, through the inhibition of CHMP3 via MAPK signaling, promotes aggressiveness and epithelial–mesenchymal transition (EMT) in TNBC [122]. Additionally, Wang et al. demonstrated, through modulation of the PI3K/Akt/mTOR/p70S6K pathway and targeting IGF1R, that hsa-mir-122-5p functions as a tumor suppressor and plays a crucial role in inhibiting the growth of new tumors. These findings indicate that hsa-mir-122-5p holds potential as a novel therapeutic or diagnostic/prognostic target for the treatment of breast cancer [123]. Finally, it has been shown that CDKN2B-AS1 regulates the expression of STK39 by acting as a sponge for hsa-mir-122-5p, thereby promoting breast cancer progression, and hsa-mir-122-5p modulates STK39 expression to regulate the impact of sh-CDKN2B-AS1 [42]. Interestingly, except for hsa-mir-10b-5p, none of these putative correlations have been previously reported in HER2+ breast cancer, opening a broad avenue of research to further understand the breast cancer biology using 3D cell culture experiments which are more approximate to the tumor microenvironment in vivo.
5. Conclusions
Our results show that the organotypic 3D cell-organization architecture of breast cancer SKBR3 cells impacts the expression pattern of the miRNAs–mRNAs network through overexpression of hsa-mir-122-5p, hsa-mir-369-3p, and hsa-mir-10b-5p. All these findings suggest that the interaction between the cell–cell and cell–extracellular matrix, as well as the change in the culture architecture impact gene expression, are pertinent to migrate breast cancer research from conventional cultures to 3D models.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ncrna9060066/s1, Video S1: Video SKBR3 Organoid.
Author Contributions
M.d.l.Á.G.-L., M.A.-M., J.L.-G., C.G.M. and G.R.-Q. conducted laboratory tests. M.B., M.A.-F., C.L.-C. and C.P.-P. collected the data and analyzed the results. M.A.-M., A.S.B. and R.R.-P. conceived and designed the study. M.A.-F., C.L.-C. and C.P.-P. confirmed the authenticity of the data. M.A.-M., A.S.B., M.B. and R.R.-P. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Consejo Nacional de Humanidades, Ciencias y Tecnologias (CONAHCYT) of Mexico, grant Programa Presupuestario F003 #51207/2020, and PROFAPI-UAS, grant PRO-A3-019.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The sequencing (Accession No. GSE239998) and Microarray (Accession No. GSE239813) data have been deposited in the Gene Expression Omnibus (GEO) under provided by NCBI, (NIH, Bethesda, MD, USA).
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Cancer Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 9 January 2023).
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef]
- Nunez-Olvera, S.I.; Aguilar-Arnal, L.; Cisneros-Villanueva, M.; Hidalgo-Miranda, A.; Marchat, L.A.; Salinas-Vera, Y.M.; Ramos-Payan, R.; Perez-Plasencia, C.; Carlos-Reyes, A.; Puente-Rivera, J.; et al. Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment. Cells 2022, 11, 3458. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Watkins, E.J. Overview of breast cancer. JAAPA 2019, 32, 13–17. [Google Scholar] [CrossRef]
- Pernas, S.; Tolaney, S.M. HER2-positive breast cancer: New therapeutic frontiers and overcoming resistance. Adv. Med. Oncol 2019, 11, 1758835919833519. [Google Scholar] [CrossRef]
- Cukierman, E.; Pankov, R.; Stevens, D.R.; Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 2001, 294, 1708–1712. [Google Scholar] [CrossRef]
- Nickerson, C.A.; Goodwin, T.J.; Terlonge, J.; Ott, C.M.; Buchanan, K.L.; Uicker, W.C.; Emami, K.; LeBlanc, C.L.; Ramamurthy, R.; Clarke, M.S.; et al. Three-dimensional tissue assemblies: Novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 2001, 69, 7106–7120. [Google Scholar] [CrossRef]
- Kelm, J.M.; Timmins, N.E.; Brown, C.J.; Fussenegger, M.; Nielsen, L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003, 83, 173–180. [Google Scholar] [CrossRef]
- Delarue, M.; Montel, F.; Vignjevic, D.; Prost, J.; Joanny, J.F.; Cappello, G. Compressive stress inhibits proliferation in tumor spheroids through a volume limitation. Biophys. J. 2014, 107, 1821–1828. [Google Scholar] [CrossRef]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Baal, N.; Widmer-Teske, R.; McKinnon, T.; Preissner, K.T.; Zygmunt, M.T. In vitro spheroid model of placental vasculogenesis: Does it work? Lab. Investig. 2009, 89, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.L.; Jiang, Q.; Han, S.; Wu, Y.; Cui Tomshine, J.; Wang, D.; Gan, Y.; Zou, G.; Liang, X.J. Multicellular tumor spheroids as an in vivo-like tumor model for three-dimensional imaging of chemotherapeutic and nano material cellular penetration. Mol. Imaging 2012, 11, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Huanwen, W.; Zhiyong, L.; Xiaohua, S.; Xinyu, R.; Kai, W.; Tonghua, L. Intrinsic chemoresistance to gemcitabine is associated with constitutive and laminin-induced phosphorylation of FAK in pancreatic cancer cell lines. Mol. Cancer 2009, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Longati, P.; Jia, X.; Eimer, J.; Wagman, A.; Witt, M.R.; Rehnmark, S.; Verbeke, C.; Toftgard, R.; Lohr, M.; Heuchel, R.L. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 2013, 13, 95. [Google Scholar] [CrossRef]
- Souza, A.G.; Silva, I.B.B.; Campos-Fernandez, E.; Barcelos, L.S.; Souza, J.B.; Marangoni, K.; Goulart, L.R.; Alonso-Goulart, V. Comparative Assay of 2D and 3D Cell Culture Models: Proliferation, Gene Expression and Anticancer Drug Response. Curr. Pharm. Des. 2018, 24, 1689–1694. [Google Scholar] [CrossRef]
- Kiss, D.L.; Windus, L.C.; Avery, V.M. Chemokine receptor expression on integrin-mediated stellate projections of prostate cancer cells in 3D culture. Cytokine 2013, 64, 122–130. [Google Scholar] [CrossRef]
- Schmidt, M.; Scholz, C.J.; Polednik, C.; Roller, J. Spheroid-based 3-dimensional culture models: Gene expression and functionality in head and neck cancer. Oncol. Rep. 2016, 35, 2431–2440. [Google Scholar] [CrossRef]
- Li, X.; Zeng, Z.; Wang, J.; Wu, Y.; Chen, W.; Zheng, L.; Xi, T.; Wang, A.; Lu, Y. MicroRNA-9 and breast cancer. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 122, 109687. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Lerner, R.G.; Petritsch, C. A microRNA-operated switch of asymmetric-to-symmetric cancer stem cell divisions. Nat. Cell Biol. 2014, 16, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Pencheva, N.; Tavazoie, S.F. Control of metastatic progression by microRNA regulatory networks. Nat. Cell Biol. 2013, 15, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Koedoot, E.; Wolters, L.; Smid, M.; Stoilov, P.; Burger, G.A.; Herpers, B.; Yan, K.; Price, L.S.; Martens, J.W.M.; Le Devedec, S.E.; et al. Differential reprogramming of breast cancer subtypes in 3D cultures and implications for sensitivity to targeted therapy. Sci. Rep. 2021, 11, 7259. [Google Scholar] [CrossRef]
- Kunigenas, L.; Stankevicius, V.; Dulskas, A.; Budginaite, E.; Alzbutas, G.; Stratilatovas, E.; Cordes, N.; Suziedelis, K. 3D Cell Culture-Based Global miRNA Expression Analysis Reveals miR-142-5p as a Theranostic Biomarker of Rectal Cancer Following Neoadjuvant Long-Course Treatment. Biomolecules 2020, 10, 613. [Google Scholar] [CrossRef]
- Salinas-Vera, Y.M.; Valdes, J.; Hidalgo-Miranda, A.; Cisneros-Villanueva, M.; Marchat, L.A.; Nunez-Olvera, S.I.; Ramos-Payan, R.; Perez-Plasencia, C.; Arriaga-Pizano, L.A.; Prieto-Chavez, J.L.; et al. Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells. Cancers 2022, 14, 2490. [Google Scholar] [CrossRef]
- Franchi, M.; Piperigkou, Z.; Riti, E.; Masola, V.; Onisto, M.; Karamanos, N.K. Long filopodia and tunneling nanotubes define new phenotypes of breast cancer cells in 3D cultures. Matrix Biol. Plus 2020, 6–7, 100026. [Google Scholar] [CrossRef]
- Han, S.J.; Kwon, S.; Kim, K.S. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021, 21, 152. [Google Scholar] [CrossRef]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef]
- Huang, H.; Ding, Y.; Sun, X.S.; Nguyen, T.A. Peptide hydrogelation and cell encapsulation for 3D culture of MCF-7 breast cancer cells. PLoS ONE 2013, 8, e59482. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Streuli, C.H.; Bailey, N.; Bissell, M.J. Control of mammary epithelial differentiation: Basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J. Cell Biol. 1991, 115, 1383–1395. [Google Scholar] [CrossRef]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef]
- Pirsko, V.; Cakstina, I.; Priedite, M.; Dortane, R.; Feldmane, L.; Nakazawa-Miklasevica, M.; Daneberga, Z.; Gardovskis, J.; Miklasevics, E. An Effect of Culture Media on Epithelial Differentiation Markers in Breast Cancer Cell Lines MCF7, MDA-MB-436 and SkBr3. Medicina 2018, 54, 11. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.Z.; Phillips, G.D.L.; Nitta, H.; Garsha, K.; Admire, B.; Kraft, R.; Dennis, E.; Vela, E.; Towne, P. Activity of trastuzumab emtansine (T-DM1) in 3D cell culture. Breast Cancer Res. Treat. 2021, 188, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Froehlich, K.; Haeger, J.D.; Heger, J.; Pastuschek, J.; Photini, S.M.; Yan, Y.; Lupp, A.; Pfarrer, C.; Mrowka, R.; Schleussner, E.; et al. Generation of Multicellular Breast Cancer Tumor Spheroids: Comparison of Different Protocols. J. Mammary Gland Biol. Neoplasia 2016, 21, 89–98. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Li, C.; Lin, Z.; Zhuang, Y.; Flemington, E.K.; Burow, M.E.; Lin, Y.I.; Shan, B. The microRNA expression associated with morphogenesis of breast cancer cells in three-dimensional organotypic culture. Oncol. Rep. 2012, 28, 117–126. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; Xu, F.; Fu, J.; Sun, J.; Gan, X.; Yang, C.; Mao, Z. ncRNAs-mediated high expression of TIMM8A correlates with poor prognosis and act as an oncogene in breast cancer. Cancer Cell Int. 2022, 22, 177. [Google Scholar] [CrossRef]
- Liu, J.; Wang, P.; Zhang, P.; Zhang, X.; Du, H.; Liu, Q.; Huang, B.; Qian, C.; Zhang, S.; Zhu, W.; et al. An integrative bioinformatics analysis identified miR-375 as a candidate key regulator of malignant breast cancer. J. Appl. Genet. 2019, 60, 335–346. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, X.; Guo, X.; Li, C.; Fan, Y.; Liu, P.; Yuan, D.; Ma, X.; Wang, J.; Zheng, J.; et al. LncRNA-BC069792 suppresses tumor progression by targeting KCNQ4 in breast cancer. Mol. Cancer 2023, 22, 41. [Google Scholar] [CrossRef]
- Xiao, B.; Zhang, W.; Chen, L.; Hang, J.; Wang, L.; Zhang, R.; Liao, Y.; Chen, J.; Ma, Q.; Sun, Z.; et al. Analysis of the miRNA-mRNA-lncRNA network in human estrogen receptor-positive and estrogen receptor-negative breast cancer based on TCGA data. Gene 2018, 658, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Pizzamiglio, S.; Cosentino, G.; Ciniselli, C.M.; De Cecco, L.; Cataldo, A.; Plantamura, I.; Triulzi, T.; El-Abed, S.; Wang, Y.; Bajji, M.; et al. What if the future of HER2-positive breast cancer patients was written in miRNAs? An exploratory analysis from NeoALTTO study. Cancer Med. 2022, 11, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, M.; Wang, S.; Xu, P.; Deng, Y.; Lin, S.; Li, N.; Liu, K.; Zhu, Y.; Zhai, Z.; et al. LncRNA MEG3 rs3087918 was associated with a decreased breast cancer risk in a Chinese population: A case-control study. BMC Cancer 2020, 20, 659. [Google Scholar] [CrossRef]
- Rawlings-Goss, R.A.; Campbell, M.C.; Tishkoff, S.A. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med. Genom. 2014, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhong, S.; Zhang, Z.; Tang, J. PHF20 is a Novel Prognostic Biomarker and Correlated with Immune Status in Breast Cancer. Biochem. Genet. 2023, 61, 1369–1386. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wang, J.; Jiang, H.; Dong, H. A Novel Necroptosis-Related miRNA Signature for Predicting the Prognosis of Breast Cancer Metastasis. Dis. Markers 2022, 2022, 3391878. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Zhang, G. miR-4458 regulates cell proliferation and apoptosis through targeting SOCS1 in triple-negative breast cancer. J. Cell. Biochem. 2019, 120, 12943–12948. [Google Scholar] [CrossRef]
- Wu, J.; Miao, J.; Ding, Y.; Zhang, Y.; Huang, X.; Zhou, X.; Tang, R. MiR-4458 inhibits breast cancer cell growth, migration, and invasiveness by targeting CPSF4. Biochem. Cell Biol. Biochim. Biol. Cell. 2019, 97, 722–730. [Google Scholar] [CrossRef]
- Elango, R.; Alsaleh, K.A.; Vishnubalaji, R.; Manikandan, M.; Ali, A.M.; Abd El-Aziz, N.; Altheyab, A.; Al-Rikabi, A.; Alfayez, M.; Aldahmash, A.; et al. MicroRNA Expression Profiling on Paired Primary and Lymph Node Metastatic Breast Cancer Revealed Distinct microRNA Profile Associated With LNM. Front. Oncol. 2020, 10, 756. [Google Scholar] [CrossRef]
- Wang, P.; Chen, W.; Zhang, Y.; Zhong, Q.; Li, Z.; Wang, Y. MicroRNA-1246 suppresses the metastasis of breast cancer cells by targeting the DYRK1A/PGRN axis to prevent the epithelial-mesenchymal transition. Mol. Biol. Rep. 2022, 49, 2711–2721. [Google Scholar] [CrossRef]
- Denkiewicz, M.; Saha, I.; Rakshit, S.; Sarkar, J.P.; Plewczynski, D. Identification of Breast Cancer Subtype Specific MicroRNAs Using Survival Analysis to Find Their Role in Transcriptomic Regulation. Front. Genet. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- Pane, K.; Zanfardino, M.; Grimaldi, A.M.; Baldassarre, G.; Salvatore, M.; Incoronato, M.; Franzese, M. Discovering Common miRNA Signatures Underlying Female-Specific Cancers via a Machine Learning Approach Driven by the Cancer Hallmark ERBB. Biomedicines 2022, 10, 1306. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Zhu, Y.; Hu, L.; Khadka, V.S.; Ai, J.; Zou, H.; Ju, D.; Jiang, B.; Deng, Y.; Hu, X. Plasma MicroRNA Pair Panels as Novel Biomarkers for Detection of Early Stage Breast Cancer. Front. Physiol. 2018, 9, 1879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, L.; Yu, G.; Sun, Z.; Wang, T.; Tian, X.; Duan, X.; Zhang, C. Exosomal miR-1246 and miR-155 as predictive and prognostic biomarkers for trastuzumab-based therapy resistance in HER2-positive breast cancer. Cancer Chemother. Pharmacol. 2020, 86, 761–772. [Google Scholar] [CrossRef]
- Zhai, L.Y.; Li, M.X.; Pan, W.L.; Chen, Y.; Li, M.M.; Pang, J.X.; Zheng, L.; Chen, J.X.; Duan, W.J. In Situ Detection of Plasma Exosomal MicroRNA-1246 for Breast Cancer Diagnostics by a Au Nanoflare Probe. ACS Appl. Mater. Interfaces 2018, 10, 39478–39486. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Z.; Lu, W.; Chen, Z.; Chen, L.; Han, S.; Wu, X.; Cai, T.; Cai, Y. Chemokines and chemokine receptors: A new strategy for breast cancer therapy. Cancer Med. 2020, 9, 3786–3799. [Google Scholar] [CrossRef]
- Yoshimura, T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: A foe or ally? Cell Mol. Immunol. 2018, 15, 335–345. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Xia, R.; Wei, Y.; Wei, X. Role of the CCL2-CCR2 signalling axis in cancer: Mechanisms and therapeutic targeting. Cell Prolif. 2021, 54, e13115. [Google Scholar] [CrossRef]
- Asem, M.S.; Buechler, S.; Wates, R.B.; Miller, D.L.; Stack, M.S. Wnt5a Signaling in Cancer. Cancers 2016, 8, 79. [Google Scholar] [CrossRef]
- Serra, R.; Easter, S.L.; Jiang, W.; Baxley, S.E. Wnt5a as an effector of TGFbeta in mammary development and cancer. J. Mammary Gland Biol. Neoplasia 2011, 16, 157–167. [Google Scholar] [CrossRef]
- Prasad, C.P.; Chaurasiya, S.K.; Guilmain, W.; Andersson, T. WNT5A signaling impairs breast cancer cell migration and invasion via mechanisms independent of the epithelial-mesenchymal transition. J. Exp. Clin. Cancer Res. CR 2016, 35, 144. [Google Scholar] [CrossRef] [PubMed]
- Borcherding, N.; Kusner, D.; Kolb, R.; Xie, Q.; Li, W.; Yuan, F.; Velez, G.; Askeland, R.; Weigel, R.J.; Zhang, W. Paracrine WNT5A Signaling Inhibits Expansion of Tumor-Initiating Cells. Cancer Res. 2015, 75, 1972–1982. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Ma, W.-M.; Shi, Z.-D.; Zhang, Z.-G.; Zhou, J.-H.; Li, Y.; Zhang, S.-Q.; Pang, K.; Li, B.-B.; Zhang, W.-D.; et al. Role of NRP1 in Bladder Cancer Pathogenesis and Progression. Front. Oncol. 2021, 11, 685980. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.; Tokunaga, T.; Hatanaka, H.; Kijima, H.; Yamazaki, H.; Abe, Y.; Osamura, Y.; Inoue, H.; Ueyama, Y.; Nakamura, M. Neuropilin 1 and neuropilin 2 co-expression is significantly correlated with increased vascularity and poor prognosis in nonsmall cell lung carcinoma. Cancer 2002, 95, 2196–2201. [Google Scholar] [CrossRef] [PubMed]
- Jubb, A.M.; Strickland, L.A.; Liu, S.D.; Mak, J.; Schmidt, M.; Koeppen, H. Neuropilin-1 expression in cancer and development. J. Pathol. 2012, 226, 50–60. [Google Scholar] [CrossRef]
- Liu, R.; Zhou, Z.; Huang, J.; Chen, C. PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppressing the Smad3/4-c-Myc-p21 Cip1 signaling pathway. J. Pathol. 2011, 223, 683–694. [Google Scholar] [CrossRef]
- Vo Nguyen, T.T.; Watanabe, Y.; Shiba, A.; Noguchi, M.; Itoh, S.; Kato, M. TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci. 2014, 105, 334–341. [Google Scholar] [CrossRef]
- Singha, P.K.; Pandeswara, S.; Geng, H.; Lan, R.; Venkatachalam, M.A.; Saikumar, P. TGF-beta induced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes Cancer 2014, 5, 320–336. [Google Scholar] [CrossRef]
- Watanabe, Y.; Itoh, S.; Goto, T.; Ohnishi, E.; Inamitsu, M.; Itoh, F.; Satoh, K.; Wiercinska, E.; Yang, W.; Shi, L.; et al. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol. Cell 2010, 37, 123–134. [Google Scholar] [CrossRef]
- Puteri, M.U.; Watanabe, Y.; Wardhani, B.W.K.; Amalia, R.; Abdelaziz, M.; Kato, M. PMEPA1/TMEPAI isoforms function via its PY and Smad-interaction motifs for tumorigenic activities of breast cancer cells. Genes Cells 2020, 25, 375–390. [Google Scholar] [CrossRef]
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.J.; Xu, J.; Zheng, W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci 2017, 13, 815–827. [Google Scholar] [CrossRef]
- Chae, Y.C.; Kim, J.Y.; Park, J.W.; Kim, K.B.; Oh, H.; Lee, K.H.; Seo, S.B. FOXO1 degradation via G9a-mediated methylation promotes cell proliferation in colon cancer. Nucleic Acids Res. 2019, 47, 1692–1705. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.G.; Lee, H.; Gupta, N.; Ramachandran, S.; Kaushik, I.; Srivastava, S.; Kim, S.-H.; Srivastava, S.K. Role of Forkhead Box Class O proteins in cancer progression and metastasis. Semin. Cancer Biol. 2018, 50, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.H.; Kollipara, R.; Chu, G.; Ji, H.; Xiao, Y.; Ding, Z.; Miao, L.; Tothova, Z.; Horner, J.W.; Carrasco, D.R.; et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 2007, 128, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi-Yamaguchi, Y.; Furuichi, T. The Homer family proteins. Genome Biol. 2007, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; He, L.; Li, S.; Li, F.; Deng, G.; Huang, X.; Yang, M.; Xiao, Y.; Chen, X.; Ouyang, Y.; et al. HOMER3 facilitates growth factor-mediated beta-Catenin tyrosine phosphorylation and activation to promote metastasis in triple negative breast cancer. J. Hematol. Oncol. 2021, 14, 6. [Google Scholar] [CrossRef]
- Cui, X.; Liang, H.; Hao, C.; Jing, X. Homer1 is a Potential Biomarker for Prognosis in Human Colorectal Carcinoma, Possibly in Association with G3BP1 Signaling. Cancer Manag. Res. 2020, 12, 2899–2909. [Google Scholar] [CrossRef]
- Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef]
- Luo, J.; Yu, F.X. GPCR-Hippo Signaling in Cancer. Cells 2019, 8, 426. [Google Scholar] [CrossRef]
- Shibata, M.; Ham, K.; Hoque, M.O. A time for YAP1: Tumorigenesis, immunosuppression and targeted therapy. Int. J. Cancer 2018, 143, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.; Itoga, K.; Okano, T.; Yonemura, S.; Sasaki, H. Hippo pathway regulation by cell morphology and stress fibers. Development 2011, 138, 3907–3914. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef]
- Codelia, V.A.; Sun, G.; Irvine, K.D. Regulation of YAP by mechanical strain through Jnk and Hippo signaling. Curr. Biol. 2014, 24, 2012–2017. [Google Scholar] [CrossRef]
- Benham-Pyle, B.W.; Pruitt, B.L.; Nelson, W.J. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science 2015, 348, 1024–1027. [Google Scholar] [CrossRef]
- Zhu, L.; Tian, Q.; Gao, H.; Wu, K.; Wang, B.; Ge, G.; Jiang, S.; Wang, K.; Zhou, C.; He, J.; et al. PROX1 promotes breast cancer invasion and metastasis through WNT/beta-catenin pathway via interacting with hnRNPK. Int. J. Biol. Sci. 2022, 18, 2032–2046. [Google Scholar] [CrossRef]
- Tocci, J.M.; Felcher, C.M.; Garcia Sola, M.E.; Kordon, E.C. R-spondin-mediated WNT signaling potentiation in mammary and breast cancer development. IUBMB Life 2020, 72, 1546–1559. [Google Scholar] [CrossRef]
- Li, S.; Mai, H.; Zhu, Y.; Li, G.; Sun, J.; Li, G.; Liang, B.; Chen, S. MicroRNA-4500 Inhibits Migration, Invasion, and Angiogenesis of Breast Cancer Cells via RRM2-Dependent MAPK Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 21, 278–289. [Google Scholar] [CrossRef]
- Qiu, X.; Li, X.; Yan, Y.; Cai, Y.; Liang, Q.; Peng, B.; Xu, Z.; Xiao, M.; Xia, F.; Peng, J. Identification of m6A-Associated Gene DST as a Prognostic and Immune-Associated Biomarker in Breast Cancer Patients. Int. J. Gen. Med. 2022, 15, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Fu, X.; Xu, S.; Qiu, P.; Lv, Z.; Cui, M.; Zhang, Q.; Xu, Y. OLR1 is a prognostic factor and correlated with immune infiltration in breast cancer. Int. Immunopharmacol. 2021, 101, 108275. [Google Scholar] [CrossRef]
- Turnbull, C.; Hines, S.; Renwick, A.; Hughes, D.; Pernet, D.; Elliott, A.; Seal, S.; Warren-Perry, M.; Gareth Evans, D.; Eccles, D.; et al. Mutation and association analysis of GEN1 in breast cancer susceptibility. Breast Cancer Res. Treat. 2010, 124, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Teng, Z.; van Duijnhoven, F.J.B.; Dong, M.; Qian, Y.; Yu, H.; Yang, J.; Han, R.; Su, J.; Du, W.; et al. Interactions between RASA2, CADM1, HIF1AN gene polymorphisms and body fatness with breast cancer: A population-based case-control study in China. Oncotarget 2017, 8, 98258–98269. [Google Scholar] [CrossRef] [PubMed]
- Damiano, J.S.; Wasserman, E. Molecular pathways: Blockade of the PRLR signaling pathway as a novel antihormonal approach for the treatment of breast and prostate cancer. Clin. Cancer Res. 2013, 19, 1644–1650. [Google Scholar] [CrossRef]
- Gerashchenko, T.S.; Zavyalova, M.V.; Denisov, E.V.; Krakhmal, N.V.; Pautova, D.N.; Litviakov, N.V.; Vtorushin, S.V.; Cherdyntseva, N.V.; Perelmuter, V.M. Intratumoral Morphological Heterogeneity of Breast Cancer As an Indicator of the Metastatic Potential and Tumor Chemosensitivity. Acta Nat. 2017, 9, 56–67. [Google Scholar] [CrossRef]
- Wang, Z.; Li, T.E.; Chen, M.; Pan, J.J.; Shen, K.W. miR-106b-5p contributes to the lung metastasis of breast cancer via targeting CNN1 and regulating Rho/ROCK1 pathway. Aging 2020, 12, 1867–1887. [Google Scholar] [CrossRef]
- Sun, W.L.; He, L.Y.; Liang, L.; Liu, S.Y.; Luo, J.; Lv, M.L.; Cai, Z.W. Ambra1 regulates apoptosis and chemosensitivity in breast cancer cells through the Akt-FoxO1-Bim pathway. Apoptosis Int. J. Program. Cell Death 2022, 27, 329–341. [Google Scholar] [CrossRef]
- Watanabe, S.; Watanabe, K.; Akimov, V.; Bartkova, J.; Blagoev, B.; Lukas, J.; Bartek, J. JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nat. Struct. Mol. Biol. 2013, 20, 1425–1433. [Google Scholar] [CrossRef]
- Nusse, R. Wnt signaling and stem cell control. Cell Res. 2008, 18, 523–527. [Google Scholar] [CrossRef]
- Schulte, G. International Union of Basic and Clinical Pharmacology. LXXX. The class Frizzled receptors. Pharmacol. Rev. 2010, 62, 632–667. [Google Scholar] [CrossRef] [PubMed]
- Roarty, K.; Serra, R. Wnt5a is required for proper mammary gland development and TGF-beta-mediated inhibition of ductal growth. Development 2007, 134, 3929–3939. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, M.; Dejmek, J.; Bendahl, P.O.; Andersson, T. Loss of Wnt-5a protein is associated with early relapse in invasive ductal breast carcinomas. Cancer Res. 2002, 62, 409–416. [Google Scholar]
- Escuin, D.; Lopez-Vilaro, L.; Mora, J.; Bell, O.; Moral, A.; Perez, I.; Arqueros, C.; Garcia-Valdecasas, B.; Ramon, Y.C.T.; Lerma, E.; et al. Circulating microRNAs in Early Breast Cancer Patients and Its Association With Lymph Node Metastases. Front. Oncol. 2021, 11, 627811. [Google Scholar] [CrossRef] [PubMed]
- Terkelsen, T.; Russo, F.; Gromov, P.; Haakensen, V.D.; Brunak, S.; Gromova, I.; Krogh, A.; Papaleo, E. Secreted breast tumor interstitial fluid microRNAs and their target genes are associated with triple-negative breast cancer, tumor grade, and immune infiltration. Breast Cancer Res. 2020, 22, 73. [Google Scholar] [CrossRef]
- Fassan, M.; Baffa, R.; Palazzo, J.P.; Lloyd, J.; Crosariol, M.; Liu, C.G.; Volinia, S.; Alder, H.; Rugge, M.; Croce, C.M.; et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009, 11, R58. [Google Scholar] [CrossRef]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.L.; Hu, G.D.; Wang, X.F.; Zhang, X.H.; Zhang, Y.K.; Yu, Z.S. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J. Int. Med. Res. 2012, 40, 859–866. [Google Scholar] [CrossRef]
- Ahmad, A.; Sethi, S.; Chen, W.; Ali-Fehmi, R.; Mittal, S.; Sarkar, F.H. Up-regulation of microRNA-10b is associated with the development of breast cancer brain metastasis. Am. J. Transl. Res. 2014, 6, 384–390. [Google Scholar]
- Liang, A.L.; Zhang, T.T.; Zhou, N.; Wu, C.Y.; Lin, M.H.; Liu, Y.J. MiRNA-10b sponge: An anti-breast cancer study in vitro. Oncol. Rep. 2016, 35, 1950–1958. [Google Scholar] [CrossRef]
- Kim, J.; Siverly, A.N.; Chen, D.; Wang, M.; Yuan, Y.; Wang, Y.; Lee, H.; Zhang, J.; Muller, W.J.; Liang, H.; et al. Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways. Cancer Res. 2016, 76, 6424–6435. [Google Scholar] [CrossRef] [PubMed]
- Bahena-Ocampo, I.; Espinosa, M.; Ceballos-Cancino, G.; Lizarraga, F.; Campos-Arroyo, D.; Schwarz, A.; Maldonado, V.; Melendez-Zajgla, J.; Garcia-Lopez, P. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016, 17, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Fang, J.; Lv, X.; Pei, Z.; Wang, Y.; Jiang, S.; Ding, K. Heparin impairs angiogenesis through inhibition of microRNA-10b. J. Biol. Chem. 2011, 286, 26616–26627. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Shehata, H.H.; Essawy, N.O. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol 2015, 36, 4487–4494. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Wang, B.; Li, J.; Han, J.; Zhao, Y.; Su, W.; Dai, Z.; Wang, X.; Ma, Q. The expression and significance of five types of miRNAs in breast cancer. Med. Sci. Monit. Basic Res. 2014, 20, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, J.; Zhang, P.Y.; Zhang, Y.; Sun, S.Y.; Yu, S.Y.; Xi, Q.S. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med. Sci. Monit. 2012, 18, BR299–BR308. [Google Scholar] [CrossRef]
- Chang, C.H.; Fan, T.C.; Yu, J.C.; Liao, G.S.; Lin, Y.C.; Shih, A.C.; Li, W.H.; Yu, A.L. The prognostic significance of RUNX2 and miR-10a/10b and their inter-relationship in breast cancer. J. Transl. Med. 2014, 12, 257. [Google Scholar] [CrossRef]
- Anfossi, S.; Giordano, A.; Gao, H.; Cohen, E.N.; Tin, S.; Wu, Q.; Garza, R.J.; Debeb, B.G.; Alvarez, R.H.; Valero, V.; et al. High serum miR-19a levels are associated with inflammatory breast cancer and are predictive of favorable clinical outcome in patients with metastatic HER2+ inflammatory breast cancer. PLoS ONE 2014, 9, e83113. [Google Scholar] [CrossRef]
- Uen, Y.; Wang, J.W.; Wang, C.; Jhang, Y.; Chung, J.Y.; Tseng, T.; Sheu, M.; Lee, S. Mining of potential microRNAs with clinical correlation—Regulation of syndecan-1 expression by miR-122-5p altered mobility of breast cancer cells and possible correlation with liver injury. Oncotarget 2018, 9, 28165–28175. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X. miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charged multivesicular body protein 3 through mitogen-activated protein kinase signaling. J. Cell Physiol. 2020, 235, 2825–2835. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, H.; Yang, Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS ONE 2012, 7, e47053. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).