Abstract
Background: Long intergenic non-coding RNA, is one type of lncRNA, exerting various cellular activities, as does ncRNA, including the regulation of gene expression and chromatin remodeling. The abnormal expression of lincRNAs can induce or suppress carcinogenesis. Main body: LincRNAs can regulate cancer progression through different mechanisms and are considered as potential drug targets. Genetic variations such as single nucleotide polymorphisms (SNPs) in lincRNAs may affect gene expression and messenger ribonucleic acid (mRNA) stability. SNPs in lincRNAs have been found to be associated with different types of cancer, as well. Specifically, LINC00511 has been known to promote the progression of multiple malignancies such as breast cancer, colorectal cancer, lung cancer, hepatocellular carcinoma, and others, making it a promising cancer prognostic molecular marker. Conclusion: LincRNAs have been proved to be associated with different cancer types through various pathways. Herein, we performed a comprehensive literature and in silico databases search listing lncRNAs, lincRNAs including LINC00511, lncRNAs’ SNPs, as well as LINC00511 SNPs in different cancer types, focusing on their role in various cancer types and mechanism(s) of action.
1. Introduction
The study of epigenetics, namely, non-protein coding RNAs (ncRNAs), has had significant increased attention recently [1]. ncRNAs are involved in a variety of physiological activities, including the regulation of gene expression and RNA splicing [2]. Long ncRNAs (lncRNA) have a length of more than 200 nucleotides and do not code for proteins [3]; at every stage of gene expression, they act as either an indicator-, decoy-, or scaffold-, and guide-lncRNA [4]. A variety of human disorders have been linked to abnormal lncRNA expression [5]. For example, Nicotinamide nucleotide transhydrogenase-antisense 1 (NNT-AS1) overexpression has been observed in a variety of malignancies, including osteosarcoma, cervical cancer (CC), breast cancer (BC), colorectal cancer (CRC) [6], gastric cancer (GC), Hepatocellular carcinoma (HCC), and non-small cell lung cancer (NSCLC). The biological roles and expression levels of lncRNA transcripts may be affected by variations in lnc gene loci [7].
1.1. Long Non-Coding RNAs (LncRNAs)
According to the Genomics Of Long non-coding RNA and Disease Lab GOLDLab [8] https://www.gold-lab.org/why-lncrnas (accessed on 31 March 2023), the number of protein-coding genes is ~19,000, and until now, <2000 lncRNAs have been investigated; therefore, 98% of lncRNAs are completely uncharacterized, making it worth studying to find new treatment targets, in other words, to find precision prognostic molecular markers. According to their different genetic origins, lncRNAs can be categorized into five groups: sense, antisense, intronic, bidirectional, and intergenic lncRNAs. Sense-lncRNA coincides with the exons of the corresponding protein-coding gene on the sense RNA strand. Antisense lncRNA is derived from the antisense (AS) RNA strand of the protein-coding gene [9]. Intronic lncRNA is the lncRNA that originates from the introns of the protein-coding gene [10]. Moreover, from the promoter of a protein-coding gene, bidirectional lncRNA is transcribed, but in the opposite direction. Finally, long intergenic non-protein coding RNAs (LincRNAs) are the lncRNAs that are situated between two protein-coding genes [11]. RNA polymerase II (Pol II) is primarily responsible for lncRNAs transcription, making them processed in a less efficient manner and more retained in the nucleus rather than the cytoplasm. Nuclear lncRNAs have a role in condensate formation or can be bound to chromatin [12].
1.2. LncRNAs and, in Particular, LINC RNA Worth Studying
LincRNAs make more than 50% of lncRNAs [13]. LincRNAs are lncRNAs that are interspersed between coding genes and do not overlap any protein-coding sequences. LincRNAs perform physiological processes like inflammation during infection [14]. LincRNAs exhibit tissue-specific expression [15], being essential for many cellular activities including the control of gene expression [16]. LincRNAs have a pathological role during cancer development [14], when the control of gene expression is perturbed after lincRNA overexpression or mutation.
1.3. Review Methodology
A manual online search into two medical e-databases, PUBMED and Google Scholar, for (“lincRNA”) AND (“lincRNA Role in Carcinogenesis”) AND (“lncRNA linc00511”) AND (“lincRNA SNPs”) AND (“future promising biomarkers”) was done in September, 2022. Priority was given to meta-analysis, randomized clinical studies, systematic review, original papers, and narrative reviews, since, but not limited to, 2010.
1.4. Review Aim
Introducing lncRNAs and their classification, briefly listing “LincRNAs in different cancer types” after identifying lncRNAs and lincRNAs via in silico databases search, mentioning LincRNAs’ role and mechanism of action in various cancer types, highlighting “LINC00511 in several cancer types”, and finally, listing published documented “Single nucleotide polymorphisms (SNPs) variants of lncRNAs and LINC00511 in different cancer types and SNPs role/mechanism of action influencing cancer risk”.
2. Long Intergenic Non-Coding RNAs (LincRNAs) and Their Involvement in Cancer
2.1. In Silico Databases Search (accessed on 22 November 2022 and Revised on 31 March 2023)
2.1.1. According to the Database by Ghent University LNCipedia
Version 5.2 [17] https://lncipedia.org/ in total, there were 127,802 transcripts and 56,946 genes for lncRNA sequence and annotation. The lncRNA class is intergenic and the Sequence Ontology term is lincRNA, where 87 lncRNA transcripts are found (Table 1).

Table 1.
Examples of lncRNAs transcripts ID, their gene ID, strand (+/−) and chromosomal location according to LNCipedia v5.2.
2.1.2. Pseudogenes-Derived LncRNAs (a Hot Area for Research; A Research Gap to Tackle)
Pseudogenes are defective copies of the genes that do not code for proteins [18]. Pseudogenes were considered as junk genes which have no functions; however, recent studies proved that pseudogene-derived lncRNAs have a role in various cancer types through being key regulators at DNA, RNA, or protein levels [19]. LincRNAs that are approved pseudogenes loci according to the National Human Genome Research Institute (NHGRI) grant HUGO gene nomenclature committee (HGNC) https://www.genenames.org/ [20] are four; LINC00265-2P [21], LINC00265-3P [22], LINC00268-2P [23] and LINC00328-2P [24] https://www.genenames.org/tools/search/#!/?query=gene_symbol:linc&rows=20&start=0&fil-ter=document_type:%22gene%22&filter=status:%22Approved%22&filter=locus_group:%22Pseudogene%22 Search (accessed on 22 November 2022 and Revised on 31st March 2023).
2.2. What Are LincRNAs?
From the intergenic spaces between two genes, polymerase enzyme II transcribes lincRNAs. The majority of annotated lincRNAs have several exons, cap, and a poly(A) tail that are similar to those of mRNA [25]. LincRNAs have a wide range of functions, including regulating epigenetic changes and regulating gene expression, as well as acting as scaffolds for protein signaling complexes [26]. LincRNA genes differ from mRNA-encoding genes in that they perform crucial roles such as chromatin remodeling and genome architectural remodeling, RNA stabilization or enhancer-associated activity [27], and finally, transcription regulation/control of neighbor genes as well [28].
2.2.1. LincRNA Role in Chromatin Remodeling
LincRNAs control gene expression through interacting with chromatin-modifying complexes to alter the later state [29]. Chromatin-modifying enzymes can be repressive or activating, or these enzymes may occasionally have bivalent domains that perform both activities (repression-activation) [30]. As shown in Figure 1a, lincRNAs can function in a cis-acting manner (cis-acting lincRNA) or trans-acting manner on gene expression. Cis-acting lincRNAs influence the expression of genes on the same chromosome close to their transcriptional location. On the contrary, trans-acting lincRNAs regulate gene expression at distinct, distant loci on a different chromosome [31]. For example, a cis-acting lincRNA HOXA transcript at the distal tip (HOTTIP) induces the expression of the HOXA gene. Trans-acting lincRNA HOX transcript antisense RNA 12 (HOTAIR12) silences HOXD gene as well as genes on other chromosomes [27]. LincRNAs interact with/via transcription factors (TFs) directly or indirectly to drive chromatin-modifying enzymes bound to RNA toward particular genomic locations [32].
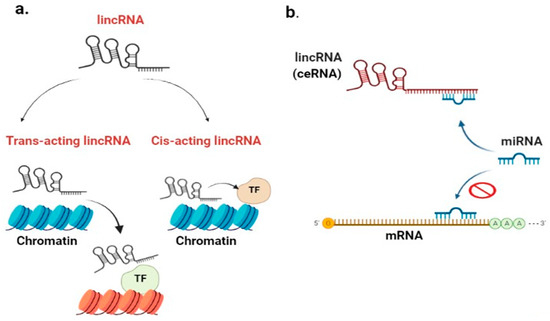
Figure 1.
LincRNA role in chromatin remodeling: (a) cis-acting lincRNA influences the expression of genes on the same chromosome that are close to its transcriptional location. Contrarily, trans-acting lincRNA can regulate gene expression at distinct loci on a different chromosome, with the involvement of TFs. LincRNA role as competitive endogenous RNA (ceRNA); (b) lincRNA competes for miR and acts as micro-RNA sponge; consequently, miR will bind to lincRNA instead of binding to mRNA, leading to an increased mRNA expression after being free. [lincRNA: long intergenic non-coding RNA; ceRNA: competitive endogenous RNA; TF: transcription factor; miR: micro-RNA; mRNA: messenger ribonucleic acid].
2.2.2. LincRNA Role in DNA Damage Repair (DDR)
LincRNAs take part in various stages of the DNA repairing process. LincRNA-p21 regulates apoptosis and cancer cell growth by blocking the translation of the target gene and activating p53 signaling [33] as the lincRNA-p21/dec. downstream target gene/p53 axis. LincRNA-p21 was found to interact with heterogeneous nuclear ribonucleoprotein-K (hnRNP-K), a protein that is directed to the tumor suppressor p53 promoters target genes, leading to transcriptional repression of p53-regulated genes [34]. LINC Regulator Of Reprogramming (Linc-ROR) regulates p53 translation, resulting in driving tumorigenesis in many cancers [35]. As a result of exposure to either exogenous or endogenous environmental stressors, DNA double-strand breaks (DSBs) abrasions occur in both DNA strands, damaging them [36]. LincRNAs have role in DSB repair through two pathways, namely, homologous recombination (HR) and the non-homologous end-joining (NHEJ) pathway. NHEJ involves ligation of the break ends without the need for a homologous template. Whereas in HR, a homologous template sequence is needed [37]. Prostate cancer associated transcript-1 (PCAT-1) is the first lincRNA known to play a role in DSBs repair [38].
2.2.3. LincRNA Role as a Competitive Endogenous RNA (ceRNA)
The interaction between lncRNAs/micro-RNAs/mRNAs makes up a complex regulatory network system, known as the competitive endogenous (ceRNA) network [39]. ceRNAs play a remarkable role in cancer and gene regulation. RNA-induced silencing complex (RISC) maintains this interaction and determines the post transcriptional regulation level of gene expression [40]. As shown in Figure 1b, lincRNAs have the potential to compete for micro-RNA (miR) and behave as a miR sponge. This later process attenuates miR activity and increases the expression of target mRNA genes [39]. LINC00691 is an example of ceRNA that competes for miR-1256 and regulates the expression or suppression of tumorigenicity 5 (ST5), leading to the suppression of sarcoma [41]. LINC00511, overexpressed in BC, has been found to sponge miR-185-3p [42].
2.2.4. LincRNAs Role as Protein Scaffold (PS)
Utilizing scaffolding molecules, which can bring several components together and direct them to enhance their activities, is one method by which the cell can overcome the difficulty of coordinating certain interactions [43]. In the nucleus, scaffold Polycomb repressive complex proteins (PRCP) lincRNA has an impact on the accessibility of chromatin, gene expression, and the structure of the nucleus [44]. Terminal differentiation-induced non-coding RNA (TINCR) lncRNA facilitates the post-transcriptional stability and accumulation of mRNAs that promote epidermal development by scaffolding staufen1, an RNA-binding protein, with the TINCR lincRNA box motif [27].
2.3. LincRNAs in Different Types of Cancer
Using RNADisease v4.0; RNA-Disease Repository; RNA-associated diseases, providing RNA-disease analysis, enrichment, and prediction; http://www.rnadisease.org/download for lncRNA-disease information (accessed on 22 November 2022 and revised on 31 March 2023).
2.3.1. LincRNAs List in Different Types of Cancer, Their Role and Mechanism(s) of Action (Table 2)
MALAT1 may represent a potential non-invasive biomarker for HCV-related hepatocellular carcinoma (HCC) prognosis, via sponging miR-204, miR-143, miR-195, miR-490, miR-216b, miR-146-5p [45]. LINC00657 can inhibit glioblastoma (GBM) through sponging miR-190a-3p and the regulation of Phosphatase and tensin homolog (PTEN) expression [46]. Moreover, LINC00707 was found to contribute to glioma cells proliferation, invasion, and migration by sponging miR-613 [47]. Elevated expression of LINC00152 sponges miR-193b-3p to induce phosphorylation and activation of the PI3K signaling pathway and downstream AKT, resulting in tongue squamous cell carcinoma (TSCC) progression [48]. LINC00662 has been discovered to promote prostate cancer tumorigenesis through sponging miR-34a [49]. LINC00657 was found to inhibit CC by sponging miR-20a-5p and the upregulation of RUNX Family Transcription Factor 3 (RUNX3) [50]. LINC01567 can regulate the proliferation of colon cancer stem cells (CSCs) through sponging miR-9, resulting in the modulation of Cyclin D2 (CCND2) and the regulation of aquaporin 3 (AQP3), which can be regulated by the CREB molecule in the cAMP–PKA pathway [51].
Also, LINC00473 contributes to the proliferation and migration of GC by acting as a ceRNA of miR-16-5p [52]. Moreover, LINC00355 can promote the progression of GC by the regulation of the wingless-INT (Wnt)/β-catenin signaling pathway [53]. In addition, Linc01555 contributes to GC cell proliferation through interacting with the Notch signaling pathway [54,55]. HOTAIR is upregulated in Laryngeal squamous cell carcinoma (LSCC), inducing invasiveness, progression, and resistance to apoptosis in LSCC cells through promoting PTEN methylation [56]. Moreover, LINC00673 promotes the progression of lung adenocarcinoma through the activation of the Wnt/B-catenin pathway [57,58]. Again, MALAT-1 contributes to the development of different types of cancer through interaction with Serine/Arginine splicing factors and changing their distribution to nuclear speckle domains [59]. Furthermore, Myocardial Infarction Associated Transcript (MIAT) can promote neuroblastoma by the modulation of MYCN and Paired-like homeobox 2b (PHOX2B) driver genes [60].

Table 2.
LincRNAs list in different types of cancer, expression level if upregulated or downregulated, sponging-miR, mechanism(s) of action, role in cancer as oncogene or tumor suppressor.
Table 2.
LincRNAs list in different types of cancer, expression level if upregulated or downregulated, sponging-miR, mechanism(s) of action, role in cancer as oncogene or tumor suppressor.
| Cancer Type | LincRNA | Expression | Sponging miR- | Mechanism of Action [Ref.] | Role in Cancer | Approach of the Study | Type of Samples Used in the Study |
|---|---|---|---|---|---|---|---|
| HCC | MALAT-1 | Upregulated | 204, 143, 195, 490, 216b, 146-5p | Promoting disease progression [45] | Oncogene | Knockdown of MALAT-1 | Human blood samples |
| GBM | LINC00657 | Downregulated | 190a-3p | Regulation of PTEN expression [46] | Tumor suppressor | Overexpression of LINC00657 | Human GBM tissues vs. adjacent normal tissues and GBM cell lines U-87 MG, LN-18, U-118 MG vs. astrocyte HA1800 |
| LINC00707 | Upregulated | 613 | Promotes progression, migration and invasion of glioma cells [47] | Oncogene | Knockdown of LINC00707 | Human Glioma tissues vs. adjacent normal tissues and glioma cell lines U87, U251, SHG-44, A172, T98G vs. normal astrocyte cell lines NHA, human embryonic kidney cell line HEK-293 | |
| TSCC | LINC00152 | Upregulated | 193b-3p | PI3K signaling pathway activation and downstream AKT enhancing cell cycle progression, tumor migration, invasion [48] | Oncogene | Knockdown of LINC00152 | Human TSCC tissues vs. adjacent normal tissues and cell lines SCC-9, CAL-27 |
| Prostate | LINC00662 | Upregulated | 34a | Promotes cancer progression [49] | Oncogene | Knockdown of LINC00662 | Human Prostate cancer tissues vs normal tissues and prostate cancer cells DU145, 22RV1, PC-3, and LNCaP vs. normal prostate epithelial cells WPMY-1 |
| CC | LINC00657 | Downregulated | 20a-5p | Upregulation of RUNX3 that targets DR5 leading to activation of NK cells [50] | Tumor suppressor | Overexpression of LINC00657 | Human CC tissues vs normal tissues and CC cell lines SiHa, HeLa, C33A, Caski vs. normal cervical squamous cell line Ect1/E6E7 |
| Colon CSCs | LINC01567 | Upregulated | 9 | CCND2 modulation and AQP3 regulation CREB/cAMP–PKA and proliferation and tumorigenesis regulation of colon CSCs [51] | Oncogene | Knockdown of LINC01567 | Human Colon cancer tissues vs. normal tissues |
| GC | LINC00473 | Upregulated | 16-5p | Modulating CCND2 expression, Promoting progression of GC and migration [52] | Oncogene | Knockdown of LINC00473 | Human GC tissues vs. normal tissues and GC cell lines BGC823, AGS, MKN-45, NCI-N87, SGC7901 vs GES-1 and female BALB/c-nude mice for implantation |
| LINC00355 | Upregulated | - | Regulating Wnt/β-catenin, promoting progression and inhibition of apoptosis [53] | Oncogene | Knockdown of LINC00355 | Human GC tissues vs normal tissues and GC cell lines BGC-823, MGC-803, AGS, SGC-7901 vs normal gastric epithelial cells GES-1 | |
| LINC01555 | Upregulated | - | Interacting with Notch signalling pathway for progression of GC [54,55] | Oncogene | Knockdown of LINC01555 | Human GC tissues vs para-carcinoma tissues and GC cell lines MGC803, MKN45, BSG823, SGC7901, vs normal human gastric mucosal epithelial cell GES-l | |
| LSCC | HOTAIR | Upregulated | - | Promoting PTEN methylation, progression, invasiveness, resistance to apoptosis [56] | Oncogene | Knockdown of HOTAIR | Human LSCC tissues vs adjacent normal tissues and and mice BALB/c for implantation |
| Lung adenocarcinoma | LINC00673 | Upregulated | - | Activation of Wnt/B-catenin for progression of the disease [57,58] | Oncogene | Knockdown of LINC00673 | Human lung adenocarcinoma cell lines HCC827, NCI-H1650, A549, NCI-H596, NCI-H1975, NCI-H1299, SK-LU-1, NCI-H358, NCI-H2009, HCC4006, NCI-H2030, PC9, and nude mice for implantation |
| Various cancer types | MALAT-1 | Upregulated | - | Interaction with Serine/Arginine splicing factors, changing distribution to nuclear speckle domains, promoting progression of the disease [59] | Oncogene | Knockdown of MALAT-1 | Human HeLa cells |
| Neuro-blastoma | MIAT | Upregulated | - | Modulation of MYCN and PHOX2B driver genes leading to progression of the disease [60] | Oncogene | Knockdown of MIAT | RNA sequencing data analysis |
| BC | Linc-ROR | Upregulated | 205, 145 | EMT induction and promoting metastasis [61] | Oncogene | Knockdown of Linc-ROR | Human BC tissues vs adjacent normal tissues and MCF10A, MDA-MB-231, BT549, BT474, MDA-MB-436, MDA-MB-435, HEK 293 and immunodeficient nude mice for implantation |
| LincRNA-BC2 | Upregulated | - | Interacting with BRCA1 and BRCA2 [62] | Oncogene | Knockdown of LincRNA-BC2 | Human BC tissues vs adjacent normal tissues | |
| TNBC | LINC00299 | Hyper-methylated | - | Hypermethylation [63] | Oncogene | Knockdown of LINC00299 | Human blood samples |
| BC | LINC00641 | Downregulated | 194-5p | Inhibition of cell growth, invasion, migration [64] | Tumor suppressor | Overexpression of LINC00641 | Human BC tissues vs adjacent normal tissues and BCAP-37, MDA-MB-453, UACC-812, MCF-7, MDA-MB-231 vs normal breast epithelial cell line MCF-10A |
| TNBC | LINC00993 | Downregulated | - | Generating G0/G1 arrest and regulation of p21 and p53 genes [65] | Tumor suppressor | Overexpression of LINC00993 | Human BC cell lines MDA-MB-231, BT-549 and female BALB/c nude mice for implantation |
| BC | LINC00885 | Upregulated | - | EGFR, EREG, FOXM1 and TP53 activation, progression of early stage BC [66] | Oncogene | Knockdown of LINC00885 | Human BC cell lines MCF10 DCIS.COM, MDA-MB-231, MCF7, T47D vs normal breast epithelial cell lines MCF10A, 184A1 |
| Linc-APOC1P1-3 | Upregulated | - | Binding tubulin to decrease α-tubulin acetylation and inactivate caspase-3, BC progression, prevention of BC cells apoptosis [67] | Oncogene | Knockdown of Linc-APOC1P1-3 | Human BC tissues vs normal tissues | |
| Linc-HOTAIR | Upregulated | - | Interaction with PRC2 and Promoting BC metastasis [68] | Oncogene | Knockdown of Linc-HOTAIR | Human BC tissues vs normal tissues, human cell lines MDA-MB-231, SK-BR-3, MCF-10A, MCF-7, HCC1954, T47D, MDA-MB-453, H16N2 and nude mice for implantation | |
| LINC00657 | Upregulated | 590-3p | GOLPH3 upregulation leading to invasion, migration, proliferation and inhibition of apoptosis of BC cells [69] | Oncogene | Knockdown of LINC00657 | Human BC tissues vs adjacent normal tissues and human BC cell lines MCF-7, MDA-MB-231, T47D, BT-549 vs normal breast epithelial cell line MCF-10A | |
| LINC00511 | Upregulated | 185-3p | - [42] | Oncogenic | Knockdown of LINC00511 | Human blood samples | |
| LINC00460 | Upregulated | 320a | MAL2 upregulation and promoting cancer cell proliferation and migration [70] | Oncogene | Knockdown of LINC00460 | MDA-MB-231, BT-549 cells | |
| LINC00922 | Upregulated | - | Promoting NKD2 methylation, promoting tumorigenesis, invasion, metastasis and regulation EMT [71] | Oncogene | Knockdown of LINC00922 | Human BC tissues vs adjacent normal tissues and human MCF-7, MDA-MB-231, SK-BR3, MCF10A | |
| CRC | LINC01088 | Upregulated | 548b-5p and 548c-5p | G3BP1 expression upregulation, enhancing CRC progression [72] | Oncogene | Knockdown of LINC01088 | Human CRC tissues vs para-cancerous tissues, human CRC cell lines, colonic epithelial cells, and mice for implantation |
[MALAT1: Metastasis Associated Lung Adenocarcinoma Transcript 1; PTEN: Phosphatase and tensin homolog; TSCC: Tongue Squamous Cell Carcinoma; SCC: Squamous Cell Carcinoma; CC: Cervical Cancer; RUNX3: RUNX Family Transcription Factor 3;CSCs: Cancer Stem Cells; CCND2: Cyclin D2; AQP3: aquaporin; 3GC: Gastric Cancer; ceRNA: competing endogenous RNA; Wnt: Wingless/Integrated; HOTAIR:HOX transcript antisense RNA; LSCC: laryngeal squamous cell carcinoma; MIAT: Myocardial Infarction Associated Transcript; PHOX2B: Paired-like homeobox 2b; Linc-ROR: Long Intergenic Non-Protein Coding RNA, Regulator Of Reprogramming; BC: Breast Cancer; EMT: Epithelial–Mesenchymal Transition; LincRNA-BC2: long intergenic non-coding RNA-Breast Cancer 2; BRCA1: Breast Cancer 1; BRCA2: Breast Cancer 2; RPISeq: RNA-Protein Interaction; TNBC: Triple Negative Breast Cancer; EGFR: epidermal growth factor receptor, EREG: epiregulin; FOXM1: forkhead box M1; Linc-APOC1P1-3: long intergenic non-coding RNA APOC1P1-3; PRC2: Polycomb Repressive Complex 2; GOLPH3: Golgi phosphoprotein 3; MAL2: Myelin And Lymphocyte protein 2; NKD2: Naked cuticle homolog 2; G3BP1: G3BP Stress Granule Assembly Factor 1].
2.3.2. LincRNAs List in Breast Cancer (BC)
Linc-ROR could promote BC metastasis through sponging miR-205, miR-145, resulting in epithelial–mesenchymal transition (EMT) induction [61]. LincRNA-BC2 is upregulated in BC, with interaction with BC associated protein antigens 1 and 2 (BRCA1 and BRCA2), which are predicted by RNA-Protein Interaction Prediction (RPISeq) [62]. Moreover, LINC00299 was found to be a promising biomarker for triple negative BC (TNBC) by its hypermethylation [63]. It has been reported that LINC00641 can be a target for BC treatment, since it inhibits proliferation, invasion, and migration of BC cells via sponging miR-194-5p [64]. LINC00993 suppresses the TNBC growth in vitro and in vivo by generating G0/G1 arrest and regulation of the genes related to cell cycle such as p21 and p53 [65]. LINC00885 was reported to act as ceRNA and regulate BC cells growth. In addition, LINC00885 is associated with early stage BC through the activation of epidermal growth factor receptor (EGFR), epiregulin (EREG), and forkhead box M1 (FOXM1) pathways and signaling pathways linked to TP53 signaling [66]. Liao et al. have found that Linc-APOC1P1-3 expression induces proliferation and inhibits apoptosis of BC cells via binding tubulin to decrease α-tubulin acetylation and, therefore, inactivates caspase-3 [67]. Researchers pointed to Linc-HOTAIR interaction with Polycomb Repressive Complex 2 (PRC2) promoting BC metastasis [68]. LINC00657 was found to play a role in the biological behavior of BC such as invasion, migration, proliferation, and apoptosis. It can sponge miR-590-3p and upregulates Golgi Phosphoprotein 3 (GOLPH3) [69]. LINC00511 was found by our group to be overexpressed in BC patient’s blood, with sponging miR-185-3p, and was considered as an early diagnostic biomarker for BC [42]. BC cell migration and proliferation was promoted by LINC00460 via sponging miR-320a and upregulation of MAL2 [70]. In addition, LINC00922 could induce BC invasion, metastasis, progression, and regulation of EMT, by NKD2 methylation [71]. All lincRNAs roles and mechanisms of action in BC are mentioned in Table 2. LINC01088 was found to bind to miR-548b-5p and miR-548c-5p to upregulate G3BP1 expression, resulting in the enhancement of CRC progression with immune scape and finally, changing cancer cell phenotypes [72].
As summarized in Table 2 “Long intergenic non-coding RNAs (lincRNAs) and their involvement in cancer”, LincRNAs have several important functions, such as chromatin remodeling, DNA damage/repair, and acting as ceRNA or protein scaffold. There are various lincRNAs that have a role in different types of cancer, leading to an increase or decrease in its risk with different mechanisms.
3. LINC00511 and Its Contribution in Different Cancer Types
3.1. LINC00511 In Silico Info (Accessed on 25 November 2022 and Revised on 31 March 2023)
The International Cancer Genome Consortium (ICGC) data portal [73,74] identified LINC00511 gene as 2265-bp, which is localized on chromosome 17q24.3 with five exons https://dcc.icgc.org/genes/ENSG00000227036 [75]. Bulk tissue gene expression for LINC00511 (ENSG00000227036.6) from the Genotype-Tissue Expression (GTEx) project helped to study tissue-specific gene expression and regulation, where 54 non-diseased tissue sites samples were collected from 1000 individuals for molecular assays. GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2) https://www.gtexportal.org/home/gene/LINC00511. Figure 2 shows the LINC00511 expression in different tissues. The highest expression of LINC00511 was found in the sun-exposed skin tissues.
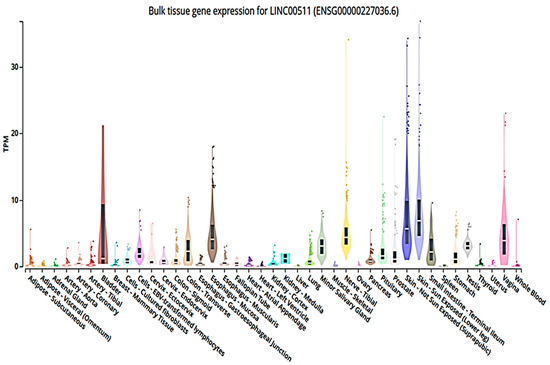
Figure 2.
Tissue gene expression graph for LINC00511 in some tissue sites. The expression graph of LINC00511 is represented in transcripts per million (TPM), as TPM is an accurate statistic used when calculating gene expression comparisons across samples. Here, Figure 2 represents the expression values of LINC00511 in non-diseased tissue sites samples collected from 1000 individuals for molecular assays. As shown, the highest expression of LINC00511 was found in the sun-exposed skin tissues from GTEx Analysis https://www.gtexportal.org/home/gene/LINC00511 (accessed on 25 November 2022 and revised on 31 March 2023).
3.2. LINC00511 in Cancer
LINC00511 was first discovered by Cabanski et al. in 2015 [76]. LINC00511 is dysregulated in multiple malignancies including glioma, BC, ovarian cancer, CC, osteosarcoma, HCC, lung cancer, TSCC, renal cell cancer, papillary thyroid carcinoma, pancreatic cancer, GC [77], and CRC [78]. This dysregulation has a role in facilitating the prognosis of cancer [79]. First, LINC00511 sponge miRNAs and alter the expression of their targets. Second, through interacting with enzymes and TFs associated to DNA methylation, LINC00511 can control tumor suppressors or oncogenes to promote tumorigenesis [77]. Additionally, LINC00511 promotes cell proliferation, cell cycle progression, tumorigenesis, invasion, and metastasis [75]. According to Lu et al., LINC00511 can promote tumor growth and metastasis and induce stemness in malignancies. In order to prevent cancer metastasis, it may be possible to employ LINC00511-modifying modalities [80]. Role of LINC00511 Expression in Different Types of Cancer (Table 3).

Table 3.
LINC00511 role in different types of cancer, expression level if upregulated or downregulated, sponging-miR, mechanism of action, role in cancer as oncogene or tumor suppressor.
3.2.1. LINC00511 Role in Colorectal Cancer (CRC)
Hypoxia Inducible Factor 1 (HIF-1) could activate LINC00511, to sponge miR-153-5p in CRC cells. MiR-153-5p targets HIF-1’s 3-UTR, forming a positive feedback loop of HIF-1/LINC00511/miR-153-5p in CRC cells [78]. LINC00511 interacts with enhancer of zeste homolog 2 (EZH2), leading to downregulation of interleukin-24 (IL-24) expression. Lu et al. found Hepatocyte Nuclear Factor4 (HNF4) could promote LINC00511 transcription to accelerate cancer progression; therefore, the LINC00511/EZH2/IL-24 axis is a potential therapeutic target [81]. Hu et al. found LINC00511 to sponge miR-29c-3p, leading to upregulation of Nuclear Factor I A (NFIA) with subsequent progression of CRC [82]. Moreover, LINC00511 promotes CRC progression by suppressing miRNA-625-5p to enhance WEE1 protein [83].
3.2.2. LINC00511 Role in Lung Cancer
Zhu et al. have found that LINC00511 promotes NSCLC through binding to lysine-specific demethylase 1 (LSD1) and EZH2, resulting in Large Tumor Suppressor Kinase 2 (LATS2) and KLF Transcription Factor 2 (KLF2) genes inhibition [84]. LINC00511 mediates oncogenesis of NSCLC via binding EZH2, silencing p57 expression. LINC00511 knockdown inhibited carcinogenesis in vivo and slowed cell proliferation via promoted apoptosis in vitro [85]. In addition, LINC00511 enhances the progression of lung adenocarcinoma by sponging miR-625-5p and regulating Pyruvate kinase M2 (PKM2) expression [86]. Zhang et al. have reported that LINC00511 promotes lung cancer progression by binding to miR-195-5p and upregulating glucosaminyl (N-acetyl) transferase 3 (GCNT3) [87]. LINC00511 can target miR-625-5p/GSPT, contributing to NSCLC proliferation and invasion [88]. However, inhibiting miR-150-5p and activating Transcriptional Adaptor 1 (TADA1), now LINC00511 can enhance the proliferation and migration of lung squamous cell carcinoma [89].
3.2.3. LINC00511 Role in Cervical Cancer (CC)
LINC00511 promotes CC by upregulating phospholipase D1 (PLD1) expression through transcription factor retinoic X receptor alpha (RXRA). Inhibition of LINC00511 induces apoptosis and decreases the progression of CC [90]. Mao et al. have found that downregulation of LINC00511 in CC cells increased sensitivity to paclitaxel, lowered cancer cell viability, proliferation, and induced apoptosis, resulting in CC recurrence prevention through regulating B-cell lymphoma 2 (Bcl-2), Bcl-2 Associated X-protein (Bax), metalloproteinases 2 and 9, multidrug resistance protein 1 (MRP1), P-glycoprotein, and cleaved caspase-3 [91]. LINC00511 can sponge miR-324-5p with regulation of the DRAM1 axis [92]. Moreover, LINC00511 enhances the proliferation and progression of CC through regulation of the miR-497-5p/MAPK1 axis [93].
3.2.4. LINC00511 Role in Gastric Cancer (GC)
LINC00511 promotes GC cells tumorigenesis and stemness by sponging miR-195-5p, then elevating SRY-box transcription factor 4 (SOX4), followed by repressing PTEN, which will activate the PI3K/AKT pathway via recruited EZH2 [94]. Moreover, LINC00511 increases GC cells proliferation by sponging miR-515-5p [95]. LINC00511 enhances GC cells growth by sponging miR-124-3p and regulating the miR-124-3p/PDK4 axis [96]. LINC00511 promotes gastric tumorigenesis by sponging miR-625-5p and targeting the nuclear factor 1/X gene (NFIX) [97].
3.2.5. LINC00511 Role in Pancreatic Cancer (PC)
LINC00511 competitively endogenously inhibits hsa-miR-29b-3p activity to upregulate vascular endothelial growth factor A (VEGFA), promoting pancreatic ductal adenocarcinoma (PDAC) stemness. Therefore, LINC00511 is considered as a promising biomarker that can be used to predict PDAC patients prognosis following surgery, and could be a therapeutic target [98].
3.2.6. LINC00511 Role in Hepatocellular Cancer (HCC)
LINC00511 could competitively interact with miR-424 to promote HCC proliferation and metastasis [99]. LINC00511 promotes HCC development by competing with miR-195 and positively correlating with Eyes absent homolog 1 (EYA1) [100]. Moreover, LINC00511 accelerate HCC progression, acting as a prognostic biomarker for the disease. However, it has an adverse interaction with miRNA-29c in HCC [101]. It is noteworthy to mention that LINC00511 promotes HCC invasion via affecting exosome secretion and invadopodia formation [102].
3.2.7. LINC00511 Role in Glioblastoma (GBM)
Du et al. discovered LINC00511 to act as a ceRNA sponging miR-524-5p, indirectly controlling the Y box binding protein 1 (YB1), boosting Zinc finger E-box-binding homeobox 1 (ZEB1) expression. This enhanced LINC00511 expression in a reverse way constructing LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop that encourage GBM cell migration and invasion [103]. Via targeting miR-15a-5p/AE Binding Protein 1 (AEBP1) axis, LINC00511 knockdown can prevent glioma cell carcinoma development [104]. LINC00511 sponges miR-126-5p and activate Wnt/β-catenin signaling, facilitating Temozolomide resistance of GBM [105].
3.2.8. LINC00511 Role in Osteosarcoma (OS)
LINC00511 induces OS via sponging miR-618 and boosting Maelstrom Spermatogenic Transposon Silencer (MAEL) expression [106]. LINC00511 sponges miR-185-3p leading to E2F transcription factor 1 (E2F1) expression regulation and promoting OS [107], or sponging miR-765, which promotes apurinic/apyrimidinic endonuclease 1 (APE1) in OS cells [108]. In contrast, another study demonstrated that LINC00511 is downregulated in OS, inhibits cell proliferation and increases apoptosis of OS cells [109].
3.2.9. LINC00511 Role in Different Types of Breast Cancer (BC)
It has been reported that the LINC00511/miR-150/MMP13 axis contributes to BC proliferation and migration. LINC00511 has the ability to sponge miR-150, leading to regulation of expression of Matrix Metallopeptidase 13 (MMP13) and promoting cell migration [110]. DNA hypomethylation induces LINC00511 expression and LINC00511 promotes BC growth by upregulating Wnt family member 10A (Wnt10A), E2F transcription factor 2 (E2F2), Transforming growth factor-alpha (TGF-A), and MET [111]. The LINC00511/miR-185-3p/E2F1/Nanog axis promotes the BC cells growth, via sponging miR-185-3p and target E2F1 protein that binds with the Nanog promoter region to activate its transcription [80]. LINC00511 has been proved to increase the expression of BC cells as well as the transcriptional control of downstream genes through an elevated LINC00511/miR-185-3p axis. Therefore, LINC00511 can be considered a marker for BC early diagnosis [42]. LINC00511 is a TNBC-specific lncRNA that functions as an oncogene to control tumor metastasis and prognosis [112]. LINC00511 enhances ER-negative BC cell growth by altering cell proliferation and apoptosis by accelerating the G1/S transition and suppressing apoptosis [113]. Zhang et al. has proved that LINC00511 downregulation enhances paclitaxel cytotoxicity in BC cells by acting as a miR-29c molecular sponge [114]. Moreover, inhibition of LINC00511 reduces its competitive binding to miR185, leading to higher STXBP4 expression and better radiation response in BC [115].
As well summarized in Table 3 “LINC00511 and its contribution in different cancer types”, different studies have proved that LINC00511 is upregulated in various types of cancer. For example, in CRC, LINC00511 promotes the progression of the disease through sponging miR-153-5p, miR-29-3p, and miR-625-5p. In addition, LINC00511 induces the proliferation of lung cancer by sponging miR-625-5p, miR-195-5p, and miR-150-5p. The progression of CC can be promoted by LINC00511 through sponging miR-324-5p and miR-497-5p. LINC00511 can stimulate GC tumorigenesis by sponging miR-195-5p, miR-515-p, miR-124-3p, and miR-625-5p. Moreover, LINC00511 can induce BC oncogenesis through sponging miR-150, miR-185, miR-185-3p, and miR-29, in addition to other discussed types of cancer that can be induced by LINC00511. Although different studies have reported that LINC00511 is upregulated in OS, one study has proved that it is downregulated in OS and inhibits cell proliferation.
4. LncRNAs SNPs in Different Cancer Types and Their Mechanism of Action
Unveiling the relationship between lncRNAs SNPs or specifically, LINC RNA SNPs and disease mechanism(s) is an important research gap to investigate, as future prospective, to relate cancer incidence or progression/remission to specific variants. This will be a step toward ncRNA precision, fulfilling big pharma’s shift toward targeting RNA for treatment instead of DNA or classical cancer-hallmarks. In the human genome, Over 10 million SNPs have been identified resulting in gene variants, which change the cell’s protein production machinery [116]. SNPs can affect RNA-RNA interaction (lncRNA-miRNA interactions) through a ceRNA mechanism, where lncRNA could competitively bind miRNAs [117]. It has been reported that SNPs within lncRNA transcripts can impact the structure and function of lncRNA, whereas SNPs in an lncRNA’s promoter region might affect its expression level [116]. Furthermore, lncRNAs SNPs that change the structure of the lncRNA influence the interaction between the RNA Binding Proteins (RBPs) and lncRNA, resulting in the regulation of several biological pathways [118]. It has been found that lincRNA SNPs in exon loci may alter the secondary structure of the lincRNA. For example, SNP rs1456315 G/A in lincRNA PRNCR1alters its secondary structure and hence, the conformation and stability of lincRNA, even causing changes in its interacting partners [117].
4.1. LncRNAs SNPs List in Breast Cancer, Their Role and Mechanism of Action (Table 4)
CDKN2B-AS1, also named ANRIL, is an lncRNA that can interfere with the expression of neighboring genes, control cell proliferation and apoptosis, and is upregulated in BC. Researchers proved that SNP rs310965215 in ANRIL altered cells’ ability to proliferate, invade, and migrate by sponging miR-4440 [119]. Another study demonstrated that SNPs rs1333045, rs1333048, rs4977574, and rs10757278 in ANRIL increase BC risk [120]. However, MALAT1 SNPs (rs3200401, rs619586, and rs7927113) have an association with BC susceptibility. Fortunately, genotypes AG and AG + GG of MALAT1 SNP rs619586 protect against BC, and CT of rs3200401 reduces BC risk [121]. Growth Arrest Specific 5 (GAS5) is a tumor suppressor and is downregulated in many cancer types including BC [122]. The GAS5 SNP rs145204276 del allele may inhibit BC development by increasing the promoter activity via binding to the TF specificity protein 1 [123]. Cancer susceptibility candidate 15 (CASC15) is a very active lncRNA in silico and is found on chromosome 6p22.3. An interaction between lncRNA CASC15 polymorphisms and susceptibility of BC has been found; rs7740084 and rs1928168 reduced BC risk, whereas, there is a correlation between rs9393266 and BC risk [124]. Similarly, HOTAIR SNP rs920778 elevates BC risk and might interact with the clinical reproductive factors [125]. MiR2052HG rs34841297 regulates miR-4456 expression, which alters BC cells proliferation and invasion, increasing BC susceptibility [126]. LINC00520 is found on human chromosome 14q22.3 [127] and is upregulated in various tumors including LSCC, nasopharyngeal carcinoma, and renal cell carcinoma. Increased TNBC susceptibility may be exploited by LINC00520 SNP rs8012083 [128]. Also, SNP rs527616 in lncRNA AQP4-AS1 increases BC susceptibility [129]. SOX2OT is an lncRNA located in the SOX2 gene in the intronic region. Via affecting the expression of SOX2OT, lncRNA SOX2OT SNP rs9839776 increases BC risk [130]. Again, lncRNA H19 SNPs (rs3741219, rs217727, and rs2839698) increase BC risk, whereas rs3741216 decreased it [131]. SRA is an lncRNA upregulated in BC and its expression correlates with levels of ER and PR. SNP rs10463297 in lncRNA SRA increased BC risk through affecting SRA mRNA expression [132]. SNPs rs11657109, rs17780195, and rs9906859 in LINC00511 may protect against BC, being related to LINC00511 secondary structure and expression [133]. Our group is currently investigating LINC00511 SNPs in BC as well as CRC patients’ blood samples (publication in progress). LincRNA-ROR SNP rs4801078 was correlated to BC risk, being affected by the interplay between linc-ROR SNPs and reproductive factors [134].

Table 4.
LncRNAs SNPs lists in Breast Cancer (BC), their mechanism of action and role in BC.
Table 4.
LncRNAs SNPs lists in Breast Cancer (BC), their mechanism of action and role in BC.
| LncRNA List | SNPs List | Mechanism of Action [Ref.] | Role in BC | Type of Samples Used in the Study |
|---|---|---|---|---|
| CDKN2B-AS1; ANRIL | rs310965215 | Sponging miR-4440 [117] | Cells’ altered ability to proliferate, invade, migrate | Extracted DNA from human blood samples |
| rs1333045, rs1333048, rs4977574, and rs10757278 | - [120] | Increased risk | ||
| MALAT1 | rs7927113 | - [121] | Association with BC susceptibility; AG, AG + GG | |
| rs619586 | - [121] | Protects against | ||
| rs3200401 | - [121] | Reduces risk | ||
| GAS5 | rs145204276 | Increasing promoter activity, binding TF specificity protein 1, raise GAS5 [123] | Inhibition of BC development | |
| CASC15 | rs7740084, rs1928168 | - [122] | Reduce risk | |
| rs9393266 | - [124] | Correlated to risk | ||
| HOTAIR | rs920778 | Interaction with reproductive factors [125] | Elevation of risk | |
| MIR2052HG | rs34841297 | Regulation of miR-4456 expression [126] | Increased susceptibility | |
| LINC00520 | rs8012083 | - [128] | Increased TNBC susceptibility | |
| AQP4-AS1 | rs527616 | - [129] | Increased susceptibility | DNA extracted from human blood samples and BC tissues vs. normal tissues |
| SOX2OT | rs9839776 | Influencing SOX2OT expression [130] | Increases risk and related to onset | Extracted DNA from human blood samples |
| H19 | rs3741219, rs217727, rs2839698 | - [128] | Increased risk | |
| rs3741216 | - [131] | Decreased risk | ||
| SRA | rs10463297 | Affecting SRA mRNA expression [132] | Increased risk | |
| LINC00511 | rs11657109, rs17780195, rs9906859 | - [133] | Protection | |
| Linc-ROR | rs4801078 | Interplay with reproductive factors [134] | Increased risk |
[CDKN2B-AS1: cyclin-dependent kinase inhibitor 2B antisense RNA 1, ANRIL: antisense non-coding RNA in the INK4 locus; BC: Breast Cancer; ER: Estrogen Receptor; PR: Progesterone Receptor; MALAT1: Metastasis Associated Lung Adenocarcinoma Transcript 1; GAS5: Growth Arrest Specific 5; CASC15: Cancer Susceptibility 15; HOTAIR: HOX transcript antisense RNA; PCAT1: Prostate Cancer Associated Transcript 1; MIR2052HG: MIR2052 Host Gene; LINC00520: Long Intergenic Non-Protein Coding RNA 520; TNBC: Triple Negative Breast Cancer; AQP4-AS1: AQP4 Antisense RNA 1; SOX2OT: SOX2 overlapping transcript; SRA: steroid receptor RNA activator; mRNA: messenger RNA; Linc-ROR: Long Intergenic Non-Protein Coding RNA, Regulator Of Reprogramming].
4.2. LncRNAs SNPs List in Lung Cancer, Their Role and Mechanism of Action (Table 5)
NEAT1 is a lncRNA located on chromosome 11q13.1, and is a component of paraspeckles. As seen in Table 5, SNP rs2239895 in lncRNA NEAT1 increases the risk of lung squamous cell carcinoma [135]. CCAT1 is located on chromosome 8q24.21 and is overexpressed in several tumors such as GC, CRC, and HCC. SNP rs1948915 in lncRNA CCAT1 is associated with decreased lung cancer susceptibility, in the study’s female population cohort [136]. Researchers have found that SNP rs219741 in lncRNA LOC105369301 elevates the risk of NSCLC, while SNP rs498238 in lncRNA LINC01833 and SNP rs16901995 in lnc-NDUFS6-5:5 all reduce the NSCLC risk [137]. Compared to people with the homozygous wild AA genotype/heterozygote GA genotype, those with the homozygous GG genotype SNP rs7248320 in lncRNA AC008392.1 had a lower chance of developing NSCLC [138]. Researchers have reached a point that SNP rs4759314 (AG genotype) in lncRNA HOTAIR can increase the risk of lung cancer development, while SNP rs12826786 (“CT” and “CT + TT” genotypes) decreased this risk [139].
Moreover, SNPs rs920778 and rs1899663 in lncRNA HOTAIR have been found to increase lung cancer susceptibility [140]. Epidermal growth factor receptor (EGFR) wild type lung adenocarcinoma, the GAS5 SNP rs145204276, may aid in tumor stage, distal metastases, and lymph node metastasis prediction [141]. PRNCR1 is an lncRNA located on chromosome 8q24.21 and is a popular oncogene in prostate cancer [142]. A study has found that the PRNCR1 SNP rs1456315 T allele compared with the C allele and the lncRNA CCAT2 SNP rs6983267 G allele, compared with the T allele, increased lung cancer risk. These SNPs affect the lncRNA secondary structure as well as the miRNAs target [143]. There has been an association between SNP rs3200401 in lncRNA MALAT1 and susceptibility of lung squamous cell carcinoma and NSCLC, via altered MALAT1’s structural properties and downstream genes contributing to the formation and progression of cancer [144]. HOXA11-AS is a lncRNA whose ectopic expression plays important roles in different cancer types. SNP rs17427875 (T allele) in HOXA11-AS increases the risk of lung adenocarcinoma, whereas SNP rs11564004 (G allele) plays a protective role, with TFs as mediators [145]. SNP rs217727 (A/A homozygous genotype) in lncRNA H19 is associated with elevated lung cancer risk, particularly adenocarcinoma and squamous cell carcinoma [146]. LncRNA LOC146880 is upregulated in NSCLC and is associated with poor prognosis of the disease. The A allele in rs140618127 SNP in lncRNA LOC146880 decreases NSCLC risk. LOC146880 offers microRNA miR-539-5p an alternative binding site, altering ENO1 phosphorylation with PI3K and Akt pathway’s activation [147].

Table 5.
LncRNAs SNPs list in lung cancer, their mechanism of action and role in lung cancer.
Table 5.
LncRNAs SNPs list in lung cancer, their mechanism of action and role in lung cancer.
| LncRNA List | SNPs List | Mechanism of Action [Ref.] | Role in Lung Cancer | Type of Samples Used in the Study |
|---|---|---|---|---|
| NEAT1 | rs2239895 | - [135] | Increased carcinoma risk | Extracted DNA from human blood samples |
| CCAT1 | rs1948915 | - [136] | Decreased cancer in females’ | |
| LOC105369301 | rs219741 | - [137] | Elevated risk | |
| LINC01833 | rs498238 | - [137] | Elevated risk | |
| lnc-NDUFS6-5:5 | rs16901995 | - [137] | Reduced risk | |
| AC008392.1 | rs7248320 | - [138] | Reduced risk in GG genotype | |
| HOTAIR | rs4759314 | - [139] | Increases cancer risk | |
| rs12826786 | - [137] | “CT” and “CT + TT” decreases risk | ||
| HOTAIR | rs920778 and rs1899663 | - [140] | Increased susceptibility | |
| GAS5 | rs145204276 | - [141] | Aiding in tumor stage, distal metastases, LN metastasis prediction, in EGFR wild type patients | |
| PRNCR1 | rs1456315 | Affecting lncRNA secondary structure and target miRNAs [143] | Increased risk in patients with T allele | |
| CCAT2 | rs6983267 | Affecting the secondary structure of lncRNA and target of miRNAs [143] | Increased risk of lung cancer in patients with G allele | |
| MALAT1 | rs3200401 | MALAT1’s structural properties alterationand cancer genes expression [144] | Increased susceptibility | |
| HOXA11-AS | rs17427875 | Associating with TFs [145] | (T allele) increases risk | |
| rs11564004 | - [143] | (G allele) play a protective role | ||
| H19 | rs217727 | - [146] | Elevated risk in A/A homozygous | |
| LOC146880 | rs140618127 | miR-539-5p alternative binding site, ENO1 phosphorylation, PI3K/Akt activation [147] | Decreased risk | Human NSCLC tissues vs. adjacent normal tissues and human NSCLC cell lines A549, PC9 vs. human lung epithelial BEAS-2B cells |
[NEAT1: Nuclear Enriched Abundant Transcript 1; CCAT1: Colon Cancer Associated Transcript 1; NSCLC: Non-Small Cell Lung Cancer; HOTAIR: HOX transcript antisense RNA; GAS5: Growth Arrest Specific 5; EGFR: Epidermal Growth Factor Receptor; PRNCR1: Prostate Cancer Associated Non-Coding RNA 1; lncRNA: long non coding RNA; CCAT2: Colon Cancer Associated Transcript 2; MALAT1: Metastasis Associated Lung Adenocarcinoma Transcript 1; HOXA11-AS: HOXA11 Antisense RNA].
4.3. LncRNAs SNPs List in Colorectal Cancer (CRC), Their Role and Mechanism of Action (Table 6)
As listed in Table 6, researchers found lncRNA MALAT1 SNPs rs619586, rs664589, and rs1194338 are associated with CRC risk through binding of various TFs [148]. rs67085638 in lncRNA CCAT1 increases risk of CRC, while rs7013433 is related to CRC late clinical stage [149]. On the other hand, rs2470151 (CT/TT genotype) in lncRNA RP11-108K3.2 could decrease the risk of CRC [150]. Linc-ROR SNP rs1942347 was associated with CRC large tumor size and mortality [151]. rs2839698 in lncRNA H19 associated with increased CRC risk, changing promotor activity and H19 function [152]. Moreover, H19 SNPs rs4930101, rs11042170, and rs27359703 were associated with increased risk of CRC [153]. HOTTIP is an antisense lncRNA that is upregulated in different tumors including CRC. SNPs rs3807598, rs2067087, and rs17427960 in HOTTIP increase susceptibility to CRC, through affecting Transcription Factor Binding Sites (TFBSs) according to SNP function prediction [154]; however, rs1859168 regulates lncRNA gene expression [155]. A positive association between rs55829688 SNP in GAS5 and CRC risk reduced GAS5 expression by altering the TF YY1′s affinity for GAS5 [156]. It has been reported that SNP rs2632159 in lncRNA PCAT1 can elevate the risk of CRC [157]. It has been demonstrated that lncRNA PUNISHER “AGAP2-AS1” has a role in inducing CRC cell proliferation, epithelial-to-mesenchymal transition, and enhancement of CRC cells’ chemoresistance to gemcitabine. PUNISHER is associated with elevated risk of CRC, tumor relapse, and short survival time, via rs12318065. This SNP could modify regulatory motifs such as MRG1, Sin3Ak-20 disc6, and HOXA9 1 that have been found to be linked to CRC, or affecting binding with the TFs POL2, ZNF263, and STAT1, associated with carcinogenesis [158]. It has been demonstrated that lncRNA MAGI2-AS3 acts as a tumor suppressor in many cancers; however, it helps in CRC progression. An increased risk for CRC is due to SNP rs7783388 in lncRNA MAGI2-AS3, via influencing the binding ability of glucocorticoid receptor (GR) to the MAGI2-AS3 promoter [159]. SNHG16 is a lncRNA located on human chromosome 17q25.1, upregulated in BC, bladder cancer, and CRC, where it affects the expression of genes associated with lipid metabolism. SNP rs7353 in lncRNA SNHG16 suppresses CRC, while rs8038 and rs15278 increase this risk [160]. Via binding to miR-4658 and impairing the expression of lncRNA CCSlnc362 “RP11-362K14.5” in CRC cells [161], SNP rs1317082 can protect against CRC. LincRNA Papillary Thyroid Carcinoma Susceptibility Candidate 3 (PTCSC3) is considered as a tumor-suppressor in thyroid cancer and glioma [162]. SNP rs944289 in PTCSC3 decreases CRC risk [163]. rs1456315 in lncRNA prostate cancer non-coding RNA (PRNCR1) increases CRC risk [164]. LncRNA MEG3 SNP rs7158663 elevates CRC risk [165]. By losing binding of miR-128-3p to LAMC2-1:1, rs2147578 in lncRNALAMC2-1:1 increased CRC risk [166]. SNP rs7958904 in lncRNA HOTAIR is associated with both CRC mortality and incidence [167]. LncRNA UCA1 is upregulated in different types of cancer including CRC, BC, bladder cancer, NSCLC, esophageal cancer, and TSCC. SNP rs12982687 in lncRNA UCA1 was proved to affect UCA1’s binding to miR-873-5p and HIF-1 signaling, resulting in a contribution to the progression of smoking-triggered CRC [168]. SNPs rs6983267 at 8q24 and HULC rs7763881 may serve as genetic indicators of a propensity towards CRC and are correlated with CCAT2 and HULC expression, respectively [169].
TINCR lncRNA SNPs rs2288947, rs8105637 are associated with CRC progression and susceptibility. The G allele of SNP rs2288947 was associated with decreased CRC risk, while the A allele of SNP rs8105637 was associated with increased CRC risk. The mechanism involves five motifs, Nanog disc3, CTCF disc9, Rad21 disc10, SP1 disc3, and SMC3 disc3, which may be affected by rs2288947. rs8105637 affects TCF12 and PITX2 expression linked to carcinogenesis [170].

Table 6.
LncRNA SNPs list in Colorectal Cancer (CRC), their mechanism of action and role in CRC.
Table 6.
LncRNA SNPs list in Colorectal Cancer (CRC), their mechanism of action and role in CRC.
| LncRNA List | SNPs List | Mechanism of Action [Ref.] | Role in CRC | Type of Samples Used in the Study |
|---|---|---|---|---|
| MALAT1 | rs619586, rs664589, rs1194338 | Affecting binding of TFs [146] | Associated with risk | Extracted DNA from human blood samples |
| CCAT1 | rs67085638 | - [149] | Increases risk | |
| rs7013433 | - [147] | Related to CRC late clinical stage | ||
| RP11-108K3.2 | rs2470151 | - [150] | Decreased risk with CT/TT genotype | |
| LINC-ROR | rs1942347 | - [151] | Associated with large tumor size and mortality | Extracted DNA from FFPE tissue samples |
| H19 | rs2839698, rs4930101, rs11042170, rs27359703 | rs2839698 change activity of promotor and H19 function [152,153] | Increased risk | Extracted DNA from human blood samples |
| HOTTIP | rs1859168 | Regulates lncRNA gene expression [154,155] | Increased susceptibility | |
| rs3807598, rs2067087, rs17427960 | Affect TFBSs [154,155] | |||
| GAS5 | rs55829688 | Reduced GAS5 expression by altering TF YY1′s affinity to GAS5 [154] | Increased risk | |
| PCAT1 | rs2632159 | - [155] | Increased risk | |
| PUNISHER “AGAP2-AS1” | rs12318065 | Modify regulatory motifs; MRG1, Sin3Ak-20_disc6, HOXA9_1, affect TFs binding POL2, ZNF263, and STAT1 [156] | Elevated risk, tumor relapse and short survival time | Extracted DNA from FFPE tissue samples |
| MAGI2-AS3 | rs7783388 | Influencing binding ability of GR to lncRNA promoter [157] | Increased risk | Extracted DNA from human blood samples |
| SNHG16 | rs7353 | Influencing lncRNA expression [158] | Suppresses susceptibility | |
| rs8038, rs15278 | Increased risk | |||
| CCSlnc362 “RP11-362K14.5” | rs1317082 | Binding miR-4658 and impairing lncRNA expression [159] | Protection | Human CRC tissues vs. adjacent normal tissues and human CRC cell lines HCT116, DLD-1, SW480, LOVO, HT29, RKO vs. the immortalized human colorectal epithelial cell line FHC |
| PTCSC3 | rs944289 | - [163] | Decreased risk | Extracted DNA from human blood samples |
| PRNCR1 | rs1456315 | - [164] | Increased risk | |
| MEG3 | rs7158663 | - [165] | Increased risk | |
| LAMC2-1:1 | rs2147578 | Losing miR-128-3p binding [164] | Increased risk | |
| HOTAIR | rs7958904 | - [165] | Associated with mortality and incidence | |
| UCA1 | rs12982687 | Affecting UCA1’s binding to miR-873-5p and HIF-1 signaling [166] | Progression of smoking-triggered CRC | |
| HULC | rs7763881 | Correlated with expression of HULC [167] | Genetic indicator for CRC | |
| TINCR | rs2288947 | Affected motifs; Nanog_disc3, CTCF_disc9, Rad21_disc10, SP1_disc3, and SMC3_disc3 [168] | Allele G associated with decreased risk | |
| rs8105637 | TCF12 and PITX2 expression linked to carcinogenesis [168] | Allele A associated with increased risk |
[MALAT1: Metastasis Associated Lung Adenocarcinoma Transcript 1; CRC: Colorectal Cancer; CCAT1: Colon Cancer Associated Transcript 1; Linc-ROR: Long Intergenic Non-Protein Coding RNA, Regulator Of Reprogramming; FFFPE: formalin-fixed, paraffin-embedded; HOTTIP: HOXA transcript at the distal tip; TFBSs: Transcription Factor Binding Sites; GAS5: Growth Arrest Specific 5; YY1: Yin Yang 1; PCAT1: Prostate cancer-associated transcript 1; AGAP2-AS1: AGAP2 Antisense RNA 1; MRG1: melanocyte-specific gene-related gene 1; HOXA9: Homeobox protein Hox-A9; POL2: RNA Polymerase 2; ZNF263: Zinc Finger Protein 263; STAT1: Signal transducer and activator of transcription 1; FFPE: Formalin-fixed, paraffin-embedded; MAGI2-AS3: MAGI2 Antisense RNA 3; GR: Glucocorticoid Receptor; SNHG16: Small Nucleolar RNA Host Gene 16; PRNCR1: Prostate Cancer Associated Non-Coding RNA 1; MEG3: Maternally Expressed 3; HOTAIR: HOX transcript antisense RNA; UCA1: Urothelial cancer associated 1; HIF-1: Hypoxia-inducible factor; CCAT2: Colon Cancer Associated Transcript 2; HULC: Highly upregulated in liver cancer; TINCR: Terminal differentiation-induced non-coding RNA; CTCF: CCCTC-binding factor; SP1: Specificity protein 1; SMC3: Structural Maintenance Of Chromosomes 3; TCF12: Transcription Factor 12; PITX2: Paired Like Homeodomain 2].
4.4. LncRNAs SNPs List in Pancreatic Cancer, Their Role and Mechanism of Action (Table 7)
ANRIL’s SNP rs1537373 may enhance pancreatic cancer susceptibility through TFs binding and Cyclin-dependent kinase inhibitor 2B (CDKN2B) expression regulation [171]. Moreover, another study reported SNP rs1412832 in ANRIL may increase PDAC risk. Several target genes are regulated by ANRIL, including CDKN2A and p16, which typically exhibits harmful somatic and germline mutations and dysregulation [172]. SNPs rs4759314 and rs200349340 in HOTAIR lncRNA can increase pancreatic cancer susceptibility. The later SNP affects HOTAIR expression by interfering with binding with miR-29a [173]. Furthermore, the rs7046076 variant in lncRNA structural maintenance of chromosomes 2 (lnc-SMC2-1) increases PDAC risk, interfering with the lncRNA’s ability to bind to miR-1256 [174]. Although LINC00673 is an oncogene in many cancer types including NSCLC, it is a tumor suppressor in Pancreatic cancer [175]. Pancreatic cancer risk increased via creating a binding site for miR-1231 upon the occurrence of SNP rs11655237 in LINC00673, via limiting degradation of protein tyrosine phosphatase non-receptor type 11 (PTPN11) [176]. On the other hand, Hu et al. have found that SNP rs1859168 in HOTTIP may reduce pancreatic cancer susceptibility by suppressing HOTTIP expression [177].

Table 7.
LncRNAs SNPs list in pancreatic cancer, their mechanism of action and role in pancreatic cancer.
Table 7.
LncRNAs SNPs list in pancreatic cancer, their mechanism of action and role in pancreatic cancer.
| LncRNA List | SNPs List | Mechanism of Action [Ref.] | Role in Pancreatic Cancer | Type of Samples Used in the Study |
|---|---|---|---|---|
| ANRIL | rs1537373 | Affecting TF binding and regulating CDKN2B expression [171] | Increased susceptibility | Extracted DNA from human blood samples |
| rs1412832 | CDKN2A, p16 exhibit harmful somatic, germline mutations and dysregulation [172] | Increased risk | ||
| HOTAIR | rs4759314 | - [173] | Increased susceptibility | |
| rs200349340 | Interfering with binding of miR-29a [173] | |||
| lnc-SMC2-1 | rs7046076 | Interfering with binding to miR-1256 [174] | Increased risk | |
| LINC00673 | rs11655237 | Binding site for miR-1231 and limits PTPN11 degradation [176] | Increased risk | Extracted DNA from human PDAC tissues vs. adjacent normal tissues |
| HOTTIP | rs1859168 | - [177] | Decreased susceptibility | Extracted DNA from human blood samples |
[ANRIL: Antisense non-coding RNA in the INK4 locus, CDKN2B: Cyclin-dependent kinase inhibitor 2B, CDKN2A: Cyclin-dependent kinase inhibitor 2A, HOTAIR: HOX transcript antisense RNA, lnc-SMC2-1: LncRNA Structural Maintenance of Chromosomes 2, PTPN11: Protein Tyrosine Phosphatase Non-receptor type 11, HOTTIP: HOXA transcript at the distal tip].
4.5. LncRNAs SNPs List in Hepatocellular Carcinoma, Their Role and Mechanism of Action (Table 8)
SNP rs7958904 in HOTAIR, SNPs rs3931282, rs1134492, and rs10589312 in Plasmacytoma variant translocation 1 (PVT1) and SNP rs84557 in Epidermal growth factor receptor-Antisense RNA 1 (EGFR-AS1) have been linked to the occurrence and prognosis of HCC through affecting their binding to different effector miRs, as listed in Table 7 [178]. HOTTIP SNPs rs2067087, rs17501292, and rs17427960 and rs4102217 in the MALAT1 lncRNA increased HCC susceptibility by the regulation of certain motifs that elevate the expression of these carcinogenic lncRNA [179]. SNP rs2839698 in lncRNA-H19 can predict the risk and prognosis of HCC [180]. LN metastasis and HCC’s increased susceptibility were related to SNP rs9914618 in LINC00673 [181].

Table 8.
LncRNAs SNPs list in HCC, their mechanism of action and role in HCC.
Table 8.
LncRNAs SNPs list in HCC, their mechanism of action and role in HCC.
| LncRNA List | SNPs List | Mechanism of Action [Ref.] | Role in HCC |
|---|---|---|---|
| HOTAIR | rs7958904 | Binding miR-615-3p [176] | Linked to incidence and prognosis |
| PVT1 | rs3931282, rs1134492, rs10589312 | Bind miR-205-5p, 34b-5p, 183-3p, 31-5p [176] | |
| EGFR-AS1 | rs84557 | Binding miR-33b-5p [176] | |
| HOTTIP | rs2067087, rs17501292, rs17427960 | Regulation of certain motifs [177] | Increased susceptibility |
| MALAT1 | rs4102217 | ||
| H19 | rs2839698 | - [178] | Prediction of risk and prognosis |
| LINC00673 | rs9914618 | - [179] | Increased susceptibility and LN metastasis |
DNA was extracted from human blood samples. [HCC: Hepatocellular Carcinoma, HOTAIR: HOX transcript antisense RNA, PVT1: Plasmacytoma variant translocation 1, EGFR-AS1: Epidermal growth factor receptor-Antisense RNA 1, HOTTIP: HOXA transcript at the distal tip, MALAT1: Metastasis Associated Lung Adenocarcinoma Transcript 1, LN: Lymph Node].
Summary of Point 4 “LncRNAs SNPs in different cancer types and their mechanism of action”: SNPs in lncRNAs can increase or decrease the risk of various cancer types such as BC, CRC, HCC, etc., through different mechanisms (summarized in Table 4, Table 5, Table 6, Table 7 and Table 8). For example, SNP rs920778 in HOTAIR increases the risk of BC, while SNP rs3200401 in MALAT1 decreases the risk of BC. These studies could help in the progress of cancer treatments.
5. Summary and Conclusions
One abundant class of lncRNAs is lincRNAs, which are involved in various important biological processes. Numerous lincRNAs have been proved to be related to cancer, either being oncogenic, increasing cancer risk, susceptibility, progression, and/or metastasis, or decreasing cancer risk, being tumor suppressors, through different mechanisms of TFs or E2F, signaling pathways, or sponging various miRs. Specifically, LINC00511 has a crucial role in various types of cancer. Different lncRNAs SNPs or particularly LINC00511 SNPs were associated with cancer risk/protection, through distinct pathways, that could be a potential target/hit for cancer treatment as presented in Figure 3.
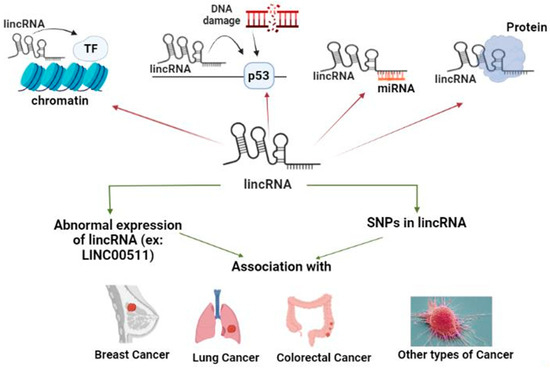
Figure 3.
LincRNAs different functions, abnormal expression, and SNPs association with various types of cancer (presented as the review conclusion summary and the graphical abstract.). LincRNAs have different functions, such as having a role in chromatin remodeling and DNA damage repair, in addition to acting as ceRNA and protein scaffold. Abnormal expression of lincRNAs, for example LINC00511, and SNPs in lincRNAs are associated with various types of cancer such as BC, lung cancer, CRC, and other types of cancer.
Implementing ncRNA measurements in blood liquid biopsy or tumor tissues will be a step toward ncRNA precision health and fulfilling both big pharma’s intention as well as the Sustainable Development Goals’ goal 3 (SDGs #3) (Better Health).
Strengths of the current review study: Our review article covered almost all lincRNAs and their role in various cancer types. LINC RNA constitute promising hit target(s) for the design of chemotherapy treatment for different types of cancer, a step toward ncRNA precision treatment.
Limitation: Pathways of some lincRNAs are still unknown or missing in different in silico/bioinformatic databases and this mandates further studies, in either clinical or in vitro experimental manners, to prove findings.
Future Prospective: More future studies are required to link lincRNAs SNPs variants and haplotypes in different types of cancer, to help pick cancer cases in an early-stage or low-grade, identifying the pre-treatment predictors of response to therapy, ensuring personalized earlier identification, improvement of patients’ survival, complementing the epigenome project, implementing ncRNA measurements in liquid biopsy or tumor tissues, a step toward ncRNA precision. Our research group “Epigenetics studies in Cancer” came into the way, at the advanced biochemistry research lab (ABRL) at the Biochemistry Dept., Faculty of Pharmacy, Ain Shams University, second to extensively studied tumor-suppressor(s) and/or oncogenic gene(s), their SNPs, variants, and haplotypes [182,183,184,185,186,187] in different cancer types.
Sustainability: as an initiative for decoding carcinogenesis from a ncRNA perspective [188], our research group are currently measuring three LINC00511 SNPs variants haplotypes in BC, HCC, and CRC clinical cohorts. Second, their link to multidrug resistance, and/or ce-miRs [189].
Author Contributions
S.E. was responsible for data curation, original draft preparation, and rewriting, first figures and tables drafts. D.N. edited, rewrote, and reviewed the manuscript. E.F.S. edited, rewrote, and careful extensive manuscript reviewing. N.M.H. was responsible for conceptualization, supervision, in silico databases search, bioinformatics software mining, review editing, rewriting, tables and figures creation, and careful extensive manuscript rewriting, reviewing from submission till acceptance. All authors approved the authorship submitted. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Figures created with subscription to BioRender.com and the partial support by ABRL #2023/9NMH8-9/10.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AEBP1 | AE Binding Protein 1 |
| AGAP2-AS1 | AGAP2 Antisense RNA 1 |
| ANRIL | Antisense non-coding RNA in the INK4 locus |
| APE1 | Apurinic/apyrimidinic endonuclease 1 |
| AQP3 | Aquaporin 3 |
| AQP4-AS1 | AQP4 Antisense RNA 1 |
| AS | Antisense |
| Bax | Bcl-2 Associated X-protein |
| BC | Breast Cancer |
| Bcl 2 | B-cell lymphoma 2 |
| BRCA1 | Breast Cancer antigen 1 |
| BRCA2 | Breast Cancer antigen 2 |
| CASC15 | Cancer Susceptibility 15 |
| CC | Cervical cancer |
| CCAT1 | Colon Cancer Associated Transcript 1 |
| CCAT2 | Colon Cancer Associated Transcript 2 |
| CCND2 | Cyclin D2 |
| CDK6 | Cyclin-dependent kinase 6 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B |
| CDKN2B-AS1 | Cyclin-dependent kinase inhibitor 2B antisense RNA 1 |
| ceRNA | competitive endogenous RNA |
| chr6 | chromosome 6 |
| CRC | Colorectal Cancer |
| CREB | cAMP response element-binding protein |
| CSCs | Cancer Stem Cells |
| CTCF | CCCTC-binding factor |
| DDR | DNA Damage Repeat |
| DRAM1 | DNA Damage Regulated Autophagy Modulator 1 |
| DSB | Double Strand Break |
| E2F1 | E2F Transcription Factor 1 |
| E2F2 | E2F Transcription Factor 2 |
| EGFR | Epidermal growth factor receptor |
| EGFR-AS1 | Epidermal growth factor receptor-Antisense RNA 1 |
| EMT | Epithelial-Mesenchymal Transition |
| ENO1 | Enolase 1 |
| ER | Estrogen Receptor |
| EREG | Epiregulin |
| EYA1 | Eyes absent homolog 1 |
| EZH2 | Enhancer of zeste homolog 2 |
| FFPE | Formalin-fixed, paraffin-embedded |
| FOXM1 | Forkhead box M1 |
| G3BP1 | G3BP Stress Granule Assembly Factor 1 |
| GACAT2 | Gastric Cancer Associated Transcript 2 |
| GAS5 | Growth Arrest Specific 5 |
| GBM | Glioblastoma |
| GC | Gastric Cancer |
| GCNT3 | Glucosaminyl (N-acetyl) transferase 3 |
| GOLPH3 | Golgi Phosphoprotein 3 |
| GR | Glucocorticoid Receptor |
| GWAS | Genome Wide Association Studies |
| HC | Hepatic Cancer |
| HCC | Hepatocellular Carcinoma |
| HCV | Hepatitis C Virus |
| HER-2 | Human epidermal growth factor receptor 2 |
| hg38 | ID used for Genome Reference Consortium Human Reference 38 |
| HIF-1 | Hypoxia Inducible Factor 1 |
| HNF4 | Hepatocyte Nuclear Factor 4 |
| hnRNP-K | heterogeneous nuclear ribonucleoprotein-K |
| HOTAIR | HOX transcript antisense RNA |
| HOTTIP | HOXA transcript at the distal tip |
| HOXA9 | Homeobox protein Hox-A9 |
| HOXA11-AS | HOXA11 Antisense RNA |
| HPV | Human papillomavirus |
| HULC | Highly upregulated in liver cancer |
| IL-24 | Interleukin 24 |
| KDM2A | Lysine-specific demethylase 2A |
| KLF2 | KLF transcription factor 2 |
| LATS2 | Large Tumor Suppressor Kinase 2 |
| LINC00511 | Long intergenic non coding RNA 00511 |
| LINC-PINT | Long Intergenic Non-Protein Coding RNA, P53 Induced Transcript |
| lincRNAs | Long intergenic non-coding RNAs |
| lincRNA-BC2 | Long intergenic non-coding RNA-Breast Cancer 2 |
| Linc-ROR | Long Intergenic Non-Protein Coding RNA, Regulator Of Reprogramming |
| LN | Lymph Node |
| LncRNA | Long non coding RNA |
| lnc-SMC2-1 | Long non coding RNA structural maintenance of chromosomes 2 |
| LSCC | Laryngeal squamous cell carcinoma |
| LSD1 | Lysine-specific demethylase 1 |
| MAEL | Maelstrom Spermatogenic Transposon Silencer |
| MAGI2-AS3 | MAGI2 Antisense RNA 3 |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 |
| MAPK1 | Mitogen-activated protein kinase 1 |
| MEG3 | Maternally Expressed 3 |
| MIR2052HG | MIR2052 Host Gene |
| miR | Micro-RNA |
| miRNA | Micro-RNA |
| MMP13 | Matrix Metallopeptidase 13 |
| MRG1 | Melanocyte-specific gene-related gene 1 |
| mRNA | Messenger ribonucleic acid |
| MRP1 | Multidrug resistance protein 1 |
| NEAT1 | Nuclear Enriched Abundant Transcript 1 |
| NFIA | Nuclear Factor 1 A |
| NFIX | Nuclear factor 1/X gene |
| NK | Natural Killer |
| NKD2 | Naked cuticle homolog 2 |
| NSCLC | Non-small cell lung cancer |
| OS | Osteosarcoma |
| PC | Pancreatic Cancer |
| PCAT-1 | Prostate Cancer Associated Transcript-1 |
| PRC2 | Polycomb Repressive Complex 2 |
| PDAC | Pancreatic ductal adenocarcinoma |
| PDK4 | Pyruvate dehydrogenase lipoamide kinase isozyme 4 |
| PHOX2B | Paired-like homeobox 2b |
| PITX2 | Paired Like Homeodomain 2 |
| PKM2 | Pyruvate kinase M2 |
| PLD1 | Phospholipase D1 |
| Pol ll | RNA Polymerase ll |
| PR | Progesterone Receptor |
| PRNCR1 | Prostate Cancer Associated Non-Coding RNA 1 |
| PS | Protein Scafold |
| PTPN11 | Protein Tyrosine Phosphatase Non-receptor type 11 |
| PTCSC3 | Papillary Thyroid Carcinoma Susceptibility Candidate 3 |
| PTEN | Phosphatase and tensin homolog |
| PVT1 | Plasmacytoma variant translocation 1 |
| RPIseq | RNA-Protein Interaction Prediction |
| RNA | Ribo Nucleic Acid |
| RUNX3 | RUNX Family Transcription Factor 3 |
| RXRA | retinoic X receptor alpha |
| SDGs #3 | Sustainable Development Goals; goal 3 |
| SMC3 | Structural Maintenance Of Chromosomes 3 |
| smORFs | Small Open Reading Frames |
| SNHG16 | Small Nucleolar RNA Host Gene 16 |
| SNPs | Single Nucleotide Polymorphisms |
| SOX2OT | SOX2 overlapping transcript |
| SOX4 | SRY-box transcription factor 4 |
| SP1 | Specificity protein 1 |
| SR | Serine/Arginine |
| SRA | Steroid Receptor RNA Activator |
| ST5 | Suppression of tumorigenicity 5 |
| STXBP4 | Syntaxin Binding Protein 4 |
| STAT1 | Signal transducer and activator of transcription 1 |
| TADA1 | Transcriptional Adaptor 1 |
| TASs | Trait-associated SNPs |
| TCF12 | Transcription Factor 12 |
| TCNR | Tumor cell necrosis rate |
| TFs | Transcription factors |
| TFBSs | Transcription Factor Binding Sites |
| TGFA | Transforming growth factor alpha |
| TINCR | Terminal differentiation-induced non-coding RNA |
| TNBC | Triple Negative Breast Cancer |
| TPM | Transcripts per million |
| TSCC | Tongue Squamous Cell Carcinoma |
| UCA1 | Urothelial cancer associated 1 |
| VEGFA | Vascular endothelial growth factor A |
| VLDLR-AS1 | VLDLR Antisense RNA 1 |
| Wnt | Wingless-INT |
| Wnt10A | Wnt Family Member 10A |
| Xist | X-inactive specific transcript |
| YB1 | Y box binding protein 1 |
| YY1 | Yin Yang 1 |
| ZEB1 | Zinc finger E-box-binding homeobox 1 |
| ZNF263 | Zinc Finger Protein 263 |
References
- Xiu, B.; Chi, Y.; Liu, L.; Chi, W.; Zhang, Q.; Chen, J.; Guo, R.; Si, J.; Li, L.; Xue, J.; et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 2019, 18, 187. [Google Scholar] [CrossRef] [PubMed]
- Luan, F.; Chen, W.; Chen, M.; Yan, J.; Chen, H.; Yu, H.; Liu, T.; Mo, L. An autophagy-related long non-coding RNA signature for glioma. FEBS Open Bio 2019, 9, 653–667. [Google Scholar] [CrossRef]
- Arun, G.; Spector, D.L. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019, 16, 860–863. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Bunch, H. Gene regulation of mammalian long non-coding RNA. Mol. Genet. Genom. 2018, 293, 1–15. [Google Scholar] [CrossRef] [PubMed]
- El-Sheikh, N.M.; Abulsoud, A.I.; Fawzy, A.; Wasfey, E.F.; Hamdy, N.M. LncRNA NNT-AS1/hsa-miR-485–5p/HSP90 axis in-silico and clinical prospect correlated-to histologic grades-based CRC stratification: A step toward ncRNA Precision. Pathol. Res. Pract. 2023, 247, 154570. [Google Scholar] [CrossRef]
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-coding RNA Gene Loci. Front. Genet. 2020, 11, 527484. [Google Scholar] [CrossRef]
- Johnson, R. What Are Long Noncoding RNAs (lncRNAs)? Goldlab. 2023. Available online: https://www.gold-lab.org/why-lncrnas (accessed on 8 February 2023).
- Sun, M.; Gadad, S.S.; Kim, D.S.; Kraus, W.L. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Mol. Cell 2015, 59, 698–711. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, K.; Yu, R.; Zhou, B.; Huang, P.; Cao, Z.; Zhou, Y.; Wang, J. From “Dark Matter” to “Star”: Insight Into the Regulation Mechanisms of Plant Functional Long Non-Coding RNAs. Front. Plant Sci. 2021, 12, 650926. [Google Scholar] [CrossRef]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef]
- Maimaitiyiming, Y.; Ye, L.; Yang, T.; Yu, W.; Naranmandura, H. Linear and Circular Long Non-Coding RNAs in Acute Lymphoblastic Leukemia: From Pathogenesis to Classification and Treatment. Int. J. Mol. Sci. 2022, 23, 4442. [Google Scholar] [CrossRef]
- Kozłowska, J.; Kolenda, T.; Poter, P.; Sobocińska, J.; Guglas, K.; Stasiak, M.; Bliźniak, R.; Teresiak, A.; Lamperska, K. Long intergenic non-coding rnas in HNSCC: From “junk dna” to important prognostic factor. Cancers 2021, 13, 2949. [Google Scholar] [CrossRef]
- Chen, H.; Shan, G. The physiological function of long-noncoding RNAs. Non-Coding RNA Res. 2020, 5, 178–184. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Louca, M.; Gkretsi, V. LincRNAs and snoRNAs in Breast Cancer Cell Metastasis: The Unknown Players. Cancers 2022, 14, 4528. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. Lncipedia 5: Towards a reference set of human long non-coding rnas. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Ding, W.; Lin, L.; Chen, B.; Dai, J. L1 elements, processed pseudogenes and retrogenes in mammalian genomes. IUBMB Life 2006, 58, 677–685. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and Their miRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 2020, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.B.E. Symbol Report for LINC00265-2P. HUGO Gene Nomencl. Comm. 2023. Available online: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:38523 (accessed on 8 February 2023).
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.B.E. Symbol Report for LINC00265-3P. HUGO Gene Nomencl. Comm. 2023. Available online: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:38536 (accessed on 8 February 2023).
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.B.E. Symbol Report for LINC00268-2P. HUGO Gene Nomencl. Comm. 2023. Available online: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:37771 (accessed on 8 February 2023).
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.B.E. Symbol Report for LINC00328-2P. HUGO Gene Nomencl. Comm. 2023. Available online: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:42027 (accessed on 8 February 2023).
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Changizian, M.; Nourisanami, F.; Hajpoor, V.; Parvaresh, M.; Bahri, Z.; Motovali-Bashi, M. LINC00467: A key oncogenic long non-coding RNA. Clin. Chim. Acta 2022, 536, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Rutenberg-Schoenberg, M.; Sexton, A.N.; Simon, M.D. The Properties of Long Noncoding RNAs That Regulate Chromatin. Annu. Rev. Genom. Hum. Genet. 2016, 17, 69–94. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Deniz, E.; Erman, B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genom. 2017, 17, 135–143. [Google Scholar] [CrossRef]
- Wongtrakoongate, P.; Riddick, G.; Fucharoen, S.; Felsenfeld, G. Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes. PLoS Genet. 2015, 11, e1005615. [Google Scholar] [CrossRef] [PubMed]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020, 10, 82. [Google Scholar] [CrossRef]
- Zhang, A.; Xu, M.; Mo, Y.-Y. Role of the lncRNA-p53 regulatory network in cancer. J. Mol. Cell Biol. 2014, 6, 181–191. [Google Scholar] [CrossRef]
- El-Sheikh, N.M.; Abulsoud, A.I.; Wasfey, E.F.; Hamdy, N.M. Insights on the potential oncogenic impact of long non-coding RNA nicotinamide nucleotide transhydrogenase antisense RNA 1 in different cancer types; integrating pathway(s) and clinical outcome(s) association. Pathol. Res. Pract. 2022, 240, 154183. [Google Scholar] [CrossRef]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Dianatpour, A.; Ghafouri-Fard, S. The Role of Long Non Coding RNAs in the Repair of DNA Double Strand Breaks. Int. J. Mol. Cell Med. 2017, 6, 1–12. [Google Scholar] [PubMed]
- Thapar, R. Regulation of DNA double-strand break repair by non-coding RNAs. Molecules 2018, 23, 2789. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Braga, E.A.; Fridman, M.V.; Moscovtsev, A.A.; Filippova, E.A.; Dmitriev, A.A.; Kushlinskii, N.E. Lncrnas in ovarian cancer progression, metastasis, and main pathways: Cerna and alternative mechanisms. Int. J. Mol. Sci. 2020, 21, 8855. [Google Scholar] [CrossRef]
- Wan, D.; Qu, Y.; Zhang, L.; Ai, S.; Cheng, L. The lncrna linc00691functions as a cerna for miRNA-1256 to suppress osteosarcoma by regulating the expression of ST5. OncoTargets Ther. 2020, 13, 13171–13181. [Google Scholar] [CrossRef]
- Mahmoud, M.M.; Sanad, E.F.; Elshimy, R.A.A.; Hamdy, N.M. Competitive Endogenous Role of the LINC00511/miR-185-3p Axis and miR-301a-3p From Liquid Biopsy as Molecular Markers for Breast Cancer Diagnosis. Front. Oncol. 2021, 11, 749753. [Google Scholar] [CrossRef]
- Spitale, R.C.; Tsai, M.C.; Chang, H.Y. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics 2011, 6, 539–543. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Toraih, E.A.; Ellawindy, A.; Fala, S.Y.; Al Ageeli, E.; Gouda, N.S.; Fawzy, M.S.; Hosny, S. Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed. Pharmacother. 2018, 102, 653–669. [Google Scholar] [CrossRef]
- Chu, L.; Yu, L.; Liu, J.; Song, S.; Yang, H.; Han, F.; Liu, F.; Hu, Y. Long intergenic non-coding LINC00657 regulates tumorigenesis of glioblastoma by acting as a molecular sponge of miR-190a-3p. Aging 2019, 11, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, K. The Long Intergenic Noncoding RNA 00707 Sponges MicroRNA-613 (miR-613) to Promote Proliferation and Invasion of Gliomas. Technol. Cancer Res. Treat. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rui, B.; Cao, Y.; Gong, X.; Li, H. Long non–coding RNA LINC00152 acts as a sponge of miRNA–193b–3p to promote tongue squamous cell carcinoma progression. Oncol. Lett. 2020, 19, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, L.-Y.; Qiao, Y.-H.; Song, R.-J. Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3688–3698. [Google Scholar]
- Qin, X.; Zhou, M.; Lv, H.; Mao, X.; Li, X.; Guo, H.; Li, L.; Xing, H. Long noncoding RNA LINC00657 inhibits cervical cancer development by sponging miR-20a-5p and targeting RUNX3. Cancer Lett. 2021, 498, 130–141. [Google Scholar] [CrossRef]
- Yu, X.; Mi, L.; Dong, J.; Zou, J. Long intergenic non-protein-coding RNA 1567 (LINC01567) acts as a “sponge” against microRNA-93 in regulating the proliferation and tumorigenesis of human colon cancer stem cells. BMC Cancer 2017, 17, 716. [Google Scholar] [CrossRef]
- Zhuo, S.; Sun, M.; Bai, R.; Lu, D.; Di, S.; Ma, T.; Zou, Z.; Li, H.; Zhang, Z. Long intergenic non-coding RNA 00473 promotes proliferation and migration of gastric cancer via the miR-16-5p/CCND2 axis and by regulating AQP3. Cell Death Dis. 2021, 12, 496. [Google Scholar] [CrossRef]
- Luan, P.-B.; Sun, X.-M.; Yao, J. LINC00355 inhibits apoptosis and promotes proliferation of gastric cancer cells by regulating Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8377–8383. [Google Scholar]
- Emam, O.; Wasfey, E.F.; Hamdy, N.M. Notch-associated lncRNAs profiling circuiting epigenetic modification in colorectal cancer. Cancer Cell Int. 2022, 22, 316. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Z. Linc01555 promotes proliferation, migration and invasion of gastric carcinoma cells by interacting with Notch signaling pathway. J. Buon 2020, 25, 1007–1012. [Google Scholar] [PubMed]
- Li, D.; Feng, J.; Wu, T.; Wang, Y.; Sun, Y.; Ren, J.; Liu, M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am. J. Pathol. 2013, 182, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Abd El Fattah, Y.K.; Abulsoud, A.I.; AbdelHamid, S.G.; Hamdy, N.M. Interactome battling of lncRNA CCDC144NL-AS1: Its role in the emergence and ferocity of cancer and beyond. Int. J. Biol. Macromol. 2022, 222, 1676–1687. [Google Scholar] [CrossRef]
- Guan, H.; Zhu, T.; Wu, S.; Liu, S.; Liu, B.; Wu, J.; Cai, J.; Zhu, X.; Zhang, X.; Zeng, M.; et al. Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 14019–14028. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Rombaut, D.; Chiu, H.S.; Decaesteker, B.; Everaert, C.; Yigit, N.; Peltier, A.; Janoueix-Lerosey, I.; Bartenhagen, C.; Fischer, M.; Roberts, S.; et al. Integrative analysis identifies lincRNAs up- and downstream of neuroblastoma driver genes. Sci. Rep. 2019, 9, 5685. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Ding, X.; Zhu, L.; Ji, T.; Zhang, X.; Wang, F.; Gan, S.; Zhao, M.; Yang, H. Long Intergenic Non-Coding RNAs (LincRNAs) Identified by RNA-Seq in Breast Cancer. PLoS ONE 2014, 9, e103270. [Google Scholar] [CrossRef]
- Bermejo, J.L.; Huang, G.; Manoochehri, M.; Mesa, K.G.; Schick, M.; Silos, R.G.; Ko, Y.D.; Brüning, T.; Brauch, H.; Lo, W.Y.; et al. Long intergenic noncoding RNA 299 methylation in peripheral blood is a biomarker for triple-negative breast cancer. Epigenomics 2019, 11, 81–93. [Google Scholar] [CrossRef]
- Mao, Q.; Lv, M.; Li, L.; Sun, Y.; Liu, S.; Shen, Y.; Liu, Z.; Luo, S. Long intergenic noncoding RNA 00641 inhibits breast cancer cell proliferation, migration, and invasion by sponging miR-194-5p. J. Cell. Physiol. 2020, 235, 2668–2675. [Google Scholar] [CrossRef]
- Guo, S.; Jian, L.; Tao, K.; Chen, C.; Yu, H.; Liu, S. Novel Breast-Specific Long Non-coding RNA LINC00993 Acts as a Tumor Suppressor in Triple-Negative Breast Cancer. Front. Oncol. 2019, 9, 1325. [Google Scholar] [CrossRef]
- Abba, M.C.; Canzoneri, R.; Gurruchaga, A.; Lee, J.; Tatineni, P.; Kil, H.; Lacunza, E.; Aldaz, C.M. Linc00885 a novel oncogenic long non-coding rna associated with early stage breast cancer progression. Int. J. Mol. Sci. 2020, 21, 7407. [Google Scholar] [CrossRef]
- Liao, X.H.; Wang, J.G.; Li, L.Y.; Zhou, D.M.; Ren, K.H.; Jin, Y.T.; Lv, L.; Yu, J.G.; Yang, J.Y.; Lu, Q.; et al. Long intergenic non-coding RNA APOC1P1-3 inhibits apoptosis by decreasing α-tubulin acetylation in breast cancer. Cell Death Dis. 2016, 7, e2236. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Shan, Q.; Qu, F.; Yang, W.; Chen, N. Effect of LINC00657 on apoptosis of breast cancer cells by regulating mir-590-3p. Cancer Manag. Res. 2020, 12, 4561–4571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, L.; Li, X.; Chen, Y.; Shi, T.; Wang, Q. LINC00460 Facilitates Cell Proliferation and Inhibits Ferroptosis in Breast Cancer Through the miR-320a/MAL2 Axis. Technol. Cancer Res. Treat. 2023, 22, 15330338231164359. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, T.; Wang, P.; Li, S.; Wu, G.; Zhou, J.; Wang, Z. LINC00922 regulates epithelial-mesenchymal transition, invasive and migratory capacities in breast cancer through promoting NKD2 methylation. Cell. Signal. 2021, 77, 109808. [Google Scholar] [CrossRef]
- Li, C.; Pan, B.; Wang, X.; Liu, X.; Qin, J.; Gao, T.; Sun, H.; Pan, Y.; Wang, S. Upregulated LINC01088 facilitates malignant phenotypes and immune escape of colorectal cancer by regulating microRNAs/G3BP1/PD-L1 axis. J. Cancer Res. Clin. Oncol. 2022, 148, 1965–1982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Baran, J.; Cros, A.; Guberman, J.M.; Haider, S.; Hsu, J.; Liang, Y.; Rivkin, E.; Wang, J.; Whitty, B.; et al. International cancer genome consortium data portal-a one-stop shop for cancer genomics data. Database 2011, 2011, 1–10. [Google Scholar] [CrossRef]
- LINC00511. Int. Cancer Genome Consort. 2023. Available online: https://dcc.icgc.org/genes/ENSG00000227036 (accessed on 23 November 2022).
- Agbana, Y.L.; Abi, M.E.; Ni, Y.; Xiong, G.; Chen, J.; Yun, F.; Yi, Z.; Zhang, Q.; Yang, Z.; Kuang, Y.; et al. LINC00511 as a prognostic biomarker for human cancers: A systematic review and meta-analysis. BMC Cancer 2020, 20, 682. [Google Scholar] [CrossRef]
- Cabanski, C.R.; White, N.M.; Dang, H.X.; Silva-Fisher, J.M.; Rauck, C.E.; Cicka, D.; Maher, C.A. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015, 12, 628–642. [Google Scholar] [CrossRef]
- Ding, J.; Cao, J.; Chen, Z.; He, Z. The role of long intergenic noncoding RNA 00511 in malignant tumors: A meta-analysis, database validation and review. Bioengineered 2020, 11, 812–823. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xia, C.; Xu, Y. HIF-1α induced lncRNA LINC00511 accelerates the colorectal cancer proliferation through positive feedback loop. Biomed. Pharmacother. 2020, 125, 110014. [Google Scholar] [CrossRef]
- Chen, M.; Qi, P.; Jiang, W. Prognostic significance of long intergenic non-protein-coding RNA 511expression in malignant tumors: A systematic review and meta-analysis. Medicine 2015, 99, e23054. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, Y.; Ma, Y.; Lu, J.; Chen, Y.; Jiang, Q.; Qin, Q.; Zhao, L.; Huang, Q.; Luo, Z.; et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 2018, 37, 289. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yu, Y.; Liu, F.; Han, Y.; Xue, H.; Sun, X.; Jiang, Y.; Tian, Z. LINC00511-dependent inhibition of IL-24 contributes to the oncogenic role of HNF4α in colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G338–G350. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, Y.; Ding, M.; Xu, R. Lncrna linc00511 acts as an oncogene in colorectal cancer via sponging mir-29c-3p to upregulate nfia. OncoTargets Ther. 2020, 13, 13413–13424. [Google Scholar] [CrossRef]
- Qian, X.; Jiang, C.; Zhu, Z.; Han, G.; Xu, N.; Ye, J.; Wang, R. Long non-coding RNA LINC00511 facilitates colon cancer development through regulating microRNA-625-5p to target WEE1. Cell Death Discov. 2022, 8, 233. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Zhang, S.R.; Wang, L.H.; Wu, W.D.; Zhao, H. LINC00511 promotes the progression of non-small cell lung cancer through downregulating LATS2 and KLF2 by binding to EZH2 and LSD1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8377–8390. [Google Scholar]
- Sun, C.C.; Li, S.J.; Li, G.; Hua, R.X.; Zhou, X.H.; Li, D.J. Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non–small-cell Lung Cancer by Binding to EZH2 and Suppressing p57. Mol. Ther.-Nucleic Acids 2016, 5, e385. [Google Scholar] [CrossRef]
- Xue, J.; Zhang, F. LncRNA LINC00511 plays an oncogenic role in lung adenocarcinoma by regulating PKM2 expression via sponging miR-625-5p. Thorac. Cancer 2020, 11, 2570–2579. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, P.; Hu, X. LINC00511 enhances LUAD malignancy by upregulating GCNT3 via miR-195-5p. BMC Cancer 2022, 22, 389. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, S.; Mu, X. Long non-coding RNA LINC00511 promotes proliferation, invasion, and migration of non-small cell lung cancer cells by targeting miR-625-5p/GSPT1. Transl. Cancer Res. 2021, 10, 5159–5173. [Google Scholar] [CrossRef]
- Wu, Y.; Li, L.; Wang, Q.; Zhang, L.; He, C.; Wang, X.; Liu, H. LINC00511 promotes lung squamous cell carcinoma proliferation and migration via inhibiting miR-150-5p and activating TADA1. Transl. Lung Cancer Res. 2020, 9, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, M.; Huang, Y.; Zhang, J.; Yin, L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA-regulated PLD1. J. Cell. Physiol. 2020, 235, 6592–6604. [Google Scholar] [CrossRef]
- Mao BDi Xu, P.; Zhong, Y.; Ding, W.W.; Meng, Q.Z. LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J. Biosci. 2019, 44, 44. [Google Scholar]
- Zhang, X.; Wang, Y.; Zhao, A.; Kong, F.; Jiang, L.; Wang, J. Long non-coding rna linc00511 accelerates proliferation and invasion in cervical cancer through targeting mir-324-5p/dram1 axis. OncoTargets Ther. 2020, 13, 10245–10256. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Gao, Q.; Wang, Y.; Ren, J.; Zhang, T. LINC00511 promotes cervical cancer progression by regulating the miR-497-5p/MAPK1 axis. Apoptosis 2022, 27, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mao, X.; Luo, F.; Wang, J. LINC00511 promotes gastric cancer progression by regulating SOX4 and epigenetically repressing PTEN to activate PI3K/AKT pathway. J. Cell. Mol. Med. 2021, 25, 9112–9127. [Google Scholar] [CrossRef]
- Wang, D.; Liu, K.; Chen, E. LINC00511 promotes proliferation and invasion by sponging miR-515-5p in gastric cancer. Cell Mol. Biol. Lett. 2020, 25, 4. [Google Scholar] [CrossRef]
- Sun, C.B.; Wang, H.Y.; Han, X.Q.; Liu, Y.N.; Wang, M.C.; Zhang, H.X.; Gu, Y.F.; Leng, X.G. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J. Gastrointest. Oncol. 2020, 12, 394–404. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, H.; Zhang, Z.; Li, G.; Liu, B. LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int. 2019, 19, 351. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Li, Z.; Zheng, S.; Wang, Z.; Li, W.; Bi, Z.; Li, L.; Jiang, Y.; Luo, Y.; et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J. Cell Mol. Med. 2018, 22, 655–667. [Google Scholar] [CrossRef]
- Wang, R.P.; Jiang, J.; Jiang, T.; Wang, Y.; Chen, L.X. Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3291–3301. [Google Scholar]
- Hu, W.Y.; Wei, H.Y.; Li, K.M.; Wang RBen Xu, X.Q.; Feng, R. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed. Pharmacother. 2020, 121, 109642. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Cui, H.; Lei, T.; Li, S.; Mai, E.; Jia, F. Linc00511 indicates a poor prognosis of liver hepatocellular carcinoma. OncoTargets Ther. 2019, 12, 9367–9376. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Li, X.; Yang, S.; Huang, M.; Wei, S.; Ma, Y.; Li, Y.; Wu, B.; Jin, H.; Li, B.; et al. LINC00511 drives invasive behavior in hepatocellular carcinoma by regulating exosome secretion and invadopodia formation. J. Exp. Clin. Cancer Res. 2021, 40, 183. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Tu, Y.; Liu, S.; Zhao, P.; Bao, Z.; Li, C.; Li, J.; Pan, M.; Ji, J. LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J. Cell. Mol. Med. 2020, 24, 1474–1487. [Google Scholar] [CrossRef]
- Liu, Z.; Tao, B.; Li, L.; Liu, P.; Xia, K.; Zhong, C. LINC00511 knockdown suppresses glioma cell malignant progression through miR-15a-5p/AEBP1 axis. Brain Res. Bull. 2021, 173, 82–96. [Google Scholar] [CrossRef]
- Lu, Y.; Tian, M.; Liu, J.; Wang, K. LINC00511 facilitates Temozolomide resistance of glioblastoma cells via sponging miR-126-5p and activating Wnt/β-catenin signaling. J. Biochem. Mol. Toxicol. 2021, 35, e22848. [Google Scholar] [CrossRef]
- Guo, W.; Yu, Q.; Zhang, M.; Li, F.; Liu, Y.; Jiang, W.; Jiang, H.; Li, H. Long intergenic non-protein coding RNA 511 promotes the progression of osteosarcoma cells through sponging microRNA 618 to upregulate the expression of maelstrom. Aging 2019, 11, 5351–5367. [Google Scholar] [CrossRef]
- Xu, J.; Chen, G.; Zhang, Y.; Huang, Z.; Cheng, X.; Gu, H.; Xia, J.; Yin, X. LINC00511 Promotes Osteosarcoma Tumorigenesis and Invasiveness through the miR-185-3p/E2F1 Axis. Biomed. Res. Int. 2020, 2020, 1974506. [Google Scholar] [CrossRef]
- Yan, L.; Wu, X.; Liu, Y.; Xian, W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J. Cell. Biochem. 2019, 120, 7248–7256. [Google Scholar] [CrossRef]
- Qiao, S.; Qi, K.; Liu, C.; Xu, C.; Ma, J.; Xu, X.; Li, C.; Wang, Z. Long intergenic non-coding RNA 511 correlates with improved prognosis, and hinders osteosarcoma progression both in vitro and in vivo. J. Clin. Lab. Anal. 2020, 34, e23164. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.H.; Cheng, Y.F.; Zhang, Y.; Guo, R.; Li, S.; Hong, X. Long non-coding RNA LINC00511/miR-150/MMP13 axis promotes breast cancer proliferation, migration and invasion. Biochim. Biophys. Acta-Mol. Basis Dis. 2021, 1867, 165957. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, Y.; Liu, X.; Fu, Y.; Zhu, K.; Niu, Z.; Liu, J.; Qian, C. Upregulation of LINC00511 expression by DNA hypomethylation promotes the progression of breast cancer. Gland Surg. 2021, 10, 1418–1430. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Kong, D.; Chen, Q.; Ping, Y.; Pang, D. Oncogenic long noncoding RNA landscape in breast cancer. Mol. Cancer 2017, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sui, S.; Wu, H.; Zhang, J.; Zhang, X.; Xu, S.; Pang, D. The transcriptional landscape of lncRNAs reveals the oncogenic function of LINC00511 in ER-negative breast cancer. Cell Death Dis. 2019, 10, 599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, B.; Wang, X.; Zhang, F.; Yu, W. LINC00511 knockdown enhances paclitaxel cytotoxicity in breast cancer via regulating miR-29c/CDK6 axis. Life Sci. 2019, 228, 135–144. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, Y.; Liu, A.M.; Feng, Y.; Chen, Y. Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7457–7468. [Google Scholar]
- Minotti, L.; Agnoletto, C.; Baldassari, F.; Corrà, F.; Volinia, S. SNPs and somatic mutation on long non-coding RNA: New frontier in the cancer studies? High-Throughput 2018, 7, 34. [Google Scholar] [CrossRef]
- Zou, H.; Wu, L.-X.; Tan, L.; Shang, F.-F.; Zhou, H.-H. Significance of Single-Nucleotide Variants in Long Intergenic Non-protein Coding RNAs. Front. Cell Dev. Biol. 2020, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, S. Effect of Single Nucleotide Polymorphisms on the structure of long noncoding RNAs and their interaction with RNA Binding Proteins. bioRxiv 2022, 2022, 501647. Available online: http://biorxiv.org/content/early/2022/07/28/2022.07.26.501647.abstract (accessed on 8 February 2023). [CrossRef]
- Sun, Q.; Chong, F.; Jiang, X.; Wang, Y.; Xu, K.; Zou, Y.; Song, C. Association study of SNPs in LncRNA CDKN2B-AS1 with breast cancer susceptibility in Chinese Han population. Int. J. Biochem. Cell Biol. 2022, 143, 106139. [Google Scholar] [CrossRef]
- Abdi, E.; Latifi-Navid, S.; Latifi-Navid, H. LncRNA polymorphisms and breast cancer risk. Pathol. Res. Pract. 2022, 229, 153729. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Luo, C.; Guo, Q.; Cao, J.; Yang, Q.; Dong, K.; Wang, S.; Wang, K.; Song, C. Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene 2018, 642, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Dai, X.; Yeung, S.C.J.; He, X. The role of long non-coding RNA GAS5 in cancers. Cancer Manag. Res. 2019, 11, 2729–2737. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Y.; Wang, X.; Liu, Y.; Zheng, K. A Genetic Variant of rs145204276 in the Promoter Region of Long Noncoding RNA GAS5 Is Associated with a Reduced Risk of Breast Cancer. Clin. Breast Cancer 2019, 19, e415–e421. [Google Scholar] [CrossRef]
- Chen, P.; Chen, R.; Guo, H.; Cheng, J.; Zhang, R.; Liu, B.; Pang, J.; Cao, W. CASC15 Polymorphisms are Correlated with Breast Cancer Susceptibility in Chinese Han Women. Clin. Breast Cancer 2021, 21, e518–e525. [Google Scholar] [CrossRef]
- Yan, R.; Cao, J.; Song, C.; Chen, Y.; Wu, Z.; Wang, K.; Dai, L. Polymorphisms in lncRNA HOTAIR and susceptibility to breast cancer in a Chinese population. Cancer Epidemiol. 2015, 39, 978–985. [Google Scholar] [CrossRef]
- Yang, H.; Sun, Q.; Chong, F.; Jiang, X.; Wang, Y.; Xu, K.; Zou, Y.; Xu, L.; Song, C. Polymorphisms in lncRNA MIR2052HG and susceptibility to breast cancer in Chinese population. Aging 2021, 13, 24360–24378. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Gao, W.; Zhang, Y.L.; Niu, M.; Cui, J.J.; Xiang, C.X.; Sang, J.W.; Wen, S.X.; Wang, B.Q. Expression and clinical significance of long non-coding RNA LINC00520 in laryngeal squamous cell carcinoma. J. Clin. Otorhinolaryngol. Head Neck Surg. 2018, 32, 91–95. [Google Scholar]
- Guo, Q.; Xu, L.; Peng, R.; Ma, Y.; Wang, Y.; Chong, F.; Song, M.; Dai, L.; Song, C. Characterization of lncRNA LINC00520 and functional polymorphisms associated with breast cancer susceptibility in Chinese Han population. Cancer Med. 2020, 9, 2252–2268. [Google Scholar] [CrossRef] [PubMed]
- Marchi, R.D.; Mathias, C.; Reiter, G.A.K.; de Lima, R.S.; Kuroda, F.; de Andrade Urban, C.; de Souza, R.L.R.; Gradia, D.F.; Ribeiro, E.M.S.F.; Cavalli, I.J.; et al. Association between snp rs527616 in lncrna aqp4-as1 and susceptibility to breast cancer in a southern brazilian population. Genet. Mol. Biol. 2021, 44, 1–7. [Google Scholar] [CrossRef]
- Tang, X.; Gao, Y.; Yu, L.; Lu, Y.; Zhou, G.; Cheng, L.; Sun, K.; Zhu, B.; Xu, M.; Liu, J. Correlations between lncRNA-SOX2OT polymorphism and susceptibility to breast cancer in a Chinese population. Biomark. Med. 2017, 11, 277–284. [Google Scholar] [CrossRef]
- Hassanzarei, S.; Hashemi, M.; Sattarifard, H.; Hashemi, S.M.; Bahari, G. Genetic polymorphisms in long noncoding RNA H19 are associated with breast cancer susceptibility in Iranian population. Meta Gene 2017, 14, 1–5. [Google Scholar] [CrossRef]
- Yan, R.; Wang, K.; Peng, R.; Wang, S.; Cao, J.; Wang, P.; Song, C. Genetic variants in lncRNA SRA and risk of breast cancer. Oncotarget 2016, 7, 22486–22496. [Google Scholar] [CrossRef] [PubMed]
- Chong, F.F.; Cao, J.J.; Wang, Y.L.; Sun, Q.Y.; Song, M.M.; Jiang, X.R.; Wang, K.J.; Xu, L.P.; Song, C.H. The Association between LINC00511 Variants and Breast Cancer Susceptibility among the Han Chinese Population. J. Nutr. Oncol. 2020, 5, 87–96. [Google Scholar] [CrossRef]
- Luo, C.; Cao, J.; Peng, R.; Guo, Q.; Ye, H.; Wang, P.; Wang, K.; Song, C. Functional Variants in Linc-ROR are Associated with mRNA Expression of Linc-ROR and Breast Cancer Susceptibility. Sci. Rep. 2018, 8, 4680. [Google Scholar] [CrossRef]
- Wang, S.; Cui, Z.; Li, H.; Li, J.; Lv, X.; Yang, Z.; Gao, M.; Bi, Y.; Zhang, Z.; Zhou, B.; et al. LncRNA NEAT1 polymorphisms and lung cancer susceptibility in a Chinese Northeast Han Population: A case-control study. Pathol. Res. Pract. 2019, 215, 152723. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, Y.; Yin, Z. Polymorphisms in lncRNA CCAT1 on the susceptibility of lung cancer in a Chinese northeast population: A case–control study. Cancer Med. 2023, 12, 500–512. [Google Scholar] [CrossRef]
- Wang, R.; Feng, N.; Wang, Y.; Gao, S.; Zhang, F.; Qian, Y.; Gao, M.; Yu, H.; Zhou, B.; Qian, B. SNPs in LncRNA genes are associated with non-small cell lung cancer in a Chinese population. J. Clin. Lab. Anal. 2019, 33, e22858. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Cui, Z.; Li, H.; Li, J.; Yang, Z.; Bi, Y.; Gao, M.; Zhou, B.; Yin, Z. Polymorphism in lncRNA AC008392.1 and its interaction with smoking on the risk of lung cancer in a Chinese population. Cancer Manag. Res. 2018, 10, 1377–1387. [Google Scholar] [CrossRef]
- Li, H.; Yang, Z.; Li, J.; Lv, X.; Gao, M.; Bi, Y.; Zhang, Z.; Wang, S.; Li, S.; Li, N.; et al. Genetic variants in LncRNA HOTAIR are associated with lung cancer susceptibility in a Chinese Han population in China: A case-control study. Cancer Manag. Res. 2018, 10, 5209–5218. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Y.; Li, Y.W.; Zhang, H.B.; Gong, H.; Yuan, Y.; Li, W.T.; Liu, H.Y.; Chen, J. HOTAIR lncRNA SNPs rs920778 and rs1899663 are associated with smoking, male gender, and squamous cell carcinoma in a Chinese lung cancer population. Acta Pharmacol. Sin. 2018, 39, 1797–1803. [Google Scholar] [CrossRef]
- Hsieh, M.H.; Wu, Y.L.; Tsao, T.C.Y.; Huang, Y.W.; Lin, J.C.; Lee, C.Y.; Hsieh, M.J.; Yang, S.F. Impact of LncRNA GAS5 Genetic Variants and the Epidermal Growth Factor Receptor Phenotypes on the Clinicopathological Characteristics of Lung Adenocarcinoma Patients. Int. J. Environ. Res. Public Health 2022, 19, 9971. [Google Scholar] [CrossRef] [PubMed]
- Bardhan, A.; Banerjee, A.; Basu, K.; Pal, D.K.; Ghosh, A. PRNCR1: A long non-coding RNA with a pivotal oncogenic role in cancer. Hum. Genet. 2022, 141, 15–29. [Google Scholar] [CrossRef]
- Yu, W.L.; Yao, J.J.; Xie, Z.Z.; Huang, Y.J.; Xiao, S. Lncrna prncr1 rs1456315 and ccat2 rs6983267 polymorphisms on 8q24 associated with lung cancer. Int. J. Gen. Med. 2021, 14, 255–266. [Google Scholar] [CrossRef]
- Tong, G.; Tong, W.; He, R.; Cui, Z.; Li, S.; Zhou, B.; Yin, Z. MALAT1 Polymorphisms and Lung Cancer Susceptibility in a Chinese Northeast Han Population. Int. J. Med. Sci. 2022, 19, 1300–1306. [Google Scholar] [CrossRef]
- Gao, M.; Li, H.; Bi, Y.; Zhang, Z.; Wang, S.; Li, J.; Yang, Z.; Lv, X.; Zhou, B.; Yin, Z. The polymorphisms of lncRNA HOXA11-AS and the risk of lung cancer in Northeastern Chinese population. J. Cancer 2020, 11, 592–598. [Google Scholar] [CrossRef]
- Li, L.; Guo, G.; Zhang, H.; Zhou, B.; Bai, L.; Chen, H.; Zhao, Y.; Yan, Y. Association between H19 SNP rs217727 and lung cancer risk in a Chinese population: A case control study. BMC Med. Genet. 2018, 19, 136. [Google Scholar] [CrossRef]
- Feng, T.; Feng, N.; Zhu, T.; Li, Q.; Zhang, Q.; Wang, Y.; Gao, M.; Zhou, B.; Yu, H.; Zheng, M.; et al. A SNP-mediated lncRNA (LOC146880) and microRNA (miR-539-5p) interaction and its potential impact on the NSCLC risk. J. Exp. Clin. Cancer Res. 2020, 39, 157. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Yan, G.; Yu, S.; Li, F.; Su, Z.; Hou, X.; Xiao, J.; Tian, T. Associations of MALAT1 and its functional single nucleotide polymorphisms with cancer. Pathol. Res. Pract. 2022, 236, 153988. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jing, F.; Ding, Y.; He, Q.; Zhong, Y.; Fan, C. Long noncoding RNA CCAT1 polymorphisms are associated with the risk of colorectal cancer. Cancer Genet. 2018, 222, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Jin, M.; Ye, D.; Li, Y.; Jing, F.; Zhang, X.; Li, Q.; Chen, K. Polymorphisms of a novel long non-coding RNA RP11-108K3.2 with colorectal cancer susceptibility and their effects on its expression. Int. J. Biol. Mark. 2020, 35, 3–9. [Google Scholar] [CrossRef]
- Shaalan, A.A.M.; Mokhtar, S.H.; Ahmedah, H.T.; Almars, A.I.; Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Salem, M.A. Prognostic Value of LINC-ROR (rs1942347) Variant in Patients with Colon Cancer Harboring BRAF Mutation: A Propensity Score-Matched Analysis. Biomolecules 2022, 12, 569. [Google Scholar] [CrossRef]
- Li, S.; Hua, Y.; Jin, J.; Wang, H.; Du, M.; Zhu, L.; Chu, H.; Zhang, Z.; Wang, M. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget 2016, 7, 25470–25477. [Google Scholar] [CrossRef]
- Qin, W.; Wang, X.; Wang, Y.; Li, Y.; Chen, Q.; Hu, X.; Wu, Z.; Zhao, P.; Li, S.; Zhao, H.; et al. Functional polymorphisms of the lncRNA H19 promoter region contribute to the cancer risk and clinical outcomes in advanced colorectal cancer. Cancer Cell Int. 2019, 19, 215. [Google Scholar] [CrossRef]
- Lv, Z.; Xu, Q.; Sun, L.; Wen, J.; Fang, X.; Xing, C.; Yuan, Y. Four novel polymorphisms in long non-coding RNA HOTTIP are associated with the risk and prognosis of colorectal cancer. Biosci. Rep. 2019, 39, BSR20180573. [Google Scholar] [CrossRef]
- Ali, M.A.; Shaker, O.G.; Ezzat, E.M.; Gaber, S.N.; Hassan, E.A.; Abdelwahed, M.Y.; AbdelHafez, M.N.; Khalil, M.A.F.; Abouelseoud, S. Association between rs1859168/HOTTIP Expression Level and Colorectal Cancer and Adenomatous Polyposis Risk in Egyptians. J. Interface Cytokine Res. 2020, 40, 279–291. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.; Yang, X.; Li, X.; Chen, R. Association between polymorphism in the promoter region of lncRNA GAS5 and the risk of colorectal cancer. Biosci. Rep. 2019, 39, BSR20190091. [Google Scholar] [CrossRef]
- Yang, M.L.; Huang, Z.; Wu, L.N.; Wu, R.; Ding, H.X.; Wang, B.G. LncRNA-PCAT1 rs2632159 polymorphism could be a biomarker for colorectal cancer susceptibility. Biosci. Rep. 2019, 39, BSR20190708. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, S.; Alshammari, E.M.; Mokhtar, S.H.; Alshanwani, A.R.; Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Maher, S.A. PUNISHER rs12318065 C>A transversion: A putative somatic driver mutation for poor prognosis in colon cancer. Biosci. Rep. 2022, 42, BSR20220465. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, S.; Li, X.; Yin, Y.; Chen, R. MAGI2-AS3 rs7783388 polymorphism contributes to colorectal cancer risk through altering the binding affinity of the transcription factor GR to the MAGI2-AS3 promoter. J. Clin. Lab. Anal. 2020, 34, e23431. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Y.; Jin, J.; Gu, X. Correlation between lncRNA SNHG16 gene polymorphism and its interaction with environmental factors and susceptibility to colorectal cancer. Medicine 2020, 99, e23372. [Google Scholar] [CrossRef]
- Shen, C.; Yan, T.; Wang, Z.; Su, H.; Zhu, X.; Tian, X.; Fang, J.; Chen, H.; Hong, J. Variant of SNP rs1317082 at CCSlnc362 (RP11-362K14.5) creates a binding site for miR-4658 and diminishes the susceptibility to CRC. Cell Death Dis. 2018, 9, 1177. [Google Scholar] [CrossRef]
- Liu, X.; Chen, M.; Liu, Q.; Li, G.; Yang, P.; Zhang, G. LncRNA PTCSC3 is upregulated in osteoporosis and negatively regulates osteoblast apoptosis. BMC Med. Genom. 2022, 15, 57. [Google Scholar] [CrossRef]
- Wang, Y.; Qiu, Z.; Tian, G.; Zhu, Q.; Zhang, Z.; Qin, R.; Peng, Y.; Tang, W.; Zhang, S.; Xi, Y. Association between long noncoding RNA rs944289 and rs7990916 polymorphisms and the risk of colorectal cancer in a Chinese population. Sci. Rep. 2022, 12, 2495. [Google Scholar] [CrossRef]
- AlMutairi, M.; Parine, N.R.; Shaik, J.P.; Aldhaian, S.; Azzam, N.A.; Aljebreen, A.M.; Alharbi, O.; Almadi, M.A.; Al-Balbeesi, A.O.; Alanazi, M. Association between polymorphisms in PRNCR1 and risk of colorectal cancer in the Saudi population. PLoS ONE 2019, 14, e0220931. [Google Scholar] [CrossRef]
- Cao, X.; Zhuang, S.; Hu, Y.; Xi, L.; Deng, L.; Sheng, H.; Shen, W. Associations between polymorphisms of long non-coding RNA MEG3 and risk of colorectal cancer in Chinese. Oncotarget 2016, 7, 19054–19059. [Google Scholar] [CrossRef]
- Gong, J.; Tian, J.; Lou, J.; Ke, J.; Li, L.; Li, J.; Yang, Y.; Gong, Y.; Zhu, Y.; Zhang, Y.; et al. A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis 2016, 37, 443–451. [Google Scholar] [CrossRef]
- Kim, J.O.; Jun, H.H.; Kim, E.J.; Lee, J.Y.; Park, H.S.; Ryu, C.S.; Kim, S.; Oh, D.; Kim, J.W.; Kim, N.K. Genetic Variants of HOTAIR Associated With Colorectal Cancer Susceptibility and Mortality. Front. Oncol. 2020, 10, 72. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, Y.; Cui, J.; Yang, G.; Peng, S.; Mi, W.; Yin, X.; Yu, Y.; Jiang, J.; Liu, Q.; et al. SNP rs12982687 affects binding capacity of lncRNA UCA1 with miR-873-5p: Involvement in smoking-triggered colorectal cancer progression. Cell Commun. Signal. 2020, 18, 37. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Senousy, M.A.; Elbaz, E.M. Association of rs6983267 at 8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. Sci. Rep. 2017, 7, 16246. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, C.; Tong, S.; Ding, Y.; Deng, W.; Song, D.; Xiao, K. Genetic variation of long non-coding RNA TINCR contribute to the susceptibility and progression of colorectal cancer. Oncotarget 2017, 8, 33536–33543. [Google Scholar] [CrossRef]
- Zhu, B.; Zhu, Y.; Tian, J.; Shen, N.; Li, J.; Lou, J.; Ke, J.; Yang, Y.; Gong, Y.; Gong, J.; et al. A functional variant rs1537373 in 9p21.3 region is associated with pancreatic cancer risk. Mol. Carcinog. 2019, 58, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Giaccherini, M.; Farinella, R.; Gentiluomo, M.; Mohelnikova-Duchonova, B.; Kauffmann, E.F.; Palmeri, M.; Uzunoglu, F.; Soucek, P.; Petrauskas, D.; Cavestro, G.M.; et al. Association between a polymorphic variant in the CDKN2B-AS1/ANRIL gene and pancreatic cancer risk. Int. J. Cancer 2023, 153, 373–379. [Google Scholar] [CrossRef]
- Jiang, D.; Xu, L.; Ni, J.; Zhang, J.; Cai, M.; Shen, L. Functional polymorphisms in LncRNA HOTAIR contribute to susceptibility of pancreatic cancer. Cancer Cell Int. 2019, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Corradi, C.; Gentiluomo, M.; Gajdán, L.; Cavestro, G.M.; Kreivenaite, E.; Di Franco, G.; Sperti, C.; Giaccherini, M.; Petrone, M.C.; Tavano, F.; et al. Genome-wide scan of long noncoding RNA single nucleotide polymorphisms and pancreatic cancer susceptibility. Int. J. Cancer 2021, 148, 2779–2788. [Google Scholar] [CrossRef]
- Qiao, K.; Ning, S.; Wan, L.; Wu, H.; Wang, Q.; Zhang, X.; Xu, S.; Pang, D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 418. [Google Scholar] [CrossRef]
- Zheng, J.; Huang, X.; Tan, W.; Yu, D.; Du, Z.; Chang, J.; Wei, L.; Han, Y.; Wang, C.; Che, X.; et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 2016, 48, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Qiao, O.; Wang, J.; Li, J.; Jin, H.; Li, Z.; Yan Jin, Y. rs1859168 A>C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J. Surg. Oncol. 2017, 15, 155. [Google Scholar] [CrossRef]
- Mo, H.; Wang, X.; Ji, G.; Liang, X.; Yang, Y.; Sun, W.; Jia, X.; Xu, L.; Qiao, Y.; Zhou, H.; et al. The effect of SNPs in lncRNA as ceRNA on the risk and prognosis of hepatocellular carcinoma. BMC Genom. 2022, 23, 769. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.G.; Xu, Q.; Lv, Z.; Fang, X.X.; Ding, H.X.; Wen, J.; Yuan, Y. Association of twelve polymorphisms in three onco-lncRNa genes with hepatocellular cancer risk and prognosis: A case-control study. World J. Gastroenterol. 2018, 24, 2482–2490. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.L.; Huang, Z.; Wang, Q.; Chen, H.H.; Ma, S.N.; Wu, R.; Cai, W.S. The association of polymorphisms in lncRNA-H19 with hepatocellular cancer risk and prognosis. Biosci. Rep. 2018, 38, BSR20171652. [Google Scholar] [CrossRef]
- Yuan, L.T.; Yang, Y.C.; Lee, H.L.; Shih, P.C.; Chen, L.H.; Tang, C.H.; Chang, L.C.; Wang, H.L.; Yang, S.F.; Chien, M.H. Genetic Polymorphisms of lncRNA LINC00673 as Predictors of Hepatocellular Carcinoma Progression in an Elderly Population. Int. J. Mol. Sci. 2022, 23, 12737. [Google Scholar] [CrossRef] [PubMed]
- Youssef, S.S.; Hamdy, N.M. SOCS1 and pattern recognition receptors: TLR9 and RIG-I.; novel haplotype associations in Egyptian fibrotic/cirrhotic patients with HCV genotype 4. Arch. Virol. 2017, 162, 3347–3354. [Google Scholar] [CrossRef]
- El Mesallamy, H.O.; Rashed, W.M.; Hamdy, N.M.; Hamdy, N. High-dose methotrexate in Egyptian pediatric acute lymphoblastic leukemia: The impact of ABCG2 C421A genetic polymorphism on plasma levels, what is next? J. Cancer Res. Clin. Oncol. 2014, 140, 1359–1365. [Google Scholar] [CrossRef]
- Aboouf, M.A.; Hamdy, N.M.; Amin, A.I.; El-Mesallamy, H.O. Genotype screening of APLN rs3115757 variant in Egyptian women population reveals an association with obesity and insulin resistance. Diabetes Res. Clin. Pract. 2015, 109, 40–47. [Google Scholar] [CrossRef]
- Kamal, A.M.; Hamdy, N.M.; Hegab, H.M.; El-Mesallamy, H.O. Expression of thioredoxin-1 (TXN) and its relation with oxidative DNA damage and treatment outcome in adult AML and ALL: A comparative study. Hematology 2016, 21, 567–575. [Google Scholar] [CrossRef]
- Ali, N.A.; Hamdy, N.M.; Gibriel, A.A.; ELMesallamy, H.O. Investigation of the relationship between CTLA4 and the tumor suppressor RASSF1A and the possible mediating role of STAT4 in a cohort of Egyptian patients infected with hepatitis C virus with and without hepatocellular carcinoma. Arch. Virol. 2021, 166, 1643–1651. [Google Scholar] [CrossRef]
- El-Derany, M.O.; Hamdy, N.M.; Al-Ansari, N.L.; El-Mesallamy, H.O. Integrative role of vitamin D related and Interleukin-28B genes polymorphism in predicting treatment outcomes of Chronic Hepatitis, C. BMC Gastroenterol. 2016, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- El-Aziz, M.K.A.; Dawoud, A.; Kiriacos, C.J.; Fahmy, S.A.; Hamdy, N.M.; Youness, R.A. Decoding hepatocarcinogenesis from a noncoding RNAs perspective. J. Cell. Physiol. 2023, 238, 1982–2009. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.M.; Sanad, E.F.; Hamdy, N.M. MicroRNAs’ role in the environment-related non-communicable diseases and link to multidrug resistance, regulation, or alteration. Environ. Sci. Pollut. Res. 2021, 28, 36984. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).