Abstract
Alternative splicing is a highly fine-tuned regulated process and one of the main drivers of proteomic diversity across eukaryotes. The vast majority of human multi-exon genes is alternatively spliced in a cell type- and tissue-specific manner, and defects in alternative splicing can dramatically alter RNA and protein functions and lead to disease. The eukaryotic genome is also intensively transcribed into long and short non-coding RNAs which account for up to 90% of the entire transcriptome. Over the years, lncRNAs have received considerable attention as important players in the regulation of cellular processes including alternative splicing. In this review, we focus on recent discoveries that show how lncRNAs contribute significantly to the regulation of alternative splicing and explore how they are able to shape the expression of a diverse set of splice isoforms through several mechanisms. With the increasing number of lncRNAs being discovered and characterized, the contribution of lncRNAs to the regulation of alternative splicing is likely to grow significantly.
1. Introduction
In the late 1970s, researchers were interested in gaining a better understanding of the mechanisms of adenoviral gene expression when they noticed something unusual, a long adenoviral transcript hybridized to the viral genome forming a three-stranded mRNA:DNA hybrid structure with an intervening DNA sequence that did not match the mature mRNA [1]. It then became apparent that these intervening sequences were in fact introns and that the primary transcript is made by a succession of exonic and intronic sequences. In what is now thought to be a mainly co-transcriptional process, introns are ‘spliced’ out from the mRNA precursor (pre-mRNA) and the exons joined together through two transesterification reactions catalysed by a complex molecular machinery consisting of five small nuclear ribonucleoproteins (snRNPs), called the spliceosome [2]. Over the past decades it has become clear that pre-mRNA splicing is a widespread phenomenon across eukaryotes and that a single gene can generate multiple transcripts often encoding different proteins by a process known as alternative splicing (AS) [3]. Many types of AS are possible, including “cassette exons”, “alternative 5′ and 3′ splice sites”, “alternative first exons” (through different promoters), “alternative last exons” (through different polyadenylation sites), “mutually exclusive exons” and “retained introns” [4]. Fundamental to the process of AS is the definition of the precise location of 5′ (donor) and 3′ (acceptor) splice sites and the assembly of the spliceosome complex. The first relies on splicing factors (SFs), a category of RNA-binding proteins (RBPs) expressed in a tissue and stage-specific way that recognize regulatory elements within exons and introns. Importantly, SF activity is in turn modified by splicing factor kinases and phosphatases activated through cell signaling mechanisms.
AS greatly enhances proteome diversity and represents an essential aspect of gene expression in development, normal physiology and disease across eukaryotes [5], from single-celled yeast to humans [6]. The advent of high-throughput sequencing technologies revealed that ~92–94% of human multi-exon genes are alternatively spliced [7], increasing the interest in understanding the mechanisms underpinning its regulation. With the discovery and growing importance of non-coding RNAs, the nature of AS regulation has become more complex. Both short (<200 nt) and long (>200 nt) non-coding RNAs can contribute to the regulation of AS in many different ways; either indirectly by regulating the activity of splice factors; or directly, by interacting with pre-mRNAs. Long non-coding RNAs (lncRNAs) are particularly well suited to these roles due to their demonstrated capacity to act as regulatory molecules that modulate gene expression at every level. Either alone, or in association with partner proteins, these long RNA polymerase II transcripts have been shown to take part in a wide range of developmental processes and disease in complex organisms [8,9,10]. Here, we review the current knowledge of the multiple mechanisms through which lncRNAs contribute to the regulation of AS (Figure 1 and Table 1).
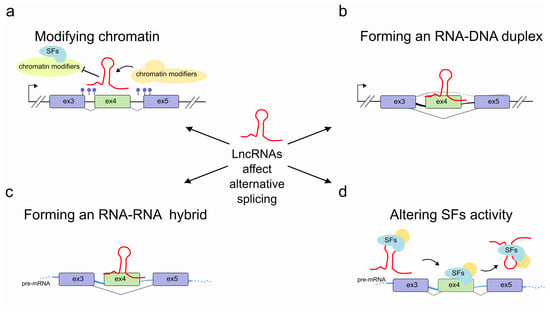
Figure 1.
Regulation of pre-mRNA splicing by lncRNAs. LncRNAs (red) are able to control pre-mRNA splicing by (a) modifying chromatin accessibility through recruiting or impeding access to chromatin modifying complexes at the transcribed genomic locus. In some cases, this might result in more drastic long-range structural changes; (b) interacting with the transcribed genomic locus through an RNA-DNA hybrid; (c) hybridizing with the pre-mRNA molecule (light blue); (d) promoting SF recruitment or by sequestering SFs into specific subnuclear compartments, thereby interfering with SF activities.
2. LncRNAs Regulate Alternative Splicing through Chromatin Modification
The eukaryotic genome is tightly packaged into chromatin fibers consisting of DNA wrapped around nucleosomes made of histone proteins. Post-translational modifications (PTMs), such as methylation, acetylation, phosphorylation, and ubiquitination, occur on the histone tails that are functionally linked to the epigenetic regulation of gene expression. By defining the accessibility to chromatin, histone modifications demarcate amenable or silenced chromatin domains which ultimately reflect the activity of gene transcription. An intimate relationship exists between lncRNAs and chromatin conformation [11,12]. LncRNAs regulate chromatin modifications by recruiting or directly interacting with histone-modifying complexes or enzymes at specific chromosomal loci; these in turn modulate gene transcription [13,14,15,16,17,18]. Histone modification signatures can also influence AS through a chromatin-reading protein which acts as an adaptor linker between the RNA polymerase II (RNAPII) and the pre-mRNA splicing machinery [19]. Several studies have also demonstrated that the local chromatin context influences the RNAPII elongation rate which in turn affects AS [20,21,22].
A possible lncRNA-mediated crosstalk between histone modifications and the pre-mRNA splicing machinery has also been proposed [23]. Cell type–specific splicing of the gene encoding the fibroblast growth factor receptor 2 (FGFR2) is now known to rely on the methylation state of the FGFR2 locus. In mesenchymal stem cells, FGFR2 is enriched in di- (me2) and tri-methylated (me3) histone H3K36, which inhibits the inclusion of the alternatively spliced exon IIIb. FGFR2 is, in contrast, devoid of H3K36 methylation in epithelial cells. The cell-specific switch in splicing is made possible by an evolutionarily-conserved nuclear antisense lncRNA (asFGFR2), transcribed within the human FGFR2 locus and exclusively expressed in epithelial cells. By recruiting Polycomb-group proteins and the histone lysine-specific demethylase 2a (KDM2a) to the locus, asFGFR2 ensures the deposition of H3K27me3 and a decrease in H3K36me2/3. This impairs both the binding of the chromatin-binding protein MRG15 for H3K36me2/3 [24] and the recruitment, via protein-protein interactions, of the negative splicing regulator PTBP1 to exon IIIb [19]. Through this combined action, the chromatin-splicing adaptor complex MRG15–PTBP1 can no longer inhibit the inclusion of exon IIIb favoring the epithelial-specific AS of FGFR2 [23] (Figure 2a).
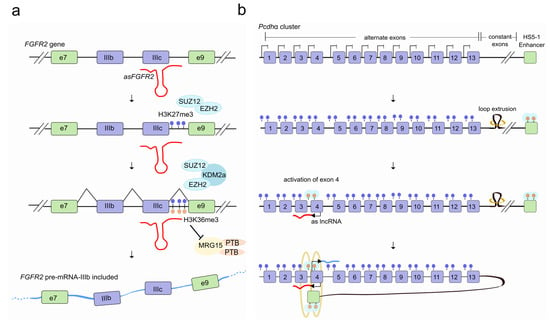
Figure 2.
LncRNAs regulate alternative splicing through chromatin modification. (a) In epithelial cells, the antisense lncRNA asFGFR2 (red), recruits the Polycomb-group proteins EZH2 and SUZ12 to the FGFR2 gene locus and allows H3K27me3 deposition (blue lollipop) and a decrease in methylation of H3K36me3 (orange lollipop) by the recruitment of the H3K36 demethylase KDM2a. As a result, the chromatin-splicing adaptor complex MRG15–PTB1 can no longer bind to exon IIIb, which is then included in the FGFR2 transcript (light blue). (b) The activation of a specific antisense lncRNA (as lncRNA; red) at the Pcdhα promoter of one (out of 13) alternate first exon promotes proximal DNA demethylation (orange lollipop) and CTCF (turquoise) recruitment and favors the interaction between the selected promoter and a distant HS5-1 enhancer by a long-range DNA loop. This ultimately triggers sense transcription (light blue) of the corresponding selected Pcdhα first-exon which is individually spliced to a downstream constant region to form a distinct transcript.
Chromatin structure is itself likely to play an important role in modulating the effects of transcription on AS [25]. In particular, the tri-dimensional chromatin organizer CCCTC-binding factor (CTCF) has been shown to bind target DNA sites located within an alternative exon creating a roadblock to transcriptional elongation that favors exon inclusion into mature mRNA [26]. Several lncRNAs appear to control important aspects of chromatin organization including chromatin looping, either remaining tethered to the site of transcription or moving over distant loci [27,28]. Interestingly, lncRNAs can efficiently remove structural roadblocks in chromatin by CTCF eviction [29,30]. A fascinating lncRNA-mediated mechanism modulates the diversity of transcripts at the complex Protocadherin (Pcdh) α gene cluster [31]. Each Pcdhα gene of the cluster functions as a ‘variable’ first exon (out of 13) that is individually spliced to a downstream constant region to form distinct transcripts, differentially expressed in individual neurons and important for neuronal self-identity. The stochastic expression of 13 alternate exons is driven by their own promoter, each of which is equally likely to be activated by a long-range DNA loop interaction between a selected Pcdhα promoter and a downstream enhancer, called “hypersensitivity site 5-1” (HS5-1) [32,33,34,35]. The Pcdhα gene choice involves the selective activation of a specific antisense lncRNA located at the promoter of the first exon of each Pcdhα alternate gene. By promoting DNA demethylation, the antisense transcript recruits CTCF at sites proximal to the relative promoter and favors the promoter-enhancer interaction which ultimately triggers the sense transcription of the corresponding selected first exon [31] (Figure 2b). Further studies will be required to understand if other clustered genes share a similar mechanism. It will also be of interest to determine how frequently mechanisms involving lncRNAs, among the thousands transcribed, mediate chromatin structure changes that result in AS.
3. LncRNAs Regulate Pre-mRNA Splicing through RNA-DNA Interactions
LncRNAs can tether DNA forming an RNA-dsDNA triplex by targeting specific DNA sequences and inserting themselves as a third strand into the major groove of the DNA duplex [30,36]. These are known as R-loops; three-stranded nucleic acid structures, composed of RNA–DNA hybrids, frequently formed during transcription. Aberrant R-loops are generally associated with DNA damage, transcription elongation defects, hyper-recombination and genome instability [37].
Recent lines of evidence indicate a potential role for R-loops in alternative pre-mRNA splicing. A class of lncRNAs, the so-called circular RNAs (circRNAs), have recently been characterized [38,39,40]. These abundant, conserved transcripts originate from a non-canonical AS process (back-splicing) leading to the formation of head-to-tail splice junctions, joined together to form circular transcripts. Recent studies suggest that they are clearly involved in multiple aspects of normal physiology, development and disease [41]. Since most circRNAs are derived from the middle exons of protein-coding genes [42], their biogenesis can itself affect splicing of their precursor transcripts and lead to altered gene expression outcomes [43]. For example, in Arabidopsis thaliana, the circular RNA derived from exon 6 of the SEPALLATA3 (SEP3) gene increases the abundance of the cognate exon-skipped alternative splicing variant (SEP3.3 isoform) which in turn drives floral homeotic phenotypes [44]. This is made possible because SEP3 exon 6 circRNA tethers to its cognate DNA locus through an R-loop promoting transcriptional pausing, which coincides with SF recruitment and AS [45,46,47] (Figure 3). Whether or not other lncRNAs are involved in similar processes in plants or other organisms remains to be investigated.
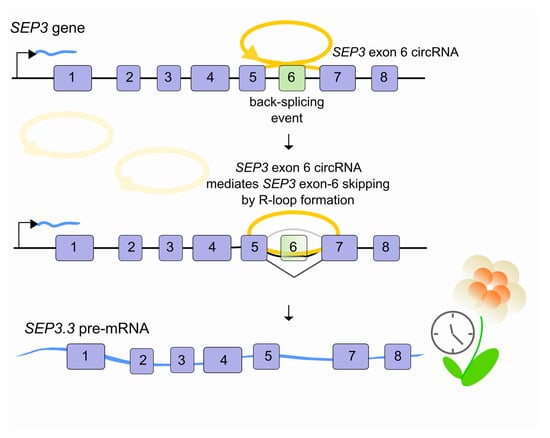
Figure 3.
LncRNAs regulate pre-mRNA splicing through an RNA-DNA interaction. In Arabidopsis thaliana, when the SEP3 gene is transcribed, exon 6 can be back-spliced into a circular RNA (SEP3 exon 6 circRNA, yellow) which interacts directly with its parental genomic locus. By forming RNA–DNA hybrids (R-loops), SEP3 exon 6 circRNA favors exon-6 skipping of its linear cognate and promotes the SEP3.3 mRNA (light blue) isoform accumulation which in turn affects flowering time.
4. LncRNAs Regulate Pre-mRNA Splicing through RNA-RNA Interactions
Over the past decades, antisense transcripts have been characterized as being widespread throughout the genomes of the vast majority of organisms [48,49,50]. It is estimated that more than 30% of annotated human transcripts have at least one cognate antisense transcript [50]. Although generally low in abundance and over 10-fold less expressed than their counterpart sense transcripts [50], antisense RNAs have been widely implicated at almost all stages of gene expression, from transcription and translation to RNA degradation [51]. A considerable proportion of genes that express multiple spliced isoforms has been associated with antisense transcription, suggesting that antisense-mediated processes could be a common mechanism to regulate AS [52]. Therapeutic strategies based on antisense-mediated exon skipping and aimed at changing the levels of alternatively spliced isoforms or at disrupting open reading frames have been also developed [20]. For example, an antisense oligoribonucleotide (AON) approach efficiently restores the open reading frame of the DMD gene and generates functional dystrophin by inducing exon skipping [53].
Identified in multiple eukaryotes, Natural Antisense Transcripts (NATs) are a class of long non-coding RNA molecules, transcribed from both coding and non-coding genes on the opposite strand of protein-coding ones [54]. Regardless of their genomic origin, NATs can hybridize with pre-mRNAs and form RNA-RNA duplexes. In some cases, a double function is also possible, and NATs can encode for proteins on one hand, while at the same time working as non-coding molecules modulating the splicing of a neighbouring gene’s transcript [55]. At the oncogene NMYC locus, for example, the cis-antisense gene NCYM located at the first NMYC intron has recently been shown to encode a protein that regulates the genesis and progression of human neuroblastomas that is associated with unfavorable prognosis [56]. However, previous studies have classified the corresponding transcript as a NAT able to modulate, via sense/antisense RNA-RNA duplexes, the processing of NMYC pre-mRNA resulting in a population of NMYC mRNA splice isoforms that retain the first intron [57] (Figure 4a).
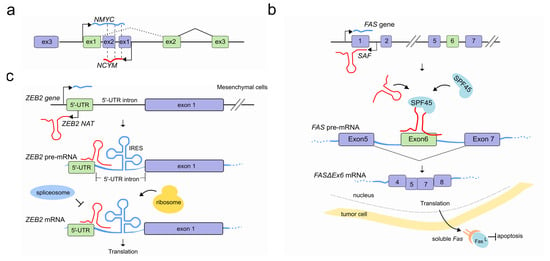
Figure 4.
LncRNAs regulate pre-mRNA splicing through an RNA-RNA interaction. (a) NAT (red) at the NCYM gene modulates splicing of the NMYC mRNA (light blue) forming a sense-antisense RNA-RNA duplex which results in an intron-retained NMYC mRNA isoform population. (b) In tumor cells the natural antisense SAF (red) is transcribed from the first intron of FAS gene and interacts with both FAS pre-mRNA (light blue) at 5–6 and 6–7 exon junctions and the human SFP45 to facilitate the AS and exclusion of exon 6. The accumulation of the exon 6-skipped alternatively spliced variant of FAS pre-mRNA (FASΔEx6 mRNA) leads to the production of a soluble Fas (sFas) protein that binds FasL and makes tumor cells resistant to FasL-induced apoptosis. (c) After EMT, Snail1 transcription factor induces the co-transcription of ZEB2 NAT (red) in mesenchymal cells. ZEB2 NAT hybridises with a region of the ZEB2 pre-mRNA (light blue) encompassing the 5′ splice site of a 3 kb-long 5′-UTR intron. This RNA-RNA duplex prevents both the binding of the spliceosome and the subsequent removal of the 5′-UTR intron. The resulting mRNA contains the full isoform of the 5′-UTR, including an internal ribosome entry site (IRES) proximal to the ZEB2 AUG, which favors translation. In absence of ZEB2 NAT (epithelial cells) instead the removal of the 5′-UTR intron results in an mRNA containing a sequence that inhibits scanning by the ribosomes and therefore prevents translation of ZEB2 protein (not shown).
Overlapping antisense transcription has been shown to modulate AS at the thyroid hormone receptor alpha (THRA) locus [58]. This locus encodes two overlapping mRNAs, α1 and α2 corresponding to TR-α1 and its splice variant TR-α2, which differ at the 3′-end because of the presence of a third overlapping mRNA, NR1D1 (also known as Rev-erbAα). The latter is transcribed in the opposite direction at the 3′-end of α2, but not α1 mRNA. It has been suggested that the relative abundance of the NR1D1 RNA prevents the splicing of α2, likely through RNA-RNA base pairing, thereby favoring the formation of the non-overlapping α1. Consistent with this hypothesis, other studies noted a positive correlation between the α2/α1 isoform ratio and the level of NR1D1 mRNA in cells [58,59]. Therefore, relatively modest changes in splice site selection of α1 and α2 caused by naturally occurring antisense RNAs might cause major changes in cellular thyroid hormone-responsiveness with a broader physiological impact.
NATs that drive AS during programmed cell death (apoptosis) have also been reported. The FAS gene encodes for a receptor protein which usually binds its Fas ligand (FasL) and triggers the apoptotic process. At the FAS locus, the lncRNA SAF is transcribed in reverse orientation and from the opposite strand of the first intron of FAS. In tumor cells, SAF transcription promotes the formation of the exon 6-skipped spliced variant of FAS pre-mRNA (FASΔEx6) by interacting with both the FAS pre-mRNA, predominantly at exon 5/6 and exon 6/7 junctions, and the human splicing factor 45 (SPF45). The resulting splicing variant lacks the transmembrane domain which gives more solubility to the isoform (sFas) and protects tumor cells against FasL-induced apoptosis [60] (Figure 4b).
Reverse transcription can affect pre-mRNA splicing by masking specific splice sites and preventing their processing. A remarkable example of how NATs can affect the splicing and in turn increase mRNA translation efficiency is the human ZEB2 gene (zinc-finger E-box-binding homeobox 2). Boosting the translation of ZEB2 repressor is one of the ways by which E-cadherin repression is initiated by the transcriptional factor Snail1 during epithelial-mesenchymal transition (EMT). Normally, the ZEB2 5′-UTR contains a structural intronic motif that works as an internal ribosome entry site (IRES) which is spliced out to hinder ZEB2 translation. However, once EMT is triggered, Snail1 induces the transcription of a ZEB2 NAT which is transcribed from the opposite strand of the ZEB2 locus, covering the 5′ splice site of the ZEB2 5′-UTR. ZEB2 NAT prevents the recognition of the splice sites by the spliceosome by RNA-RNA duplex interaction with ZEB2 mRNA and promotes the subsequent inclusion of the intron present in the ZEB2 5′ UTR, thereby promoting ZEB2 translation [61] (Figure 4c).
Masking canonical splicing sites has also been linked with the most common form of dementia, Alzheimer’s disease (AD). Sortilin-related receptor 1 (SORL1) expression is generally reduced in brain tissues from individuals with AD [62] suggesting a potential role in AD pathogenesis [63,64]. The importance of this receptor is underlined by the recent demonstration that SORL1 downregulation promotes amyloid precursor protein (APP) secretion and subsequently an increase of neurotoxic β-amyloid peptide (Aβ) [65,66]. A 300 nt antisense non-coding RNA transcribed by RNA polymerase III, called 51A, maps to the intron 1 of the SORL1 gene and, by pairing with the SORL1 pre-mRNA, drives a splicing shift of SORL1 from the canonical full-length protein variant A to an alternatively spliced shorter protein form (variant B). This process results in the decreased synthesis of SORL1 variant A and is associated with impairing processing of APP, leading to increase of Aβ formation [67].
Antisense transcripts that cause a shift in isoform balance occur also at the GPR51 locus, hosting the antisense lncRNA 17A on its intron 3. LncRNA 17A expression is induced by inflammatory molecules and leads to the production of the GABAB R2 protein isoform devoid of transduction activity and the concomitant down-regulation of the canonical full-length GABAB R2 variant, which impairs GABAB signaling. The change in the ratio of the two isoforms was found to be linked to AD. Increased levels of 17 A expression have been found in patient brains, suggesting a role of this lncRNA in GPR51 splicing regulation to preserve cerebral function [68].
Alternative isoform expression can also be controlled by antisense transcription via transcription attenuation (transcription RNAPII pausing and/or premature termination). A recent study shows that during specific differentiation stages in mouse embryonic stem cells (mESCs), the expression of two novel antisense enhancer-associated RNAs, Zmynd8as and Brd1as, is associated with shorter overlapping sense transcript isoforms with alternative termination sites [69], a phenomenon similarly found affecting the length of sense mRNAs of genes in a single operon in some bacteria [70]. Whereas the mechanism through which isoform specificity is achieved via enhancer-associated antisense RNAs has not been totally elucidated, this example enhances the corollary of antisense-mediated splicing mechanisms. A similar transcription attenuation mechanism mediating splicing is likely to occur at other genomic loci occupied by overlapping coding and non-coding genes [52].
5. LncRNAs Regulate Pre-mRNA Splicing by Modulating the Activity of Splicing Factors
As well as modifying AS by altering the chromatin landscape, through transcription, or through direct nucleic acid interactions, lncRNAs also interact in a dynamic network with many SFs and their pre-mRNA target sequences to modulate transcriptome reprogramming in eukaryotes.
LncRNAs that are notoriously associated with pre-mRNA splicing are the nuclear MALAT1/NEAT2 and NEAT1, both known to regulate the localization and phosphorylation status of SFs, and differentially expressed in a wide range of tissues in human and mouse. They are localized to specific subnuclear domains mainly in the nuclear speckle periphery, also known as paraspeckles (NEAT1); while MALAT1/NEAT2 is part of the polyadenylated component of nuclear speckles [71].
MALAT1/NEAT2 regulates splicing by modulating the activity of the conserved family of serine/arginine (SR) splicing factors by modifying their localization and phosphorylation [72] through shuttling between speckles and the sites of transcription, where splicing occurs [73]. In human cells, MALAT1/NEAT2 knockdown enhances the phosphorylated pool of SR proteins, displaying a more homogeneous nuclear distribution resulting in the mislocalization of speckle components and altered patterns of AS of pre-mRNAs [74,75,76]. MALAT1/NEAT2 binds to the SRSF1 splice factor through its RRM domain [77,78]. A correct phosphorylation/dephosphorylation cycle of SR proteins is fundamental to ensure the proper nucleocytoplasmic transport of mRNA–protein complexes (mRNPs). When SRSF1 is phosphorylated, it accumulates in nuclear speckles; while its dephosphorylation favors the interaction with mRNAs, transport and accumulation in the cytoplasm [79,80]. Although the exact mechanisms through which MALAT1/NEAT2-interacting with SRSF1 modulates the phosphorylated/dephosphorylated ratio of SR proteins remains unclear, it might occur through interaction with PP1/2A phosphatases or with the SRPK1 splice factor kinase [81,82,83] or alternatively, by the direct interaction with MALAT1/NEAT2 [73] (Figure 5a). Beyond AS, controlled levels of phosphorylated SR proteins are also likely to regulate other SR-dependent post-transcriptional regulatory events such as RNA export, nonsense mediated decay, and translation [77,81]. Interestingly, additional studies have also shown that MALAT1/NEAT2 can hybridize with many nascent pre-mRNAs at active gene loci and participate in pre-mRNA splicing of such actively transcribed genes by recruiting SFs to the pre-mRNAs [84]. Furthermore, according to the psoralen analysis of RNA interactions and structures (PARIS) [85] and to the more recent developed RIC-seq application [86], multiple interaction sites exist between MALAT1 and the spliceosomal RNA, U1snRNA, raising the possibility that MALAT1/NEAT2 influences RNA processing through the recruitment or modification of other proteins localized to these sites.
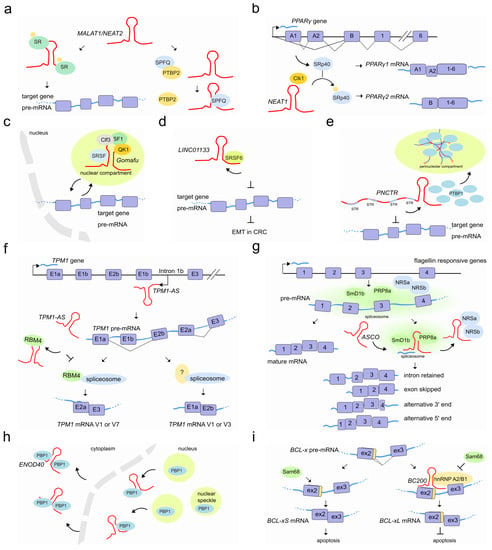
Figure 5.
LncRNAs regulate pre-mRNA splicing by recruiting or sequestering splicing factors into subnuclear compartments. (a) Left, MALAT1/ NEAT2 (red) is responsible of phosphorylated/dephosphorylated SFs shuttle from nuclear speckles to target mRNAs and cytoplasm. Right, MALAT1/ NEAT2 in colon cancer. The binding of SFPQ with MALAT1/ NEAT2 causes the disruption of the splicing regulator complex SFPQ-PTBP2 and the release of PTBP2. (b) During adipogenesis, the lncRNA NEAT1 (red) interacts with the CLK1 splicing factor kinase (orange) and regulates PPARγ gene splicing by modulating SRp40 (light blue, also known as SRSF5) phosphorylation status (light orange). When Srp40 is phosphorylated, the PPARγ pre-mRNA is mainly processed into the PPARy2 mRNA, whereas when dephosphoryled, Srp40 promotes the accumulation of the PPARy1 isoform. (c) Gomafu (red) sequesters multiple splicing factors (e.g., QKI, SRSF1, SF1, Clf3) in nuclear compartments and after specific stimuli/conditions it releases them in the nucleus to then direct the alternative splicing of pre-mRNA target genes (light blue) such as the schizophrenia-associated genes. (d) The lncRNA LINC01133 (red), by sequestering the splicing factor SRSF6, impairs the alternative splicing events on target pre-mRNA genes which ultimately lead to the inhibition of EMT and metastasis in colorectal cancer (CRC). (e) PNCTR (red), contains hundreds of short tandem repeats (STR) to bind and sequester a substantial fraction of PTBP1 in the perinucleolar compartment. (f) Sense and antisense TPM1 gene cotrascription results in both TPM1 pre-mRNA (light blue) and lncRNA TPM1-AS (red). The latter is then able to sequester RBM4 protein, forcing the splicing of TPM1 pre-mRNA (likely in cooperation with other protein partners) toward RBM4-deprived specific isoforms (V1 or V3). (g) LncRNA ASCO (red) associates with the two core components of the spliceosome SmD1b and PRP8a (green) and concomitantly sequesters NSRa and b proteins (light blue). By this mechanism ASCO enhances transcriptome diversity in response to flagellin, resulting in a variety pool of isoforms. (h) ENOD40 is recognized by MtRBP1 (here RBP1 for simplicity) and is responsible of its nucleocytoplasmic trafficking and accumulation into cytoplasmic granules, likely modulating RBP1-dependent splicing. (i) Left, BCL-x pre-mRNA interacts with Sam68 that promotes pre-mRNA splicing in the apoptotic isoform BCL-xS. Right, the presence of BC200 lncRNA and the recruitment of the hnRNP A2/B1 splicing factor interferes with the association of Sam68 and promote BCL-x splicing into the anti-apoptotic BCL-xL.
MALAT1/NEAT2 is abundantly expressed and widely associated with a variety of cancers. In hepatocellular carcinoma, MALAT1/NEAT2 acts as a proto-oncogene through Wnt pathway activation and transcriptional induction of SRSF1. The latter leads to the over accumulation of its active form in the cell nucleus and the modulation of SRSF1 splicing targets, including the anti-apoptotic AS isoforms of S6K1 [87]. In colorectal cancer, instead, MALAT1/NEAT2 triggers tumor growth and metastasis by binding to the splicing factor SFPQ causing the subsequent disruption of the splicing regulator complex SFPQ-PTBP2 and the release of the oncogene PTBP2 [88].
During adipocyte differentiation, the 4 kb lncRNA NEAT1 tethers the SR protein SRp40 (now known as SRSF5) and retains it in paranuclear bodies to fine-tune the relative abundance of mRNA isoforms of the major transcription factor driving adipogenesis, PPARγ. It has been observed that the NEAT1-SRp40 association enhances SRp40 phosphorylation by CLK1 splicing factor kinase activity [89]. Conversely, depletion of NEAT1 upon drug or siRNAs treatment, causes a decrease of both PPARγ isoforms (PPARγ1 and especially PPARγ2) and SRp40 phosphorylation impairment, respectively. Furthermore, while SRp40 depletion resulted in deregulation of both PPARγ isoforms and, predominantly of PPARγ2 mRNA levels, its overexpression increased exclusively PPARγ2. Therefore, fluxes in NEAT1 levels during adipogenesis seem to modulate AS events likely by controlling the availability of phosphorylated SRp40 thereby affecting PPARγ splicing [90] (Figure 5b).
Another lncRNA abundantly localized to nuclear bodies is the lncRNA Gomafu/RNCR2/MIAT which is expressed in a distinct set of neurons in the mouse retina [91,92] and implicated in retinal cell specification [93,94] brain development [95] and post-mitotic neuronal function [92,96]. Gomafu was found to interact directly with the splicing factors QKI and SRSF1 and its dysregulation leads to aberrant AS patterns that resemble those observed in schizophrenia-associated genes (DISC1 and ERBB4) [97]. In addition, Gomafu harbors a conserved tandem sequence of UACUAAC motifs that binds the splicing factor SF1, an early stage player of spliceosome assembly [98]. Furthermore, the splicing factor Clf3 was found to interact specifically with Gomafu in RNA–protein complexes containing the splicing factors SF1 and localize in specific nuclear bodies named CS bodies in the neuroblastoma cell line Neuro2A [99,100]. It has been proposed that Gomafu regulates splicing efficiency by changing the local concentration of SFs by sequestering them to separate regions of the nucleus [98] (Figure 5c).
An additional example of how lncRNAs may hijack SFs to fine-tune AS is the lncRNA LINC01133. This lncRNA binds the AS factor SRSF6, which induces EMT in colorectal cancer. By sequestering SRSF6 from other mRNA substrates, LINC01133 modulates SRSF6 activity and reshapes the population of AS isoforms of SRSF6 mRNA targets which finally leads to the inhibition of EMT and metastasis [101] (Figure 5d). Similarly, the lncRNA PNCTR, over-expressed in a variety of cancer cells, contains hundreds of short tandem repeats to bind and sequester a consistent fraction of PTBP1 in the perinucleolar compartment [102]. This prevents PTBP1 from influencing splicing and therefore PTBP1-dependent pro-apoptotic events [103,104,105] (Figure 5e).
LncRNAs that act as sponge molecules can extensively rewire post-transcriptional gene regulatory networks by uncoupling the protein–RNA interaction landscape in a cell-type-specific manner. A recent study showed that the loss of 39 lncRNAs causes many thousands of skipped exons and retained intron splicing events affecting a total of 759 human genes at the post-transcriptional level. Interestingly, the alternatively spliced events were found associated with RBPs binding in proximal intron–exon junctions in a cell-type-specific manner [106]. Similarly, the natural antisense TPM1-AS, reverse-transcribed from the fourth intronic region of the tropomyosin I gene (TPM1), regulates TPM1 alternative splicing through interaction with RNA-binding motif protein 4 (RBM4). The interaction prevents the binding of RBM4 to TPM1 pre-mRNA and inhibits TPM1 exon 2a inclusion (Figure 5f) [107]. Plant lncRNAs are also able to modulate AS by hijacking RBPs from their targets. In A. thaliana, an important number of intron retention events and a differential 5′ or 3′-end have been observed in a subset of genes in the plant-specific AS regulators (NSRa and NSRb) mutant compared to wild type plants [108]. In vitro experiments suggested that the lncRNA ASCO competes with other mRNA-target for its binding to these NSR regulators [109]. More recently, researchers analyzed the genome-wide effect of the knock-down and overexpression of ASCO and found a large number of deregulated and differentially spliced genes related to flagellin responses and biotic stress [110]. During this splicing process, ASCO interacts with multiple SFs including the highly conserved core spliceosome component PRP8a and another spliceosome component, SmD1b (Figure 5g). The NSR’s closest homolog in the model legume Medicago truncatula, MtRBP1/MtNSR1, has been characterized as a protein partner of the highly conserved and structured lncRNA ENOD40, which participates in root symbiotic nodule development [111]. ENOD40 appears to re-localize MtRBP1 from nuclear speckles into cytoplasmic granules during nodulation thereby modulating MtRBP1-dependent splicing events [112] (Figure 5h).
SF-associated lncRNAs might also influence a specific splicing outcome depending on a given cellular context. For example, the prostate-specific lncRNA PCGEM1 can mutually bind the splicing factors hnRNP A1 (silencer) and U2AF65 (enhancer) with opposite effects. While its interaction with hnRNP A1 suppresses the expression of androgen receptor (AR) splice variants such as AR3 by exon skipping, the interaction of PCGEM1 with U2AF65 promotes AR3 by exonization and favors castration resistance [113]. In the brain, the cytoplasmic 200 long non-coding RNA BC200 (BCYRN1) prevents apoptosis by modulating AS of a member of the Bcl-2 family proteins, the BCL-x gene [114]. AS of BCL-x leads to opposite effects on apoptosis when processed in either BCL-xL (anti-apoptotic) or BCL-xS (pro-apoptotic) [115]. Whereas BC200 overexpression promotes BCL-xL, its depletion induces BCL-xS formation. A 17-nucleotide complementary sequence to BCL-x pre-mRNA in BC200 appears to facilitate its binding to the pre-mRNA and promotes the recruitment of the hnRNP A2/B1 splicing factor. HnRNP A2/B1 binding interferes with association of BCL-x pre-mRNA with the BCL-xS-promoting factor Sam68 [116], leading to a blockade of Bcl-xS expression and anti-apoptotic conditions [117] (Figure 5i). Another example of a cellular context that causes isoform switching through lncRNAs is that of fibroblast growth factor receptors. FGF-2-sensitive cells arise following lnc-Spry1 depletion. This lncRNA acts as an early mediator of TGF-β signaling-induced EMT and regulates the expression of TGF-β-regulated gene targets. However, lnc-Spry1 has also been found to interact with the U2AF65 pyrimidine-tract binding splicing factor suggesting a dual role in affecting both transcriptional and post-transcriptional gene regulation in epithelial cells promoting a mesenchymal-like phenotype [118]. Recently, a link between stress-induced lncRNAs and AS has also been shown. The lncRNA LASTR, elevated in hypoxic breast cancer, is upregulated through the stress-induced JNK/c-JUN pathway. It interacts with SART3, a U4/U6 snRNP recycling factor, and promotes splicing efficiency. Depletion of LASTR leads to increased intron retention, with the resulting downregulation of essential genes to the detriment of cancer cells [119].
Ribosomal and RNA splicing complexes components, including YBX1, PCBP1, PCBP2, RPS6 and RPL7, have been shown to bind LINC-HELLP, a lncRNA implicated in the pregnancy-specific HELLP syndrome, through a splicing-mediated mechanism that is largely unknown. HELLP patient mutations within LINC-HELLP, alter the binding with these proteins depending to their location and negatively affect trophoblast differentiation. While mutations occurring from the 5′-end up to the middle of the LINC-HELLP are likely to cause loss of partner protein interactions, those at the far 3′-end increase their binding [120]. Among a cohort of breast cancer-associated and oestrogen-regulated lncRNAs, DSCAM-AS1 has been recently found to be associated with tumor progression and tamoxifen resistance [121]. Researchers found over 2085 splicing events regulated by DSCAM-AS1, including alternative polyadenylation sites, 3′ UTR shortening and exon skipping events. DSCAM-AS1 affects target gene expression and causes changes in the AS by interacting with hnRNPL which appears to mediate the exon skipping and 3′ UTR usage by a mechanism not yet fully elucidated [121].
Canonical splicing of the linear pre-mRNA can compete for SFs with circularization of exons in circRNAs by mechanisms that are tissue specific and conserved in animals [122]. In flies and humans, the second exon of the SF muscleblind (Mbl (fly)/MBNL1 (human)) is circularized in circMbl. The introns flanking this circRNA as well as the circRNA itself contain highly conserved Mbl/MBNL1 binding sites, which are strongly and specifically bound by Mbl. Modulation of Mbl levels regulates the splicing of its own pre-mRNA into circMbl, and this in turn relies on Mbl binding sites [123] (Figure 6a). A circRNA proposed to act as an angiogenesis regulator by sponging SFs, is circSMARCA5. CircSMARCA5 interacts with SRSF1 and promotes the switching from pro- to anti-angiogenic splicing isoforms of VEGF-A in glioblastoma multiforme, representing an opportunity to develop a novel anti-angiogenic cancer therapy [124]. Interestingly, circRNAs have been also found associated with the splicing factor QKI during human EMT [123], and correlate with exon skipping throughout the genome in human endothelial cells [125].
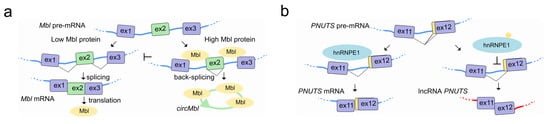
Figure 6.
LncRNAs regulate pre-mRNA splicing by competing for splicing factors during their own splicing. (a) Left, In the presence of low amounts of Mbl (orange), the Mbl transcript is canonically spliced into a translatable mRNA encoding the Mbl protein. Right, when Mbl levels are high, Mbl binds to the pre-mRNA at the intronic regions flanking exon 2 and causes the exon2 back-splicing into circMbl (green), thereby preventing linear splicing and translation of the Mbl protein. CircMbl can also sequester Mbl protein, lowering its free cellular concentration, thereby providing a feedback mechanism to regulate Mbl levels. (b) The PNUTS gene can encode either the PNUTS mRNA or the lncRNA PNUTS depending on the usage of the 3′ alternative splice site located at the 5′-end of exon 12 which leads to the change of the ORF and the generation of a premature stop codon. Left, upon the binding of hnRNP E1 to a BAT consensus element located in the alternative splicing site that mask and prevents its usage, PNUTS pre-mRNA is spliced into PNUTS mRNA then translated into the PNUTS protein. Right, loss of hnRNP E1 binding to the alternative splice site uncovers the consensus element and allows its usage by the spliceosome machinery to achieve the splicing to yield the lncRNA PNUTS transcript.
LncRNAs can also interact with SFs to regulate their own splicing as is the case with the lncRNA PNUTS, also known as a competitive endogenous RNA (ce-RNA). The PNUTS gene can express a regular PNUTS mRNA encoding for the protein phosphatase 1 binding protein, but also to an alternatively spliced non-coding isoform called lncRNA-PNUTS with a distinct biological function. While PNUTS mRNA is ubiquitously expressed, the lncRNA-PNUTS one is more tumor-relevant and generally serves as a competitive sponge for miR-205 during EMT. The splicing decision to produce either mRNA or lncRNA relies on the binding of hnRNP E1 to a structural element located in exon 12 of PNUTS pre-RNA. Once released from this structural element, hnRNP E1 translocates from the nucleus to cytoplasm, allowing the AS and generation of the non-coding isoform of PNUTS to take place [126] (Figure 6b).
6. Concluding Remarks and Future Perspectives
Growing evidence suggests that lncRNAs control the regulation of AS in response to several physiological stimuli or during disease processes through changes in chromatin conformation, or by interfering with the overlapping antisense genes, genomic loci or SF activity. LncRNA antisense transcription pausing and elongation, as well as the capability of sponging RBPs, can also result in altered mRNA splice isoform expression patterns. The recent discovery of the circRNAs has also shown how a special class of lncRNAs can wholly integrate with the splicing process itself, affecting the splicing outcome of their linear cognates.
Some aspects of lncRNA-mediated AS regulation remain mostly unexplored. For instance most lncRNA sequences are not conserved across species, suggesting that most of their functionality might relies on their RNA structure. The role played by lncRNA secondary structure in determining their ability to regulate AS remains poorly investigated. Moreover, mRNA methylation is known to impact on AS by affecting the accessibility of hnRNPs to pre-mRNAs. Specifically, N6-methyladenosine (m6A) can serve as a switch to regulate gene expression and RNA maturation [127]. The existence of an interplay between RNA methylation and long non-coding RNA also raises the question of whether or not lncRNAs play a role in recruiting or reading mRNA methylation during AS processes. Furthermore, m6A modifications that occur on lncRNAs and circRNAs might change their function in AS regulation by providing a binding site for the m6A reader proteins or by modulating their structure–all of these questions remain unanswered.
Over the past years, our understanding of the mechanisms through which lncRNAs affect gene expression has been limited by their intrinsic properties (mainly length and low expression) and the lack of powerful experimental assays. With the increasing prevalence of splicing events and the discovery of over a hundred thousand lncRNAs, it is likely that the involvement of lncRNAs in regulating AS is far greater than the currently known. Further research is needed to gain a deeper understanding of how lncRNAs contribute to the regulation of AS in development and disease.

Table 1.
List of lncRNAs involved in splicing regulation.
Table 1.
List of lncRNAs involved in splicing regulation.
| LncRNA Name | Splicing Target | Splicing Mechanism | Regulatory Effect or Associated Disease | Ref |
|---|---|---|---|---|
| LncRNAs regulating AS by chromatin modifications | ||||
| asFGFR2 | FGFR2 | Recruiting Polycomb complexes and KDM2a to modify histone methylation and favor exon IIIb inclusion | Epithelial development | [23] |
| Antisense transcripts at each Pcdhα first exon | Pcdhα | First exon selection by histone modifications and distant DNA loop | Neuronal self-identity | [31] |
| LncRNAs regulate AS through DNA-RNA interactions | ||||
| SEP3 exon6 circRNA (plant) | SEP3 | Exon skipping through R-loop formation at exon 6 | Flowering time | [44] |
| LncRNAs regulate AS through RNA-RNA interactions | ||||
| NCYM NAT | NMYC | Intron I retention via antisense-sense RNA-RNA duplex | Cancer | [57] |
| NR1D1 | THRA | Favoring α1 isoform by forming antisense-sense RNA-RNA duplex with the α2 mRNA | Thyroid hormone-responsiveness | [58,59] |
| SAF | FAS | Exon 6 skipping by forming RNA-RNA duplex with the target pre-mRNA and recruiting SPF45 | Cancer Apoptosis | [60] |
| ZEB2 NAT | ZEB2 | Preventing splicing of the IRES-containing intron through RNA-RNA interaction with the mRNA | EMT | [61] |
| 51A | SORL1 | Splicing shift from A to variant B by antisense-sense RNA-RNA duplex with an intronic sequence of the pre-mRNA | Alzheimer | [67] |
| 17A | GPR51 | Splicing shift from full-length to shorter GABAB R2 variant by antisense-sense RNA-RNA duplex | Alzheimer | [68] |
| LncRNAs regulate AS by modulating the activity of splicing factors | ||||
| MALAT1/NEAT2 | Modulation of SR localization and phosphorylation Uncoupling PTBP2 from SFPQ-PTBP2 | Cancer | [73,88] | |
| NEAT1 | PPARγ | By interacting with CLK1 kinase to modulate SRp40 phosphorylation status | Adipocyte differentiation | [71,89,90] |
| Gomafu/RNCR2/ MIAT | Interaction with QKI and SRSF1 Association with SF1 Localization of SF1 and Clf3 in CS bodies | Schizophrenia Retinal cell and brain development Post-mitotic neuronal function | [97,98,99,100] | |
| LINC01133 | Interaction and titration of SRSF6 splicing factor from target genes | EMT | [101] | |
| PNCTR | Hijacking PTBP1 in the perinucleolar compartment | Cell survival | [102] | |
| TPM1-AS | TPM1 | Splicing shift to V1 or V3 isoforms by sequestering RBM4 | Cancer | [107] |
| ASCO (plant) | Association with SmD1b and PRP8a and hijacking NSRa/b from the spliceosome | Lateral root formation | [109,110] | |
| ENOD40 (plant) | Control nucleocytoplasmic of MtRBP1 | Symbiotic nodule development | [111,112] | |
| PCGEM1 | Mutual bond with either hnRNP A1 or U2AF65 to promote or suppress specific AR splice variants | Castration resistance | [113] | |
| BC200 | BCL-x | Interaction with pre-mRNA and recruitment of the hnRNP A2/B1 which prevent Sam68 association | Apoptosis | [115,117] |
| Lnc-Spry1 | Interaction with U2AF65 | EMT | [118] | |
| LASTR | Promoting splicing efficiency by interacting with SART3 | Stress-induced JNK/c-JUN pathway | [119] | |
| LINC-HELLP | Interaction with ribosomal and splicing complex components (eg: YBX1, PCBP1, PCBP2, RPS6 and RPL7) | HELLP syndrome | [120] | |
| DSCAM-AS1 | Exon skipping and 3′ UTR usage by interaction with hnRNPL | Tumor progression and anti-estrogen resistance | [121] | |
| CircMbl | Mbl | Competing with the linear cognate by sequestering Mbl protein | Neuron Development | [123] |
| CircSMARCA5 | Interaction with SRSF1 and promotion of the anti-angiogenic splicing isoforms of VEGF-A | Angiogenesis | [124] | |
| PNUTS | PNUTS | Self-splicing regulation modulating the activity of hnRNP E1 | EMT | [126] |
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
Alternative splicing (AS); splicing factor (SF); RNA binding proteins (RBPs); long non-coding RNAs (lncRNAs); RNA polymerase II (RNAPII); CCCTC-binding factor (CTCF); Natural Antisense Transcripts (NATs); epithelial-mesenchymal transition (EMT); Alzheimer’s disease (AD); amyloid precursor protein (APP); circular RNAs (circRNAs); N6-methyladenosine (m6A).
References
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175. [Google Scholar] [CrossRef]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670. [Google Scholar] [CrossRef]
- Rosenfeld, M.G.; Amara, S.G.; Roos, B.A.; Ong, E.S.; Evans, R.M. Altered expression of the calcitonin gene associated with RNA polymorphism. Nature 1981, 290, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Ward, A.J.; Cooper, T.A. The pathobiology of splicing. J. Pathol. 2010, 220, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Magen, A.; Ast, G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007, 35, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. RNA regulation: A new genetics? Nat. Rev. Genet. 2004, 5, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- David Wang, X.Q.; Crutchley, J.L.; Dostie, J. Shaping the Genome with Non-coding RNAs. Curr. Genom. 2011, 307–321. [Google Scholar] [CrossRef]
- Flynn, R.A.; Chang, H.Y. Active chromatin and noncoding RNAs: An intimate relationship. Curr. Opin. Genet. Dev. 2012, 22, 172–178. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Goff, L.A.; Rinn, J.L. Linking RNA biology to lncRNAs. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef]
- Schor, I.E.; Rascovan, N.; Pelisch, F.; Alió, M.; Kornblihtt, A.R. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 4325–4330. [Google Scholar] [CrossRef]
- Batsché, E.; Yaniv, M.; Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct Mol. Biol. 2006. [Google Scholar] [CrossRef] [PubMed]
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26. [Google Scholar] [CrossRef]
- Gonzalez, I.; Munita, R.; Agirre, E.; Dittmer, T.A.; Gysling, K.; Misteli, T.; Luco, R.F. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015, 22, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Du, J.; Sun, B.; Dong, X.; Xu, G.; Zhou, J.; Huang, Q.; Liu, Q.; Hao, Q.; Ding, J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006, 34, 6621–6628. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R. Chromatin, transcript elongation and alternative splicing. Nat. Struct. Mol. Biol. 2006, 13, 5–7. [Google Scholar] [CrossRef]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef]
- Amaral, P.P.; Leonardi, T.; Han, N.; Viré, E.; Gascoigne, D.K.; Arias-Carrasco, R.; Büscher, M.; Pandolfini, L.; Zhang, A.; Pluchino, S.; et al. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018, 19, 32. [Google Scholar] [CrossRef]
- Pisignano, G.; Pavlaki, I.; Murrell, A. Being in a loop: How long non-coding RNAs organise genome architecture. Essays Biochem. 2019, 63, 177–186. [Google Scholar] [CrossRef]
- Lefevre, P.; Witham, J.; Lacroix, C.E.; Cockerill, P.N.; Bonifer, C. The LPS-Induced Transcriptional Upregulation of the Chicken Lysozyme Locus Involves CTCF Eviction and Noncoding RNA Transcription. Mol. Cell. 2008, 32, 129–139. [Google Scholar] [CrossRef]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffié, R.; Monahan, K.; O’Keeffe, S.; Simon, M.D.; et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 2019, 177, 639–653. [Google Scholar] [CrossRef]
- Guo, Y.; Monahan, K.; Wu, H.; Gertz, J.; Varley, K.E.; Li, W.; Myers, R.M.; Maniatis, T.; Wu, Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl. Acad. Sci. USA 2012, 109, 21081–21086. [Google Scholar] [CrossRef] [PubMed]
- Kehayova, P.; Monahan, K.; Chen, W.; Maniatis, T. Regulatory elements required for the activation and repression of the protocadherin-á gene cluster. Proc. Natl. Acad. Sci. USA 2011, 108, 17195–17200. [Google Scholar] [CrossRef] [PubMed]
- Monahan, K.; Rudnick, N.D.; Kehayova, P.D.; Pauli, F.; Newberry, K.M.; Myers, R.M.; Maniatis, T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of Protocadherin-α gene expression. Proc. Natl. Acad. Sci. USA 2012, 109, 9125–9130. [Google Scholar] [CrossRef] [PubMed]
- Ribich, S.; Tasic, B.; Maniatis, T. Identification of long-range regulatory elements in the protocadherin-α gene cluster. Proc. Natl. Acad. Sci. USA 2006, 103, 19719–19724. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Lee, E.C.S.; Elhassan, S.A.M.; Lim, G.P.L.; Kok, W.H.; Tan, S.W.; Leong, E.N.; Tan, S.H.; Chan, E.W.L.; Bhattamisra, S.K.; Rajendran, R.; et al. The roles of circular RNAs in human development and diseases. Biomed. Pharmacother. 2019, 111, 198–208. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang HBin Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Alexander, R.D.; Innocente, S.A.; Barrass, J.D.; Beggs, J.D. Splicing-Dependent RNA polymerase pausing in yeast. Mol. Cell. 2010, 40, 582–593. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; Webb, S.; Kerr, A.; Tollervey, D. Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria. PLoS Genet. 2014, 10, e1004716. [Google Scholar] [CrossRef] [PubMed]
- Wongsurawat, T.; Jenjaroenpun, P.; Kwoh, C.K.; Kuznetsov, V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012. [Google Scholar] [CrossRef] [PubMed]
- Sugino, A.; Hirose, S.; Okazaki, R. RNA-linked nascent DNA fragments in Escherichia coli. Proc. Natl. Acad. Sci. USA 1972. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989. [Google Scholar] [CrossRef]
- Williams, J.S.; Kunkel, T.A. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair (Amst.) 2014. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Morrissy, A.S.; Griffith, M.; Marra, M.A. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Van Ommen, G.J.B. Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA 2007. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Myers, A.J.; Hsiao, J.; Wahlestedt, C. Natural antisense transcripts. Hum. Mol. Genet. 2014. [Google Scholar] [CrossRef]
- Bardou, F.; Merchan, F.; Ariel, F.; Crespi, M. Dual RNAs in plants. Biochimie 2011. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, Y.; Islam, S.M.; Alagu, J.; Kaneko, Y.; Kato, M.; Tanaka, Y.; Kawana, H.; Hossain, S.; Matsumoto, D.; Yamamoto, M.; et al. NCYM, a Cis-Antisense Gene of MYCN, Encodes a De Novo Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef]
- Krystal, G.W.; Armstrong, B.C.; Battey, J.F. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol. Cell Biol. 1990. [Google Scholar] [CrossRef] [PubMed]
- Munroe, S.H.; Lazar, M.A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol Chem. 1991, 266, 22083–22086. [Google Scholar] [CrossRef]
- Chassande, O.; Fraichard, A.; Gauthier, K.; Flamant, F.; Legrand, C.; Savatier, P.; Laudet, V.; Samarut, J. Identification of Transcripts Initiated from an Internal Promoter in the c-erbAα Locus That Encode Inhibitors of Retinoic Acid Receptor-α and Triiodothyronine Receptor Activities. Mol. Endocrinol. 1997, 11, 1278–1290. [Google Scholar] [CrossRef][Green Version]
- Villamizar, O.; Chambers, C.B.; Riberdy, J.M.; Persons, D.A.; Wilber, A. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget 2016. [Google Scholar] [CrossRef]
- Beltran, M.; Puig, I.; Peña, C.; García, J.M.; Alvarez, A.B.; Peña, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.L.; Galasko, D.R.; Ringman, J.M.; Vinters, H.V.; Edland, S.D.; Pomakian, J.; Ubeda, O.J.; Rosario, E.R.; Teter, B.; Frautschy, S.A.; et al. Reduction of SorLA/LR11, a sorting protein limiting β-amyloid production, in alzheimer disease cerebrospinal fluid. Arch. Neurol. 2009, 66, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Reitz, C.; Cheng, R.; Rogaeva, E.; Lee, J.H.; Tokuhiro, S.; Zou, F.; Bettens, K.; Sleegers, K.; Tan, E.K.; Kimura, R.; et al. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch. Neurol. 2011, 68, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Rogaeva, E.; Meng, Y.; Lee, J.H.; Gu, Y.; Kawarai, T.; Zou, F.; Katayama, T.; Baldwin, C.T.; Cheng, R.; Hasegawa, H.; et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007, 39, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Reiche, J.; Schmidt, V.; Gotthardt, M.; Spoelgen, R.; Behlke, J.; von Arnim, C.A.; Breiderhoff, T.; Jansen, P.; Wu, X.; et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA 2005, 102, 13461–13466. [Google Scholar] [CrossRef]
- Small, S.A.; Kent, K.; Pierce, A.; Leung, C.; Kang, M.S.; Okada, H.; Honig, L.; Vonsattel, J.P.; Kim, T.W. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann. Neurol. 2005, 58, 909–919. [Google Scholar] [CrossRef]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. DMM Dis Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Onodera, C.S.; Underwood, J.G.; Katzman, S.; Jacobs, F.; Greenberg, D.; Salama, S.R.; Haussler, D. Gene isoform specificity through enhancer-associated antisense transcription. PLoS ONE 2012, 7, e43511. [Google Scholar] [CrossRef]
- Stork, M.; Di Lorenzo, M.; Welch, T.J.; Crosa, J.H. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J. Bacteriol. 2007. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007, 8, 39. [Google Scholar] [CrossRef]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.G.; Porro, E.B.; Galceran, J.; Tempst, P.; Nadal-Ginard, B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993. [Google Scholar] [CrossRef]
- Gozani, O.; Patton, J.G.; Reed, R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994. [Google Scholar] [CrossRef]
- Misteli, T.; Cáceres, J.F.; Clement, J.Q.; Krainer, A.R.; Wilkinson, M.F.; Spector, D.L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998. [Google Scholar] [CrossRef] [PubMed]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009. [Google Scholar] [CrossRef]
- Cao, W.; Jamison, S.F.; Garcia-Blanco, M.A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 1997, 3, 1456–1467. [Google Scholar] [PubMed]
- Sanford, J.R.; Ellis, J.D.; Cazalla, D.; Cáceres, J.F. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15042–15047. [Google Scholar] [CrossRef]
- Huang, Y.; Yario, T.A.; Steitz, J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 2004. [Google Scholar] [CrossRef]
- Stamm, S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol Chem. 2008. [Google Scholar] [CrossRef]
- Shi, Y.; Manley, J.L. A Complex Signaling Pathway Regulates SRp38 Phosphorylation and Pre-mRNA Splicing in Response to Heat Shock. Mol. Cell 2007. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Ding, J.H.; Adams, J.A.; Ghosh, G.; Fu, X.D. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014, 159, 188–199. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Cao, C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z.; Hu, N.; Yu, X.; et al. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature 2020, 582, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017, 77, 155–1167. [Google Scholar] [CrossRef]
- Ji, Q.; Zhang, L.; Liu, X.; Zhou, L.; Wang, W.; Han, Z.; Sui, H.; Tang, Y.; Wang, Y.; Liu, N.; et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer. 2014, 111, 736–748. [Google Scholar] [CrossRef]
- Jiang, K.; Patel, N.A.; Watson, J.E.; Apostolatos, H.; Kleiman, E.; Hanson, O.; Hagiwara, M.; Cooper, D.R. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCβJII messenger ribonucleic acid. Endocrinology 2009, 150, 2087–2097. [Google Scholar] [CrossRef]
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARγ2 splicing during adipogenesis in 3T3-L1 cells. Genes 2014, 5, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Blackshaw, S.; Harpavat, S.; Trimarchi, J.; Cai, L.; Huang, H.; Kuo, W.P.; Weber, G.; Lee, K.; Fraioli, R.E.; Cho, S.H.; et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004, 2. [Google Scholar] [CrossRef] [PubMed]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120, 2498–2506. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Blackshaw, S. New meaning in the message: Noncoding RNAs and their role in retinal development. Dev. Dyn. 2009. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Blackshaw SRapicavoli, N.A.; Poth, E.M.; Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008. [Google Scholar] [CrossRef]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef]
- Tsuiji, H.; Yoshimoto, R.; Hasegawa, Y.; Furuno, M.; Yoshida, M.; Nakagawa, S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 2011, 16, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Ladd, A.N. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol. Cell Neurosci. 2013, 56, 456–464. [Google Scholar] [CrossRef]
- Ishizuka, A.; Hasegawa, Y.; Ishida, K.; Yanaka, K.; Nakagawa, S. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cells. 2014, 19, 704–721. [Google Scholar] [CrossRef]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial–mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.C.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540. [Google Scholar] [CrossRef]
- Bushell, M.; Stoneley, M.; Kong, Y.W.; Hamilton, T.L.; Spriggs, K.A.; Dobbyn, H.C.; Qin, X.; Sarnow, P.; Willis, A.E. Polypyrimidine Tract Binding Protein Regulates IRES-Mediated Gene Expression during Apoptosis. Mol. Cell 2006, 23, 401–412. [Google Scholar] [CrossRef]
- Bielli, P.; Bordi, M.; Di Biasio, V.; Sette, C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res. 2014, 42, 12070–12081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bahi, N.; Llovera, M.; Comella, J.X.; Sanchis, D. Polypyrimidine tract binding proteins (PTB) regulate the expression of apoptotic genes and susceptibility to caspase-dependent apoptosis in differentiating cardiomyocytes. Cell Death Differ. 2009, 16, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- Porto, F.W.; Daulatabad, S.V.; Janga, S.C. Long non-coding RNA expression levels modulate cell-type-specific splicing patterns by altering their interaction landscape with RNA-binding proteins. Genes 2019, 10, 593. [Google Scholar] [CrossRef]
- Huang, G.W.; Zhang, Y.L.; Liao LDi Li, E.M.; Xu, L.Y. Natural antisense transcript TPM1-AS regulates the alternative splicing of tropomyosin I through an interaction with RNA-binding motif protein 4. Int. J. Biochem. Cell Biol. 2017, 90, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.; Crespi, M. Long Noncoding RNA Modulates Alternative Splicing Regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef]
- Tran, V.D.T.; Souiai, O.; Romero-Barrios, N.; Crespi, M.; Gautheret, D. Detection of generic differential RNA processing events from RNA-seq data. RNA Biol. 2016, 13, 59–67. [Google Scholar] [CrossRef]
- Rigo, R.; Bazin, J.; Romero-Barrios, N.; Moison, M.; Lucero, L.; Christ, A.; Benhamed, M.; Blein, T.; Huguet, S.; Charon, C.; et al. The Arabidopsis lncRNA ASCO modulates the transcriptome through interaction with splicing factors. EMBO Rep. 2020, 21, e48977. [Google Scholar] [CrossRef]
- Crespi, M.D.; Jurkevitch, E.; Poiret, M.; d’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E.; Kondorosi, A. Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099–5112. [Google Scholar] [CrossRef]
- Campalans, A.; Kondorosi, A.; Crespi, M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant. Cell 2004, 16, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, N.; Huang, J.; Ho, T.T.; Zhu, Z.; Qiu, Z.; Zhou, X.; Bai, C.; Wu, F.; Xu, M.; et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget 2016, 7, 15481–15491. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C. BCL-2 protein family. Essential regulators of cell death. Preface. Adv. Exp. Med. Biol. 2010. [Google Scholar]
- Minnt, A.J.; Boise, L.H.; Thompson, C.B. Bcl-XS Antagonizes the protective effects of Bcl-xL. J. Biol. Chem. 1996, 271, 6306–6312. [Google Scholar] [CrossRef] [PubMed]
- Paronetto, M.P.; Achsel, T.; Massiello, A.; Chalfant, C.E.; Sette, C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 2007, 176, 929–939. [Google Scholar] [CrossRef]
- Singh, R.; Gupta, S.C.; Peng, W.X.; Zhou, N.; Pochampally, R.; Atfi, A.; Watabe, K.; Lu, Z.; Mo, Y.Y. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016, 7, e2262. [Google Scholar] [CrossRef]
- Rodríguez-Mateo, C.; Torres, B.; Gutiérrez, G.; Pintor-Toro, J.A. Downregulation of Lnc-Spry1 mediates TGF-β-induced epithelial-mesenchymal transition by transcriptional and posttranscriptional regulatory mechanisms. Cell Death Differ. 2017, 24, 785–797. [Google Scholar] [CrossRef]
- De Troyer, L.; Zhao, P.; Pastor, T.; Baietti, M.F.; Barra, J.; Vendramin, R.; Dok, R.; Lechat, B.; Najm, P.; Van Haver, D.; et al. Stress-induced lncRNA LASTR fosters cancer cell fitness by regulating the activity of the U4/U6 recycling factor SART3. Nucleic Acids Res. 2020, 48, 2502–2517. [Google Scholar] [CrossRef]
- van Dijk, M.; Visser, A.; Buabeng, K.M.L.; Poutsma, A.; van der Schors, R.C.; Oudejans, C.B.M. Mutations within the LINC-HELLP non-coding RNA differentially bind ribosomal and RNA splicing complexes and negatively affect trophoblast differentiation. Hum. Mol. Genet. 2015, 24, 5475–5485. [Google Scholar] [CrossRef] [PubMed]
- Elhasnaoui, J.; Miano, V.; Ferrero, G.; Doria, E.; Leon, A.E.; Fabricio, A.S.C.; Annaratone, L.; Castellano, I.; Sapino, A.; De Bortoli, M. DSCAM-AS1-driven proliferation of breast cancer cells involves regulation of alternative exon splicing and 3′-end usage. Cancers 2020, 12, 1453. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell. Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Xie, L.; Mao, M.; Xiong, K.; Jiang, B. Circular RNAs: A novel player in development and disease of the central nervous system. Front. Cell Neurosci. 2017, 11, 354. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef]
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. J. Mol. Biol. 2015, 427, 2414–2417. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6 -methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).