Abstract
The first reference to the “C-value paradox” reported an apparent imbalance between organismal genome size and morphological complexity. Since then, next-generation sequencing has revolutionized genomic research and revealed that eukaryotic transcriptomes contain a large fraction of non-protein-coding components. Eukaryotic genomes are pervasively transcribed and noncoding regions give rise to a plethora of noncoding RNAs with undeniable biological functions. Among them, long noncoding RNAs (lncRNAs) seem to represent a new layer of gene expression regulation, participating in a wide range of molecular mechanisms at the transcriptional and post-transcriptional levels. In addition to their role in epigenetic regulation, plant lncRNAs have been associated with the degradation of complementary RNAs, the regulation of alternative splicing, protein sub-cellular localization, the promotion of translation and protein post-translational modifications. In this review, we report and integrate numerous and complex mechanisms through which long noncoding transcripts regulate post-transcriptional gene expression in plants.
1. Introduction
Unlike in prokaryotes, genomes in eukaryotes exhibit a large variability in their size [1,2], which does not always correlate with the number of protein-coding genes nor the developmental complexity of organisms. This paradox of an apparent imbalance between organismal genome size and morphological complexity, dubbed the “C-value paradox” [3,4], was in part solved by the extraordinary progress made in next-generation sequencing technologies. Indeed, eukaryotic transcriptomes include a large fraction of non-protein-coding components [5]. Although up to 90% of eukaryotic genomes is estimated to be transcribed during development, only an estimated 2% of transcribed RNAs will code for proteins [6,7]. The noncoding genome, long considered silent and declared as “junk DNA” due to its high content in pseudogenes, simple repeats, and transposons [8,9], encodes a plethora of noncoding RNAs (ncRNAs) with unarguable biological functions. These comprise housekeeping RNAs (small nuclear and nucleolar RNAs, transfer RNAs, ribosomal RNAs, telomerase RNAs, tRNA-derived fragments, and tRNA halves), small regulatory RNAs (micro RNAs, small interfering RNAs, piwi-interacting RNAs, and Y RNAs), and long noncoding RNAs (lncRNAs), also including enhancer RNAs, transposon-derived RNAs, and circular RNAs [10].
LncRNAs form the most diversified group of ncRNAs, exhibiting a large range of sizes varying from 200 bases to over 100 kb in length. They are expressed in various tissues, cell-types, and cell-states, and function in the nucleus or cytoplasm [11,12]. Given their vast diversity, lncRNAs are commonly classified according to their location and orientation relative to neighboring protein-coding transcripts. Long intronic RNAs are transcribed exclusively from intronic regions, whereas long intergenic ncRNAs lie outside of genes and include promoter-, enhancer-, and transposable element-derived lncRNAs and sometimes give rise to double-stranded RNAs. Sense and antisense double-stranded lncRNAs are transcribed from the sense and antisense strands, respectively, while natural antisense transcripts (NATs) initiate in the reverse strand of sense protein coding regions (cis-NATs) or are complementary to a sense transcript located in a distinct genomic locus (trans-NATs) [6,13]. CircRNAs constitute a novel class of lncRNAs consisting in covalently closed molecules of single-stranded RNA, resulting from back-splicing, a non-canonical form of alternative splicing [14]. Alternatively, lncRNAs can be further categorized depending on their molecular functions and interactions with additional regulatory molecules such as proteins, DNA, or other RNAs [15,16].
It is increasingly clear that lncRNAs participate in virtually every aspect of gene expression. In plants, although the functional characterization of ncRNAs is still in its early stages, several lncRNAs have been described as regulators of gene transcription, capable of conditioning the epigenetic environment of their genomic targets and of modulating the activity of transcriptional complexes [17]. In addition, at the post-transcriptional level, various lncRNAs have been associated with complementary target-RNA degradation, alternative splicing, promotion of translation, protein sub-cellular localization and post-translational modifications. Notably, lncRNA-mediated post-translational modifications of histones are related to the transcriptional regulation of target genes, which has been recently reviewed [17]. Here, we report and integrate recent discoveries about plant lncRNA-mediated regulations of post-transcriptional gene expression.
2. Long Noncoding RNAs Mediating Complementary Target-RNA Degradation
Natural antisense transcripts (NATs) constitute an important class of lncRNAs, exerting a wide variety of molecular functions in eukaryotes [18,19]. They are complementary to sense mRNAs and can be classified into cis-NATs generated from a single locus showing sequence complementarity with their corresponding sense transcript or trans-NATs that are transcribed from different distant loci and typically display imperfect complementarities with their target endogenous RNA [20,21] (Figure 1). In silico analyses performed in several plant species have led to the identification of a large number of NATs [20,22,23,24,25,26]. In particular, a genome-wide analysis using a custom-designed NAT array revealed that up to 70% of annotated mRNAs have complementary NATs in Arabidopsis thaliana [27].
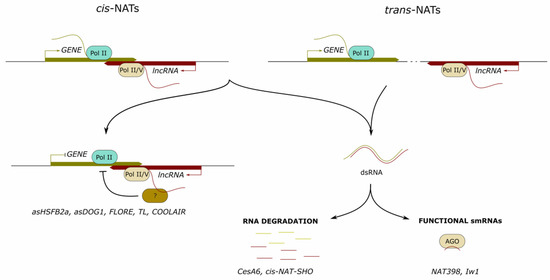
Figure 1.
Long noncoding RNAs forming RNA–RNA pairs in the nucleus. Long noncoding (lnc) RNAs can form RNA pairs (dsRNA) with complementary mRNAs in cis (cis-NATs) or in trans (trans-NATs), leading to RNA degradation or to the formation of functional small (sm) RNAs. In addition, antisense transcripts can locally recruit protein partners that modulate the transcriptional activity of overlapping protein-coding genes. Examples of characterized lncRNAs are indicated at the bottom.
2.1. LncRNAs Involved in Discordant Regulation
NATs can affect positively (concordant regulation) or negatively (discordant regulation) the expression of sense transcripts. An example of discordant regulation is provided by the NAT-lncRNA asHSFB2a which counteracts the expression of the HEAT SHOCK FACTOR B2a (HSFB2a) mRNA in A. thaliana female gametophytes [28]. The overexpression of asHSFB2a in transgenic plants leads to the absence of HSFB2a RNA while the overexpression of HSFB2a results in a complete loss of asHSFB2a expression, suggesting that HSFB2a and asHSFB2a are mutually repressive [28]. Similarly, the NAT-lncRNA DELAY OF GERMINATION 1 (asDOG1) was found to be a negative regulator of DOG1 expression, a gene involved in the control of germination [29]. Down-regulation of asDOG1 transcription increases the levels of DOG1 sense mRNAs, enhancing seed dormancy [29]. As a last example in A. thaliana, a screening of lncRNAs using a custom-made array led to the identification of the circadian-regulated lncRNA CDF5 LONG NONCODING RNA (FLORE), a NAT of the CYCLING DOF FACTOR 5 (CDF5) transcript, which likely connects the circadian clock to the photoperiodic flowering pathway [30]. FLORE is specifically expressed in the vasculature and regulates CDF5 in cis as well as CDF1 and CDF3 in trans. Interestingly, FLORE and CDF5 show a mutual inhibition behavior, suggesting that the CDF5/FLORE NAT pair constitutes a circadian regulatory module, which buffers its own circadian oscillation and photoperiodic flowering [30]. In rice, the NAT-lncRNA TWISTED LEAF (TL) is transcribed from the opposite strand of the OsMYB60 locus encoding an R2R3 MYB transcription factor [31]. Downregulation of TL by RNA interference leads to a significant increase in OsMYB60 expression levels and twisted leaf blades. It was suggested that TL may play a cis-regulatory role on OsMYB60 by affecting H3K27me3, H3K36me2, and H3K36me3 histone mark deposition [31].
2.2. LncRNAs as Precursors of Small Regulatory RNAs
In addition, various NAT-lncRNAs have been reported to cause post-transcriptional silencing through the production of regulatory small interfering (si) RNAs derived from NAT pairs. This mechanism was first described in the regulation of salt tolerance in A. thaliana [32]. Under salt stress, the induction of SRO5 mRNA allows the production of 24 nucleotides (nt) siRNAs from the region overlapping with Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) transcripts. Subsequently, P5CDH transcripts are cleaved to generate 21nt siRNAs [32]. Similar mechanisms have been described in plant responses to pathogens and sperm cell development [33,34]. In barley, an increase in the NAT-lncRNA CesA6 transcript levels leads to the production of 21 and 24nt siRNAs that correlates with the down-regulation of CesA6 gene and several loci in trans involved in the regulation of cellulose rates and in the modulation of cell wall biosynthesis [35]. Similarly, in Petunia hybrida, the Sho gene involved in the production of cytokinin phytohormones contains an antisense ORF partially overlapping with the ORF of the Sho sense transcript that encodes the SHO protein [36]. The tissue specific transcription of cis-NAT SHO leads to the association of Sho sense and antisense transcripts in a double-stranded RNA likely targeted by a DICER complex for degradation into 24nt siRNAs [36]. Another mechanism, related to thermotolerance, was reported in A. thaliana. The NAT-lncRNAs NAT398b and NAT398c are cis-NATs of the MIRNA genes MIR398b and MIR398c, respectively. Knock-down of NAT398b/c promotes the accumulation of MIR398b and MIR398c, while the overexpression of NAT398b and NAT398c represses the processing of miR398. Notably, the overexpression of siRNAs derived from NAT398 overlapping transcripts, so-called nat-siR398, reduces the levels of pri-miR398b and pri-miR398c [37].
Interestingly, computational analyses performed for A. thaliana revealed that antisense transcription is associated with micro (mi) RNA-targeted mRNAs [38]. In wheat, the lncRNA INHIBITOR of WAX1 (Iw1) contains an inverted repeat showing more than 80% identity to the WAX1-COE gene, encoding a carboxylesterase-like protein that controls glaucousness [39]. The Iw1 transcript is able to form a miRNA precursor-like long hairpin which produces small RNAs, including the 21nt-miRNA miRW1. The accumulation of miRW1 is linked to the down-regulation of W1-COE and its paralog W2-COE, the cleavage of W1-COE transcripts and glaucous repression [39].
3. Long Noncoding RNAs Involved in the Regulation of Alternative Splicing
In addition to capping and polyadenylation, the production of mature mRNAs from pre-mRNAs relies on the prior removal of introns and the ligation of the majority of exons in the order in which they appear in a gene, a process known as RNA splicing. Under certain circumstances, some exons can be skipped, generating various isoforms of mature mRNAs from a single pre-mRNA. This process, called alternative splicing (AS), is mediated by the spliceosome and involves a subclass of small nuclear RNAs referred to as nuclear uridine-rich RNAs. They function in collaboration with core small nuclear ribonucleoprotein complex subunits (snRNPs) and non-snRNPs splicing factors (SFs) whose interaction with lncRNAs likely condition their stability and sub-cellular localization [40,41,42,43]. LncRNAs mainly regulate AS through interactions with specific SFs, by the regulation of chromatin remodeling that fine-tunes the splicing of specific targets and via the formation of lncRNA-pre-mRNA duplexes [43] (Figure 2).
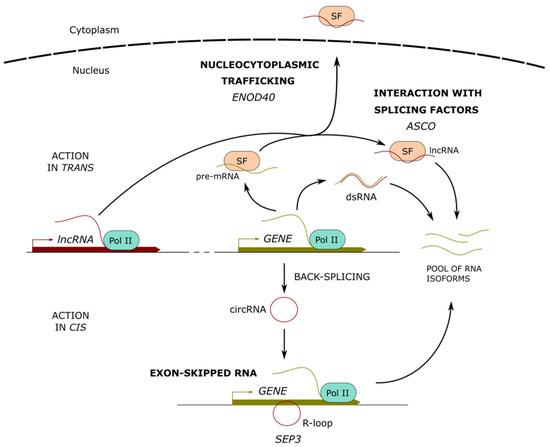
Figure 2.
Long noncoding RNAs modulating alternative splicing. Long noncoding (lnc) RNAs can form RNA interactions (dsRNA) with pre-mRNAs, fine-tuning their splicing output. In addition, lncRNAs can interact with splicing factors (SF), affecting their recognition of pre-mRNA targets or their sub-cellular localization. Protein-coding transcripts can suffer back-splicing, leading to the formation of circular RNAs (circRNA), which can interact with the parent gene to form DNA-RNA duplexes (R-loops) and modulate the alternative splicing of the nascent transcripts. Examples of characterized lncRNAs are indicated below each mechanism.
3.1. LncRNAs Interacting with Splicing Factors
In plants, AS plays crucial functions in the control of gene expression, boosting the protein-coding capacity and contributing to developmental plasticity [44,45]. In Arabidopsis, the lncRNA ALTERNATIVE SPLICING COMPETITOR (ASCO) interacts in vivo with the plant-specific SFs NUCLEAR SPECKLE RNA-BINDING PROTEINS (NSRa and b), which localize in nuclear speckles and are involved in splicing [46]. NSRs participate in the regulation of molecular and growth responses to auxin. After an auxin treatment, the double mutant nsra/b exhibited over 2200 genes differentially regulated in comparison to wild-type plants, as well as a reduced number of lateral roots suggesting a decreased sensitivity to auxin. Interestingly, the identification of RNA processing events in the nsra/b mutant revealed an important number of intron retention events and differential 5′ start or 3′ ends in a subset of genes, including a high number of auxin-related genes that behave accordingly in the ASCO overexpressing lines [46,47]. Remarkably, in vitro binding assays additionally showed that ASCO competes with mRNA-targets for the binding to NSRs, suggesting that ASCO regulates the AS of pre-mRNAs in response to auxin by hijacking NSRs [46]. More recently, a NSRa-directed RNA immunoprecipitation (RIP)-Seq approach in A. thaliana revealed that lncRNAs are overrepresented among NSRa targets [48]. As NSRa targets are mainly enriched for genes related to biotic stress responses, the interplay between lncRNAs and AS mRNAs in NSR-containing complexes was suggested to integrate the auxin and immune response pathways [48]. In agreement with this expectation, both knock-down and overexpression of ASCO led to the deregulation of expression and splicing of a large number of genes related to biotic stress and flagellin response in A. thaliana [49]. Remarkably, RNAi-ASCO plants and the double mutant nsra/b were found to exhibit a different response to flagellin, suggesting that ASCO also modulates AS in an NSR-independent manner. Consistently, an ASCO-directed chromatin isolation by RNA purification (ChIRP) coupled to mass spectrometry and RIP assays allowed the identification of other putative ASCO protein partners, including the pre-mRNA-processing-splicing factor 8A (PRP8a) and the spliceosome-core component SmD1b [49,50,51]. As previously observed for NSRs [46], ASCO overexpression also competes for PRP8a binding to particular mRNA targets [49].
3.2. LncRNAs Regulating Splicing Through Chromatin Remodeling
An additional mechanism of AS regulation involving circular non-coding RNAs (circRNAs) was reported in A. thaliana [52]. The overexpression of a circRNA comprising the entire exon 6 of the SEPALLATA 3 (SEP3) gene increases the abundance of the naturally occurring exon-skipped AS variant SEP3.3, which lacks exon 6 [52]. SEP3 is a member of the plant MADS (MCM1-AGAMOUS-DEFICIENS-SRF)-box transcription factor superfamily involved in flower development, and modifications of SEP3 splicing gives rise to floral homeotic phenotypes [52,53]. Remarkably, SEP3 exon 6 circRNA can directly interact with its cognate DNA locus, forming an RNA:DNA hybrid (R-loop), which results in transcriptional pausing and correlates with the recruitment of splicing factors and AS. This mechanism suggests that circRNAs expressed from distant loci may increase the splicing efficiency of their cognate exon-skipped messenger RNAs and that chromatin conformation and R-loop formation are critical modulators of splicing patterns [52].
3.3. LncRNA-RNA Duplexes Regulating Alternative Splicing
The analysis of transcription data for overlapping gene pairs in A. thaliana revealed a large proportion of convergently overlapping pairs (COPs) with the potential to form double-stranded RNAs [23]. Interestingly, intron-containing genes and genes with alternatively spliced transcripts are over-represented among COPs. In addition, the loci where antisense transcripts overlap with sense transcript introns mostly show AS and/or variation of polyadenylation, suggesting that the formation of NAT lncRNA-RNA pairs may regulate the AS of protein-coding genes [23]. Consistently, a genome-wide screen of trans-NATs in Arabidopsis led to the identification of 1320 putative trans-NAT pairs [24]. Most of them are predicted to form extended double-stranded RNA duplexes if sense and anti-sense are expressed in the same sub-cellular compartment, and they may lead to gene silencing and indirect AS regulation [24]. Taken together, these studies suggest that lncRNAs integrate a dynamic splicing network to control transcriptome reprogramming through AS.
4. Long Noncoding RNAs as Molecular Cargos for Protein Re-Localization
Short open reading frame (sORF) mRNAs are atypical mRNAs that contain only sORFs (shorter than 100 amino acids) and accumulate in the cytoplasm where they can be translated into oligopeptides acting as signal molecules [54,55]. Remarkably, in legumes, the highly conserved EARLY NODULIN 40 (ENOD40) genes known to participate in root symbiotic nodule organogenesis, contain only sORFs whose transcripts may encode short peptides [56,57,58]. In soybean, the lncRNA GmENOD40 encodes two oligopeptides of 12 and 24 aa residues that may have a transport function and specifically bind to sucrose synthase subunit nodulin 100 to control the use of sucrose in nitrogen-fixing nodules [58]. In M. truncatula, MtENOD40 is rapidly induced by symbiotic rhizobial bacteria in the root pericycle and is also detected in the differentiating cells of the nodule primordia [56,57]. MtENOD40 has been described as highly structured and not associated to polysomes [56,59]. Yeast three-hybrid assays revealed that the structured MtENOD40 RNA directly interacts with the constitutive RNA Binding Protein 1 (MtRBP1), a close homolog of lncRNA-interacting AtNSRs [60], located in the nuclear speckles where the splicing machinery is also hosted [61]. During nodulation, MtRBP1 is exported by MtENOD40 to cytoplasmic granules. Hence, while ENOD40-encoded peptides are likely involved in sugar metabolism, the highly structured ENOD40 RNA contributes to nucleocytoplasmic trafficking [61] (Figure 2).
5. Long Noncoding RNAs Promoting Translation
The translation process of mature mRNAs into proteins can be divided into four phases, namely initiation, elongation, termination, and ribosome recycling. The regulation of translation, so-called translational control, is a mechanism that allows a rapid modulation of gene expression through the activation or repression of pre-synthesized mRNA translation without requiring de novo transcription [62,63]. The global regulation of translation of most cellular mRNAs mainly relies on the modification of translation-initiation factors, while specific control targeting certain mRNAs likely involves regulatory protein complexes, microRNA-containing ribonucleoprotein complexes, and lncRNAs [64,65,66,67,68,69,70]. LncRNAs can be recruited to polysomes to regulate the translation of target mRNAs positively or negatively or indirectly regulate translation by sequestering miRNAs that direct the cleavage of target mRNAs (Figure 3).
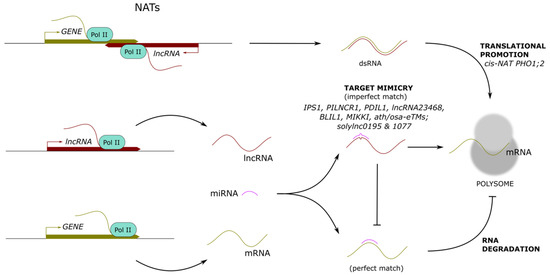
Figure 3.
Long noncoding RNAs modulating the translation of protein-coding genes. Long noncoding (lnc) RNAs can form RNA–RNA interactions (dsRNA), promoting the shuttle to polysomes and enhancing translation. In addition, lncRNAs can act as miRNA target mimicry, titrating active miRNA abundance and boosting mRNA translation. Examples of characterized lncRNAs are indicated below each mechanism.
5.1. LncRNA-mRNA Pairs into Polysomes
In rice, the PHOSPHATE1;2 (PHO1;2) gene is involved in the export of phosphate to the apoplastic space of xylem vessels [71,72,73]. The complementary strand of PHO1;2 encodes the associated cis-NAT PHO1;2. Both genes are controlled by promoters active in the vascular cylinders of roots and leaves, but while PHO1;2 promoter is unresponsive to phosphate, cis-NAT PHO1;2 promoter is strongly activated under phosphate deficiency [74]. In phosphate-deficient plants, cis-NAT PHO1;2 transcripts and PHO1;2 protein amount increase, although PHO1;2 mRNA levels remain unchanged. In addition, the downregulation or constitutive overexpression of cis-NAT PHO1;2 leads, respectively, to a decrease or strong increase in PHO1;2 protein levels, whereas the level of expression and nuclear export of PHO1;2 mRNA are not affected. Notably, the expression of cis-NAT PHO1;2 is associated with the shuttle of the PHO1;2–cis-NAT PHO1;2 sense-antisense pair towards the polysomes, supporting a role for cis-NAT PHO1;2 in the promotion of PHO1;2 translation through polysomal recruitment, to regulate phosphate homeostasis [74].
More recently, global analyses of polysome-associated RNAs and ribosome footprints in A. thaliana led to the identification of novel lncRNAs controlling cognate mRNA translation [70,75]. Under phosphate deficiency, five ribosome associated cis-NATs showed an induction correlated with the enhanced translation of their cognate sense transcripts, including two ATP BINDING CASSETTE SUBFAMILY G transporters (ABCG2 and ABCG20) and a POLLEN-SPECIFIC RECEPTOR-LIKE KINASE 7 (PRK7) family member, associated with nutrient uptake, lateral root formation, and root cell elongation, respectively [70]. In addition, five trans-NATs showed a positive correlation between their expression and their target mRNA levels, and the expression of four trans-NATs was found to correlate with a change in target mRNA polysome association under low phosphate conditions [75].
5.2. LncRNAs as Target Mimics for miRNAs
MiRNAs are ncRNAs of 20–22 nucleotides that play key regulatory roles in various biological processes in plants [76]. They are processed by Dicer-like proteins from imperfectly paired stem-loop precursors and repress gene expression by directing the cleavage or the translational arrest of target mRNAs [77,78,79]. Some lncRNAs with highly similar target sites as miRNA targets (miRNA recognition elements) can act as inhibitors of miRNA activity. They function as competing endogenous RNAs (ceRNAs), binding to miRNAs with imperfect base complementarity and blocking their interaction with authentic targets [80,81]. This regulatory mechanism is known as “target mimicry”. In plants, ceRNAs are named “target mimics” (TMs), also referred to as miRNA sponges or miRNA decoys in mammals.
In A. thaliana, the lncRNA INDUCED BY PHOSPHATE STARVATION 1 (IPS1) is a functional endogenous target mimic (eTM) of miR399 involved in inorganic phosphate (Pi) homeostasis [82]. The Pi starvation-responsive AtmiR399 directs the cleavage of the mRNA AtPHO2 (Phosphate 2), encoding an E2 ubiquitin conjugase-related protein, which negatively regulates Pi remobilization and Pi content in shoots. The sequences of the mRNA AtPHO2 and lncRNA IPS1 contain a similar motif of 23 nucleotides complementary to AtmiR399. However, in contrast to AtPHO2, IPS1 pairing with AtmiR399 is interrupted by a mismatched loop in the expected AtmiR399 cleavage site, which prevents its degradation. When IPS1 sequesters AtmiR399, the authentic AtmiR399-target AtPHO2 is accumulated, leading to a decrease in shoot Pi content [82]. More recently, a very similar mechanism was reported in maize. The lncRNA PI-DEFICIENCY-INDUCED LONG NONCODING RNA 1 (PILNCR1) functions as an eTM for ZmmiR399, thwarting the ZmmiR399-guided posttranscriptional repression of ZmPHO2 and favoring maize adaptation to Pi deficiency [83]. Additionally, in Medicago truncatula, the PI-DEFICIENCY-INDUCED LNCRNA 1 (PDIL1) was reported to regulate Pi transport by inhibiting the degradation of MtPHO2, also acting as an eTM for MtmiR399 [84]. Another example of lncRNA functioning as eTM is the tomato lncRNA23468 involved in the resistance to Phytophthora infestans [85]. Overexpression of lncRNA23468 induces a significant decrease in miR482b accumulation and an increase in the miR482b target genes NBS-LRR (nucleotide binding sites-leucine-rich repeat) expression. It was thus proposed that lncRNA23468 may decoy miR482b for targeted cleavage, thereby increasing the expression levels of NBS-LRRs genes, enhancing tomato resistance to P. infestans [85].
Computational analyses also led to the identification of putative eTMs originating from intergenic or noncoding genes for 20 highly conserved miRNAs in Arabidopsis thaliana and rice [86]. The identified TMs for miR160 (ath-eTM160-1 and osa-eTM160-3) and miR166 (ath-eTM166-1 and osa-eTM166-2) were proven to be functional target mimics, their overexpression leading to diverse altered phenotypes such as smaller and serrated leaves, spoon-shaped cotyledons, curled rosette leaves, or accelerated flowering. The effectiveness of TMs for miR156, miR159, and miR172 was also confirmed by transient agroinfiltration assay [86]. In tomato, the lncRNAs slylnc0195 and slylnc1077 involved in the tomato yellow leaf curl virus response were predicted to be eTMs of miR166 and miR399, respectively, and the functionality of slylnc0195 was also validated using a transient agro-infiltration assay [87]. Recently, 407 competing endogenous (ce)RNA pairs were constructed in A. thaliana to identify lncRNAs involved in blue light-directed plant photomorphogenesis and acting as ceRNAs. The lncRNA BLUE LIGHT-INDUCED LNCRNA 1 (BLIL1) was found to inhibit hypocotyl elongation under blue light and in response to mannitol stress by serving as a ceRNA to sequester miR167 [88].
Interestingly, the mechanism of target mimicry can be engineered and exploited to inhibit specific miRNAs via artificial miRNA TMs (aTMs) in order to establish their functionality. In A. thaliana, a collection of transgenic plants expressing aTMs predicted to reduce the activity of most of the miRNA families was generated, leading to morphological abnormalities in the aerial part for ~20% of the miRNAs targeted [89].
Finally, transposable element (TE)-derived transcripts that contain binding sites for miRNAs can also function as eTMs. In rice, the retrotransposon-derived transcript MIKKI (“decoy” in Korean) was identified as an eTM for miR171, known to target mRNAs encoding SCARECROW-Like (SCL) transcription factors for cleavage [90]. MIKKI is a TE-derived locus including Osr29 Long Terminal Repeat (LTR) retrotransposon, and its mature transcript contains an imperfect binding site for miR171, generated by a splicing event and likely attenuating the cleavage activity of miR171. In roots, MIKKI transcripts bind to miR171, stabilizing SCL mRNAs, which play an important role in root development [90].
6. Long Noncoding RNAs Mediating Post-Translational Modifications: Impact on Chromatin Remodeling and Transcription
In mammals, several examples illustrate the action of lncRNAs in protein post-translational modifications. By bringing together target proteins and specific kinases, phosphatases, or ubiquitin-ligases, lncRNAs can regulate post-translational modifications that will modulate the activity of enzymes [91,92], the stability of proteins [93], or their sub-cellular localization [94]. Intriguingly, the only known post-translational modifications modulated by plant lncRNAs are related to histones, thus affecting the epigenetic profile of target genes and their transcriptional status. The epigenetic regulation of gene expression by lncRNAs has been recently reviewed [17]. Here, we focus on the histone post-translational modifications modulated by lncRNAs in plants.
Polycomb Group (PcG) proteins are critical regulators of gene expression, essential for development in many organisms. They form complexes that modify post-translationally histones tails of target genes. In plants, the histone H3K27 trimethyltransferase CURLY LEAF (CLF) functions as a catalytic subunit of the Polycomb Repressive Complex 2 (PRC2) complex [95,96,97]. H3K27me3 then assists to recruit the PRC1-like components LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) and AtRING1 [98]. Additionally, the Trithorax H3K4 methyltransferase ARABIDOPSIS TRITHORAX-LIKE PROTEIN 1 (ATX1) mediates the establishment of H3K4me3 [99]. Interestingly, various lncRNAs have been associated with the post-translational modifications of histones at target loci, mediated by the recruitment or removal of PcG and Trithorax proteins (Figure 4).
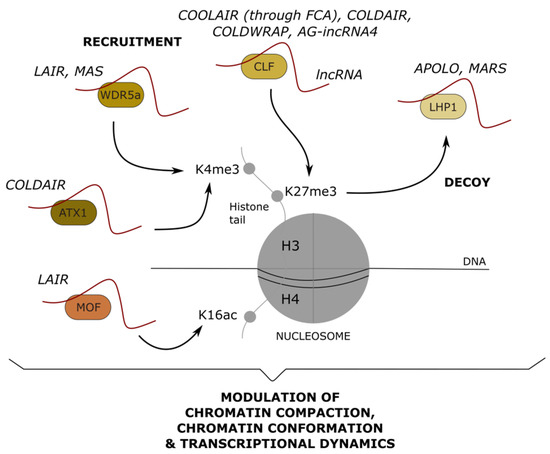
Figure 4.
Long noncoding RNAs modulating post-translational modifications of histone proteins. Long noncoding (lnc) RNAs can recruit or decoy nuclear protein complexes that modify histone tails. H3K4 trimethylation (me3) can be modulated by the lncRNA-mediated recruitment of WDR5a (COMPASS-like complex) or ATX1 (Trithorax). H3K27 trimethylation (me3) can be modulated by the lncRNA-mediated recruitment of CLF (PRC2) or the decoy of LHP1 (PRC1). Finally, H4K16 acetylation (ac) can be modulated by the recruitment of MOF. The molecular output of histone post-translational modifications on chromatin and transcription is indicated below. Examples of characterized lncRNAs are indicated above each chromatin-related player.
In A. thaliana, the FLOWERING LOCUS C (FLC) gene encodes a MADS-box-containing transcription factor (TF) that acts as a critical repressor of flowering [100]. FLC transcription is antagonistically regulated not only by the active histone modifications H3K4me3 and H3K36me3 but also by the repressive histone modification H3K27me3 [101]. Upon transition to flowering, H3K4me3 is removed, while H3K27me3 is deposited, leading to a decrease in FLC expression [102]. Remarkably, FLC transcriptional regulation depends on cis-acting lncRNAs, including COOLAIR, COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), and COLDWRAP [103,104,105,106]. COOLAIR is a set of antisense transcripts physically associated with the FLC locus, linked to the synchronized replacement of H3K36 methylation with H3K27me3 during the early stages of vernalization, independent of Polycomb complexes [107]. COOLAIR directly binds to the RNA binding protein FLOWERING CONTROL LOCUS A (FCA), which further interacts with the PRC2 component CLF. This allows the recruitment of CLF at FLC for H3K27me3 deposition [108]. COLDAIR is transcribed from the first intron of FLC and cooperates with COLDWRAP, derived from FLC proximal promoter, to facilitate the establishment of H3K27me3 during the late stage of vernalization, through the formation of a repressive intragenic chromatin loop that retains CLF at the FLC promoter [104,105]. Strikingly, ectopic overexpression of COLDAIR suppresses H3K27me3 and induces H3K4me3 at the FLC locus depending on the recruitment of ATX1 and removal of CLF, leading to enhanced FLC expression [109]. Remarkably, the overexpression of intronic lncRNAs derived from several other H3K27me3-enriched MADS-box genes also led to the activation of their corresponding genes by suppressing H3K27me3 and promoting H3K4me3 deposition [109]. The NAT-lncRNA MADS AFFECTING FLOWERING4 (MAS), transcribed from the MADS AFFECTING FLOWERING 4 (MAF4) locus, is also involved in the regulation of flowering [110]. MAS is induced by cold and activates MAF4, encoding a MADS-box containing TF, by interacting with WDR5a, a structural core component of a COMPASS-like H3K4 histone methylation complex. MAS mediates the recruitment of WDR5a to MAF4 for H3K4me3 deposition and activation of MAF4 [110]. In rice, the lncRNA LRK Antisense Intergenic RNA (LAIR) is transcribed from the antisense strand of its neighboring LRK (leucine-rich repeat receptor kinase) gene cluster and can interact with OsWDR5 as well as with the histone H4K16 acetyltransferase OsMOF [111]. LAIR overexpression is associated with higher H3K4me3 and H4K16ac levels at the LRK1 chromatin region and with the upregulation of RLK1, leading to increased grain yield [111]. AGAMOUS (AG) is another MADS TF, involved in the specification of stamens and carpels, in a tissue-specific manner [112,113,114]. The AG second intron encodes several ncRNAs, including AGAMOUS INTRONIC RNA 4 (AG-incRNA4), which recruits CLF and represses AG transcription likely through the deposition of H3K27me3 [115].
In A. thaliana, the PRC1 protein LHP1 recognizes H3K27me3 deposition and ensures the spreading of this repressive mark, controlling global genome topology [116]. In response to auxin, the lncRNA AUXIN REGULATED PROMOTER LOOP (APOLO) is transcribed from the promoter region of its neighboring gene PINOID, a key regulator of polar auxin transport, and interacts with LHP1 in vivo [117]. APOLO recognizes a subset of auxin-related genes in trans, through sequence complementarity and DNA–RNA hybrid formation (R-loops). Remarkably, overexpression of APOLO leads to the decoy of LHP1 from common target genes over the genome, and is associated with a decrease in H3K27me3 deposition as well as with modifications of chromatin conformation [118]. Similarly, the lncRNA MARNERAL SILENCING (MARS) is transcribed in response to abscisic acid (ABA) from the marneral cluster, which includes the marneral synthase MRN1 gene and the two P450 cytochrome-encoding genes CYP705A12 and CYP71A16 [119]. MARS over-accumulation is associated with the decoy of LHP1 and a decrease in H3K27me3 distribution throughout the marneral cluster. Loss of H3K27me3 likely allows the formation of a chromatin loop bringing together an enhancer element enriched in ABA-related TF binding sites and MRN1 proximal promoter, resulting in the transcriptional activation of MRN1 and a delay in seed germination [119].
7. Conclusions and Future Perspectives
Compelling evidence supports the involvement of lncRNAs in diverse and numerous aspects of post-transcriptional gene regulations in plants. Future research will likely shed light on the basis governing lncRNA interaction with diverse molecular partners, including DNA, proteins, or transcripts. The noncoding transcriptome has been shown to differ across ecotypes of the same species, notably in response to the environment [120]. This observation suggests that lncRNAs may be the key players in natural variation, contributing to plant adaptation during evolution. The conserved role of divergent lncRNAs across species likely depends on the presence of specific short sequences as well as on their secondary structure. Remarkably, the growing number of identified cold-responsive lncRNAs participating not only in the post-transcriptional but also in the transcriptional regulation of gene expression [121], e.g., the lncRNA SVALKA [122], suggests that the noncoding transcriptome is a central actor in responses to the environment. Notably, as plants cannot regulate their corporal temperature, the structure of lncRNAs and mRNAs, as well as their interactions, is most likely affected by this environmental cue. In agreement with this hypothesis, it has been recently demonstrated that the secondary structure of plant mRNAs in response to warm temperatures may modulate their translational activity, acting as thermosensor tools [123]. Similarly, structured regions in bacterial mRNAs, named RNA thermometers (RNATs), can function as thermosensors and regulate translation [124]. The advent of cutting-edge technologies, including SHAPE-seq (selective 2’-hydroxyl acylation analyzed by primer extension sequencing) to characterize RNA folding [125], will likely allow one to determine whether plant lncRNAs adopt alternative structures in response to temperature, and might function as new emerging regulators fine-tuning the protein-coding genome in response to climate change.
Funding
This research was funded by Saclay Plant Sciences-SPS (ANR-10-LABX-40) and PIOSYM (ANR-19-CE20-0011-03) ANR projects, and Agencia Nacional de Promoción Científica y Tecnológica (PICT). Both laboratories are involved in the International Associated Laboratory (LIA) NOCOSYM from CNRS-CONICET.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We thank Olivier Martin (IPS2) for the careful reading of this manuscript. M.C. is a member of CNRS (France) and C.F.F. is a post-doctoral fellow of the same institution. F.A. is a member of CONICET (Argentina).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Corradi, N.; Pombert, J.-F.; Farinelli, L.; Didier, E.S.; Keeling, P.J. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. 2010, 1, 77. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Fay, M.F.; Leitch, I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010, 164, 10–15. [Google Scholar] [CrossRef]
- Thomas, C. The Genetic Organization of Chromosomes. Annu. Rev. Genet. 1971, 5, 237–256. [Google Scholar] [CrossRef]
- Eddy, S.R. The C-value paradox, junk DNA and ENCODE. Curr. Biol. 2012, 22, R898–R899. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant. Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Comings, D.E. The Structure and Function of Chromatin. Adv. Hum. Genet. 1972, 3, 237–431. [Google Scholar] [CrossRef]
- Ohno, S. So much “junk” DNA in our genome. Brookhaven Symp. Biol. 1972, 23, 366–370. [Google Scholar] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 1–12. [Google Scholar] [CrossRef]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lucero, L.; Ferrero, L.; Fonouni-Farde, C.; Ariel, F. Functional classification of plant long noncoding RNAs: A transcript is known by the company it keeps. New Phytol. 2021, 229, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Lucero, L.; Fonouni-Farde, C.; Crespi, M.; Ariel, F. Long noncoding RNAs shape transcription in plants. Transcription 2020, 11, 160–171. [Google Scholar] [CrossRef]
- Lapidot, M.; Pilpel, Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006, 7, 1216–1222. [Google Scholar] [CrossRef]
- Britto-Kido, S.D.A.; Ferreira Neto, J.R.C.; Pandolfi, V.; Marcelino-Guimarães, F.C.; Nepomuceno, A.L.; Vilela Abdelnoor, R.; Benko-Iseppon, A.M.; Kido, E.A. Natural antisense transcripts in plants: A re-view and identification in soybean infected with phakopsora pachyrhizi supersage library. Sci. World J. 2013, 2013, 219798. [Google Scholar] [CrossRef]
- Wang, X.-J.; Gaasterland, T.; Chua, N.-H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005, 6, R30. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Qin, L.; Guo, Z.-M.; Liu, L.; Xu, H.; Hao, P.; Su, J.; Shi, Y.; He, W.-Z.; Li, Y.-X. In silico discovery of human natural antisense transcripts. BMC Bioinform. 2006, 7, 18. [Google Scholar] [CrossRef]
- Osato, N.; Yamada, H.; Satoh, K.; Ooka, H.; Yamamoto, M.; Suzuki, K.; Kawai, J.; Carninci, P.; Ohtomo, Y.; Murakami, K.; et al. Antisense transcripts with rice full-length cDNAs. Genome Biol. 2003, 5, R5. [Google Scholar] [CrossRef] [PubMed]
- Jen, C.-H.; Michalopoulos, I.; Westhead, D.R.; Meyer, P. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 2005, 6, R51. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chua, N.-H.; Wang, X.-J. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 2006, 7, R92. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sunkar, R.; Jin, H.; Zhu, J.K.; Zhang, W. Genome-wide identification and analysis of small RNAs origi-nated from natural antisense transcripts in Oryza sativa. Genome Res. 2009, 19, 70–78. [Google Scholar] [CrossRef]
- Lu, T.; Zhu, C.; Lu, G.; Guo, Y.; Zhou, Y.; Zhang, Z.; Zhao, Y.; Li, W.; Lu, Y.; Tang, W.; et al. Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genom. 2012, 13, 721. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long non-coding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2004, 24, 444–453. [Google Scholar] [CrossRef]
- Wunderlich, M.; Gross-Hardt, R.; Schöffl, F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 2014, 85, 541–550. [Google Scholar] [CrossRef]
- Fedak, H.; Palusinska, M.; Krzyczmonik, K.; Brzezniak, L.; Yatusevich, R.; Pietras, Z.; Kaczanowski, S.; Swiezewski, S. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA 2016, 113, E7846–E7855. [Google Scholar] [CrossRef]
- Henriques, R.; Wang, H.; Liu, J.; Boix, M.; Huang, L.-F.; Chua, N.-H. The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 2017, 216, 854–867. [Google Scholar] [CrossRef]
- Liu, X.; Li, D.; Zhang, D.; Yin, D.; Zhao, Y.; Ji, C.; Zhao, X.; Li, X.; He, Q.; Chen, R.; et al. A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol. 2018, 218, 774–788. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.-K. Endogenous siRNAs Derived from a Pair of Natural cis-Antisense Transcripts Regulate Salt Tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.-K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Saez, M.A.; Eshed-Williams, L.; Fletcher, J.C.; McCormick, S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010, 24, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Held, M.A.; Penning, B.; Brandt, A.S.; Kessans, S.A.; Yong, W.; Scofield, S.R.; Carpita, N.C. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc. Natl. Acad. Sci. USA 2008, 105, 20534–20539. [Google Scholar] [CrossRef]
- Zubko, E.; Meyer, P. A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an anti-sense mechanism in cytokinin regulation. Plant J. 2007, 52, 1131–1139. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Luo, Q.-J.; Samanta, M.P.; Koksal, F.; Janda, J.; Galbraith, D.W.; Richardson, C.R.; Ou-Yang, F.; Rock, C.D. Evidence for Antisense Transcription Associated with MicroRNA Target mRNAs in Arabidopsis. PLoS Genet. 2009, 5, e1000457. [Google Scholar] [CrossRef]
- Huang, D.; Feurtado, J.A.; Smith, M.A.; Flatman, L.K.; Koh, C.; Cutler, A.J. Long noncoding miRNA gene repres-ses wheat β-diketone waxes. Proc. Natl. Acad. Sci. USA 2017, 114, E3149–E3158. [Google Scholar] [CrossRef]
- Gilbert, W. Why genes in pieces? Nat. Cell Biol. 1978, 271, 501. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-Scale Proteomic Analysis of the Human Spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef]
- Yan, K.; Liu, P.; Wu, C.-A.; Yang, G.-D.; Xu, R.; Guo, Q.-H.; Huang, J.-G.; Zheng, C.-C. Stress-Induced Alternative Splicing Provides a Mechanism for the Regulation of MicroRNA Processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef]
- Palusa, S.G.; Reddy, A.S.N. Differential recruitment of splice variants from SR Pre-mRNAs to polysomes during development and in response to stresses. Plant Cell Physiol. 2015, 56, 421–427. [Google Scholar] [CrossRef]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.S.; Crespi, M. Long Noncoding RNA Modulates Alternative Splicing Regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef]
- Tran, V.D.T.; Souiai, O.; Romero-Barrios, N.; Crespi, M.; Gautheret, D. Detection of generic differential RNA processing events from RNA-seq data. RNA Biol. 2016, 13, 59–67. [Google Scholar] [CrossRef]
- Bazin, J.; Romero-Barrios, N.; Rigo, R.; Charon, C.; Blein, T.; Ariel, F.; Crespi, M. Nuclear Speckle RNA Binding Proteins Remodel Alternative Splicing and the Non-coding Arabidopsis Transcriptome to Regulate a Cross-Talk Between Auxin and Immune Responses. Front. Plant. Sci. 2018, 9, 1209. [Google Scholar] [CrossRef] [PubMed]
- Rigo, R.; Bazin, J.; Romero-Barrios, N.; Moison, M.; Lucero, L.; Christ, A.; Benhamed, M.; Blein, T.; Huguet, S.; Charon, C.; et al. The Arabidopsis lnc RNA ASCO modulates the transcriptome through interaction with splicing factors. EMBO Rep. 2020, 21, e48977. [Google Scholar] [CrossRef]
- Grainger, R.J.; Beggs, J.D. Prp8 protein: At the heart of the spliceosome. RNA 2005, 11, 533–557. [Google Scholar] [CrossRef]
- Elvira-Matelot, E.; Bardou, F.; Ariel, F.; Jauvion, V.; Bouteiller, N.; Le Masson, I.; Cao, J.; Crespi, M.D.; Vaucheret, H. The Nuclear Ribonucleoprotein SmD1 Interplays with Splicing, RNA Quality Control, and Posttranscriptional Gene Silencing in Arabidopsis. Plant Cell 2016, 28, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Severing, E.I.; Van Dijk, A.D.J.; Morabito, G.; Busscher-Lange, J.; Immink, R.G.H.; Van Ham, R.C.H.J. Predicting the Impact of Alternative Splicing on Plant MADS Domain Protein Function. PLoS ONE 2012, 7, e30524. [Google Scholar] [CrossRef]
- Maclntosh, G.; Wilkerson, C.; Green, P. Identification and analysis of Arabidopsis expressed sequence tags cha-racteristic of non-coding RNAs. Plant Physiol. 2001, 3, 765–776. [Google Scholar] [CrossRef]
- Lindsey, K.; Casson, S.; Chilley, P. Peptides: New signalling molecules in plants. Trends Plant Sci. 2002, 7, 78–83. [Google Scholar] [CrossRef]
- Crespi, M.; Jurkevitch, E.; Poiret, M.; D’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E. Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099–5112. [Google Scholar] [CrossRef] [PubMed]
- Compaan, B.; Yang, W.-C.; Bisseling, T.; Franssen, H. ENOD40 expression in the pericycle precedes cortical cell division in Rhizobium-legume interaction and the highly conserved internal region of the gene does not encode a peptide. Plant Soil 2001, 230, 1–8. [Google Scholar] [CrossRef]
- Röhrig, H.; Schmidt, J.; Miklashevichs, E.; Schell, J.; John, M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. USA 2002, 99, 1915–1920. [Google Scholar] [CrossRef]
- Asad, S.; Fang, Y.; Wycoff, K.L.; Hirsch, A.M. Isolation and characterization of cDNA and genomic clones of MsENOD40; transcripts are detected in meristematic cells of alfalfa. Protoplasma 1994, 183, 10–23. [Google Scholar] [CrossRef]
- Lucero, L.; Bazin, J.; Rodriguez Melo, J.; Ibañez, F.; Crespi, M.D.; Ariel, F. Evolution of the small family of alter-native splicing modulators nuclear speckle RNA-binding proteins in plants. Genes 2020, 11, 207. [Google Scholar] [CrossRef]
- Campalans, A.; Kondorosi, A.; Crespi, M. Enod40, a short open reading frame-containing mRNA, induces cyto-plasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 2004, 16, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Von Arnim, A.G. Translational Regulation of Cytoplasmic mRNAs. Arab. Book 2013, 11, e0165. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Meyerowitz, E.M. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 2010, 6, 419. [Google Scholar] [CrossRef]
- Juntawong, P.; Sorenson, R.; Bailey-Serres, J. Cold shock protein 1 chaperones mRNAs during translation inArabidopsis thaliana. Plant J. 2013, 74, 1016–1028. [Google Scholar] [CrossRef]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef]
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B.; et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 2013, 153, 562–574. [Google Scholar] [CrossRef]
- Li, S.; Le, B.; Ma, X.; Li, S.; You, C.; Yu, Y.; Zhang, B.; Liu, L.; Gao, L.; Shi, T.; et al. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 2016, 5, 1–24. [Google Scholar] [CrossRef]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA sequestration by OLIGOURIDYLATEBINDING PROTEIN 1 con-tributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2373–2378. [Google Scholar] [CrossRef]
- Bazin, J.; Baerenfaller, K.; Gosai, S.J.; Gregory, B.D.; Crespi, M.; Bailey-Serres, J. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E10018–E10027. [Google Scholar] [CrossRef]
- Hamburger, D.; Rezzonico, E.; Petétot, J.M.-C.; Somerville, C.; Poirier, Y. Identification and Characterization of the Arabidopsis PHO1 Gene Involved in Phosphate Loading to the Xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef]
- Stefanovic, A.; Arpat, A.B.; Bligny, R.; Gout, E.; Vidoudez, C.; Bensimon, M.; Poirier, Y. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 2011, 66, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Arpat, A.B.; Magliano, P.; Wege, S.; Rouached, H.; Stefanovic, A.; Poirier, Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012, 71, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Jabnoune, M.; Secco, D.; Lecampion, C.; Robaglia, C.; Shu, Q.; Poirier, Y. A Rice cis-Natural Antisense RNA Acts as a Translational Enhancer for Its Cognate mRNA and Contributes to Phosphate Homeostasis and Plant Fitness. Plant Cell 2013, 25, 4166–4182. [Google Scholar] [CrossRef] [PubMed]
- Deforges, J.; Reis, R.S.; Jacquet, P.; Sheppard, S.; Gadekar, V.P.; Hart-Smith, G.; Tanzer, A.; Hofacker, I.L.; Iseli, C.; Xenarios, I.; et al. Control of Cognate Sense mRNA Translation by cis-Natural Antisense RNAs. Plant Physiol. 2019, 180, 305–322. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA Targets Identified by Sequencing of the Arabidopsis Degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread Translational Inhibition by Plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Du, Q.; Wang, K.; Zou, C.; Xu, C.; Li, W.X. The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 2018, 177, 1743–1753. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen-Hao, Z.; Mysore, K.S.; Zhang, W.-H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Cui, J.; Shi, Y.; Yang, G.; Zhou, X.; Hou, X.; Meng, J.; Luan, Y. Tomato lncRNA23468 functions as a com-peting endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic. Res. 2019, 6, 28. [Google Scholar] [CrossRef]
- Wu, H.-J.; Wang, Z.-M.; Wang, M.; Wang, X.-J. Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2005, 5, 1–16. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, K.; Han, Z.; Wang, P.; Fang, Y. Genome-wide identification of Arabidopsis long noncoding RNAs in response to the blue light. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Paszkowski, J. Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife 2017, 6, e30038. [Google Scholar] [CrossRef]
- Jiang, K.; Patel, N.A.; Watson, J.E.; Apostolatos, H.; Kleiman, E.; Hanson, O.; Hagiwara, M.; Cooper, D.R. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternati-ve splicing of PKCβJII messenger ribonucleic acid. Endocrinology 2019, 150, 2087–2097. [Google Scholar] [CrossRef]
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARγ2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 2014, 5, 1050–1063. [Google Scholar] [CrossRef]
- Taniue, K.; Kurimoto, A.; Sugimasa, H.; Nasu, E.; Takeda, Y.; Iwasaki, K.; Nagashima, T.; Okada-Hatakeyama, M.; Oyama, M.; Kozuka-Hata, H.; et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. USA 2016, 113, 1273–1278. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Goodrich, J.; Puangsomlee, P.; Martín, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nat. Cell Biol. 1997, 386, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Schubert, D.; Primavesi, L.; Bishopp, A.; Roberts, G.; Doonan, J.; Jenuwein, T.; Goodrich, J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006, 25, 4638–4649. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Al-Abdallat, A.; Ndamukong, I.; Alvarez-Venegas, R.; Avramova, Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 2007, 35, 6290–6296. [Google Scholar] [CrossRef]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis Homolog of Trithorax, Activates Flower Homeotic Genes. Curr. Biol. 2003, 13, 627–637. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a re-pressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Yang, H.; Howard, M.; Dean, C. Antagonistic Roles for H3K36me3 and H3K27me3 in the Cold-Induced Epigenetic Switch at Arabidopsis FLC. Curr. Biol. 2014, 24, 1793–1797. [Google Scholar] [CrossRef]
- Pien, S.; Fleury, D.; Mylne, J.S.; Crevillen, P.; Inzé, D.; Avramova, Z.; Dean, C.; Grossniklaus, U. ARABIDOPSIS TRITHORAX1 Dynamically Regulates FLOWERING LOCUS C Activation via Histone 3 Lysine 4 Trimethylation. Plant Cell 2008, 20, 580–588. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2010, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312.e4. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 recruitment and H3K27me3 deposi-tion at FLC require FCA binding of COOLAIR. Sci. Adv. 2019, 5, 7246–7270. [Google Scholar] [CrossRef]
- Liu, Z.-W.; Zhao, N.; Su, Y.-N.; Chen, S.-S.; He, X.-J. Exogenously overexpressed intronic long noncoding RNAs activate host gene expression by affecting histone modification in Arabidopsis. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Sieburth, L.E.; Meyerowitz, E.M. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 1997, 9, 355–365. [Google Scholar]
- Deyholos, M.K.; Sieburth, L.E. Separable whorl-specific expression and negative regulation by enhancer ele-ments within the AGAMOUS second intron. Plant Cell 2000, 12, 1799–1810. [Google Scholar] [CrossRef]
- Busch, W.; Miotk, A.; Ariel, F.D.; Zhao, Z.; Forner, J.; Daum, G.; Suzaki, T.; Schuster, C.; Schultheiss, S.J.; Leibfried, A.; et al. Transcriptional Control of a Plant Stem Cell Niche. Dev. Cell 2010, 18, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-W.; Deng, S.; Xu, H.; Mao, H.-Z.; Liu, J.; Niu, Q.-W.; Wang, H.; Chua, N.-H. A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves. New Phytol. 2018, 219, 1480–1491. [Google Scholar] [CrossRef]
- Veluchamy, A.; Jégu, T.; Ariel, F.; Latrasse, D.; Mariappan, K.G.; Kim, S.-K.; Crespi, M.; Hirt, H.; Bergounioux, C.; Raynaud, C.; et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS ONE 2016, 11, e0158936. [Google Scholar] [CrossRef]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcrip-tion by alternative rna polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef] [PubMed]
- Roulé, T.; Ariel, F.; Hartmann, C.; Crespi, M.; Blein, T. The lncRNA MARS modulates the epigenetic reprogram-ming of the marneral cluster in response to ABA. BioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.08.10.236562v1.full (accessed on 26 January 2021). [CrossRef]
- Blein, T.; Balzergue, C.; Roulé, T.; Gabriel, M.; Scalisi, L.; François, T.; Sorin, C.; Christ, A.; Godon, C.; Delannoy, E.; et al. Landscape of the Noncoding Transcriptome Response of Two Arabidopsis Ecotypes to Phosphate Starvation. Plant. Physiol. 2020, 183, 1058–1072. [Google Scholar] [CrossRef]
- Kindgren, P.; Ivanov, M.; Marquardt, S. Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis. Nucleic Acids Res. 2020, 48, 2332–2347. [Google Scholar] [CrossRef]
- Kindgren, P.; Ard, R.; Ivanov, M.; Marquardt, S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nat. Cell Biol. 2020, 585, 256–260. [Google Scholar] [CrossRef]
- Kortmann, J.F.; Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Genet. 2012, 10, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Watters, K.E.; Yu, A.M.; Strobel, E.J.; Settle, A.H.; Lucks, J.B. Characterizing RNA structures in vitro and in vivo with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Methods 2016, 103, 34–48. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).