Abstract
In this study, twelve terrestrial hysteriaceous saprobic fungi growing on different pieces of dead wood were collected from Yunnan Province, China. All hysteriaceous strains isolated in this study tallied with the general characteristics associated with Rhytidhysteron. Detailed morphological characteristics and combined multigene phylogeny of LSU, ITS, SSU, and TEF showed that the twelve hysteriaceous fungi strains represent four distinct new species, and seven new host or geographical records of Rhytidhysteron. Based on morphological and phylogenetic evidence, the four new species (Rhytidhysteron bannaense sp. nov., R. coffeae sp. nov., R. mengziense sp. nov., and R. yunnanense sp. nov.) expand the number of species of Rhytidhysteron from thirty-three to thirty-seven, while seven new geographical records expand the records of Rhytidhysteron in China from six to thirteen. In addition, 10 new Rhytidhysteron host records are reported for the first time, thus expanding the known hosts for Rhytidhysteron from 52 to 62. Full descriptions, images of the morphology, and phylogenetic analyses to show the position of the Rhytidhysteron taxa are provided. In addition, the present study summarizes the main morphological characteristics, host associations, and locations of this genus.
1. Introduction
The Dothideomycetes O.E. Erikss. & Winka is the largest class in the Ascomycota Caval.-Sm [1,2,3]. Currently, it is made up of the Dothideomycetidae P.M. Kirk, P.F. Cannon, J.C. David & Stalpers (three orders with 25 families), and Pleosporomycetidae C.L. Schoch, Spatafora, Crous & Shoemaker (four orders with 94 families) [2,4,5]. This highly diverse class is mainly characterized by bitunicate asci (asci with two-wall layers), and often with fissitunicate dehiscence [2,6].
The Hysteriales Lindau belong to the subclass Pleosporomycetidae [4,5]. This monotypic order is characterized by its thick-walled, navicular ascomata which dehisce by an invaginated slit or sulcus [4,7].
The Hysteriaceae Chevall., the only family in Hysteriales, has been classified under the Pseudosphaeriales E. Müll. & Arx, Dothiorales Luttrell, Dothideales Lindau, and the Pleosporales Luttr. ex M.E. Barr, previously [4,8,9,10,11,12]. The Hysteriaceae include the hysteriaceous fungi, which currently contain 13 genera [5]. Hysteriaceous fungi are characterized by hysterithecioid or apothecioid ascomata, semi-immersed to superficial, carbonaceous, thick-walled, and distinctly navicular with a pronounced, longitudinal slit, ascospores that are hyaline to pigmented, muriform, and one to multi-septate in bitunicate asci [2,12,13,14,15,16,17,18]
The genus Rhytidhysteron Speg. was introduced by Spegazzini [19] to accommodate two species (R. brasiliense Speg. and R. viride Speg.), but no type species was designated. Subsequently, Clements and Shear [20] designated R. brasiliense as the type species [12,16,21,22]. The genus was transferred by Boehm et al. [13] from the Patellariaceae Corda to Hysteriaceae, based on molecular data. Currently, 33 records of Rhytidhysteron are listed in the Index Fungorum [23]. The sexual morph is described as having large ascomata, conspicuous, navicular, usually with a perpendicular striae margin, and pigmented, septate and muriform to sub-muriform ascospores [16,18]. Currently, only four species (R. hysterinum [Dufour] Samuels & E. Müll., R. rufulum [Spreng] Speg., R. thailandicum Thambug. & K.D. Hyde, and R. xiaokongense G.C. Ren & K.D. Hyde) have been described with an asexual morph, and conidia are classified into two types—“Aposphaeria-like” and “Diplodia-like” [16,18,21]. The main characteristics used to distinguish some species in this genus are the shape and border of the hysterothecium, the type of the exciple, the color and reaction of the epithecium, and the size of the ascospores [24]. Species of Rhytidhysteron are widely distributed in 33 countries and on 52 hosts [6,18,25,26]. Members of Rhytidhysteron play an indispensable role as saprobes, endophytes, and weak pathogens on woody plants in both terrestrial and marine habitats, and some are rarely found as human pathogens [6,16,18,25,27].
In this study, we collected 12 strains of hysteriaceous fungi from Yunnan Province, China, and based on molecular phylogenetic analyses (LSU, ITS, SSU, and TEF) and morphological characteristic comparisons, they were identified as four distinct new species (Rhytidhysteron bannaense sp. nov., R. coffeae sp. nov., R. mengziense sp. nov., and R. yunnanense sp. nov.) and seven new host records (R. bruguierae Dayar., R. camporesii Ekanayaka & K.D. Hyde, R. hongheense Wanas., R. magnoliae N.I. de Silva & K.D. Hyde, R. neorufulum Thambug. & K.D. Hyde, R. tectonae Doilom & K.D. Hyde, and R. thailandicum).
2. Materials and Methods
2.1. Sample Collection, Morphological Identification, and Single Spore Isolation
Dead plant specimens with fungal fruiting bodies were collected from Yunnan Province, China, between 2020 and 2021. Specimens were placed in plastic bags, important information such as collection date, location, and host name was recorded, and then the specimens were brought them back to the lab for isolation and morphological observation.
Senanayake et al. [28] was followed for the morphological study and single spore isolation. Morphological structures were examined under an OPTEC SZ650 dissecting stereomicroscope. An OLYMPUS optical microscope (Tokyo, Japan) was used to observe microscopic fungal structures and an OLYMPUS DP74 (Tokyo, Japan) digital camera fitted to the microscope was used to take photographs. All micro-morphological structures were measured with the Tarosoft ® Image Framework program and photo plates were made using Adobe Photoshop CS3 Extended version 10.0 software (Adobe Systems, San Jose, CA, USA).
Single spore isolation was carried out for all the specimens and the pure cultures were grown on potato dextrose agar (PDA). Germinated spores were transferred to new PDA plates under sterile conditions and incubated at 28 °C. Culture characteristics (mycelia color, shape, and edge feature) were observed after one week.
The specimens were deposited in the Kunming Institute of Botany, Academia Sinica (HKAS), Kunming, China. Living cultures were deposited in the Kunming Institute of Botany Culture Collection (KUMCC), China. Facesoffungi (FoF) numbers were registered as described in Jayasiri et al. [29], and MycoBank number (MB) was registered as outlined in MycoBank (http://www.MycoBank.org, accessed on 24 October 2022).
2.2. DNA Extraction, PCR Amplification, and Sequencing
Dissanayake et al. [30] was followed for molecular studies. Fresh mycelia which grew on PDA plates for two weeks were scraped off the plates, and then DNA was extracted using DNA Extraction Kit-BSC14S1 (BioFlux, Hangzhou, P.R. China), following the manufacturer’s protocol. Polymerase chain reaction (PCR) was used to amplify four gene regions and the primers and protocols were used for the amplification following Wanasinghe et al. [25]. The LSU gene was amplified by using the primers LR0R and LR5 [31], the ITS gene was amplified by using the primers ITS5 and ITS4 [32], the SSU gene was amplified using the primers NS1 and NS4 [32], and the TEF gene was amplified using the primers EF1-983F and EF1-2218R [33]. The total volume of the PCR mixture for amplifications was 25 μL, which consisted of 12.5 μL 2xMaster Mix (mixture of Easy Taq TM DNA Polymerase, dNTPs, and optimized buffer (Beijing Trans Gen Biotech Co., Chaoyang District, Beijing, China)), 8.5 μL ddH2O, 2 μL of DNA template, and 1 μL of each forward and reverse primer (10 pM) [34]. Purification and sequencing of PCR products were carried out by Qinke Biotech Co., Kunming, China.
2.3. Phylogenetic Analyses
The sequences of all strains obtained in this study were checked in BioEdit v.7.2.6.1 [35] for quality. Geneious 9.1.8 was used to splice forward and reverse sequences. The combined sequences were searched for similar taxa via Blast in NCBI (http://blast.ncbi.nlm.nih.gov/ (accessed on 10 November 2022)), and the most closely related taxa were put together for the phylogenetic analyses. Phylogenetic analyses were carried out with 65 sequences (Table 1) and two outgroup taxa—Gloniopsis calami S. Konta & K.D. Hyde (MFLUCC 15-0739) and G. praelonga (Schwein.) Underw. & Earle (CBS 112415). All four gene sequences were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/ (accessed on 10 November 2022)) and aligned by MAFFT v.7 (http://mafft.cbrc.jp/alignment/server/ (accessed on 10 November 2022)) [36]. TrimAl.v1.2rev59 was used to optimize the alignment of sequences [37], sequences were combined in BioEdit v.7.2.6.1. FASTA alignment formats were converted to PHYLIP and NEXUS formats in ALTER (http://www.sing-group.org/ALTER/ (accessed on 10 November 2022)) [38].

Table 1.
Taxa name, strain numbers, and GenBank accession numbers included in the phylogenetic analyses carried out in the present study.
In phylogenetic analyses, Randomized Accelerated Maximum Likelihood (RAxML) and Bayesian inference analyses (BI) were carried out in the CIPRES Science Gateway (https://www.phylo.org/portal2/login!input.action (accessed on 10 November 2022)) [39]. The RAxML trees are performed using RAxML-HPC2 on XSEDE (8.2.12) [40,41] with the GTR + I + G model of evolution. Additionally, Bayesian analyses were conducted using the Markov Chain Monte Carlo (MCMC) method in MrBayes on XSEDE (3.2.7a) [42] to evaluate posterior probabilities [43,44]: the best model of LSU and ITS is GTR + I+G, the best model of SSU is HKY + I, and the best model of TEF is GTR + G. Six simultaneous Markov chains were run for 1,000,000 generations, and trees were sampled at every 100th generation. Max-trees was set to 5000 and clade robustness was assessed using a bootstrap (BT) analysis of 1000 replicates. Phylogenetic trees were visualized with FigTree v.1.4.2 [45], bootstrap values showing at the nodes, and edited by Microsoft Office PowerPoint 2010. The newly obtained alignments and phylogenetic trees were deposited in TreeBASE, submission ID: 30049 (https://treebase.org/treebase-web/user/submissionList.html, accessed on 28 November 2022).
3. Results
3.1. Phylogenetic Analyses
The phylogenetic trees obtained from RAxML and BI analyses provided essentially similar topologies. The RAxML analyses of the combined dataset yielded the best scoring tree (Figure 1), with a final ML optimization likelihood value of −12081.382479. The matrix had 761 distinct alignment patterns, with 23.87% being undetermined characters or gaps. Parameters for the GTR + I + G model of the combined LSU, ITS, SSU, and TEF were as follows: estimated base frequencies A = 0.239683, C = 0.247793, G = 0.276186, T = 0.236338; substitution rates AC = 1.225201, AG = 2.493063, AT = 1.186723, CG = 0.732384, CT = 4.977839, GT = 1.0; proportion of invariable sites I = 0.675821; and gamma distribution shape parameter α = 0.554013. The final RAxML tree is shown in Figure 1.
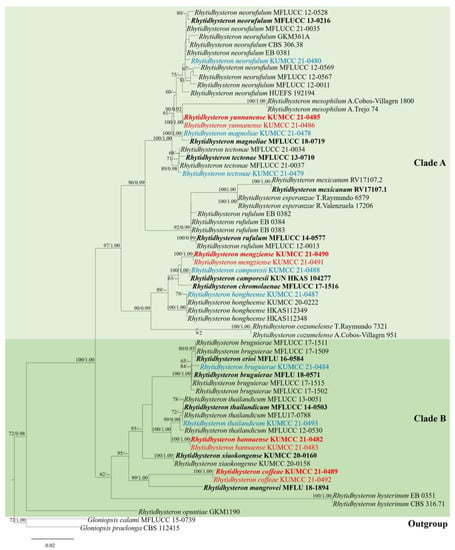
Figure 1.
Phylogenetic tree generated from RAxML analyses based on combined LSU, ITS, SSU, and TEF sequence data for Rhytidhysteron. The tree was rooted with Gloniopsis calami (MFLUCC 15-0739) and G. praelonga (CBS 112415). Bootstrap support values equal to or higher than 60% ML and posterior probability values equal to or higher than 0.90 Bayesian PP are indicated on the nodes. New species are in red, new records are in blue, and ex-type strains are in bold.
The final RAxML tree was divided into two clades of Rhytidhysteron and the results are similar to those reported by Wanasinghe et al. [25]. In this study, two new species R. mengziense (KUMCC 21-0490, KUMCC 21-0491) and R. yunnanense (KUMCC 21-0485, KUMCC 21-0486), and five new records for R. camporesii (KUMCC 21-0488), R. hongheense (KUMCC 21-0487), R. magnoliae (KUMCC 21-0478), R. neorufulum (KUMCC 21-0480), and R. tectonae (KUMCC 21-0479) were clustered within clade A. Two new species R. bannaense (KUMCC 21-0482, KUMCC 21-0483) and R. coffeae (KUMCC 21-0489, KUMCC 21-0492), along with two new records, R. bruguierae (KUMCC 21-0484) and R. thailandicum (KUMCC 21-0493), were included in clade B.
The four new species formed separate branches in the phylogenetic tree. Rhytidhysteron bannaense was well separated from R. thailandicum in an independent lineage with relatively good statistical support (100% ML/1.00 PP). Rhytidhysteron coffeae was separated from R. mangrovei Vinit & K.D. Hyde with good statistical support (99% ML/1.00 PP). Rhytidhysteron mengziense was well separated from R. camporesii with low statistical support. Rhytidhysteron yunnanense was separated from R. mesophilum Cobos-Villagrán, R. Valenz, Hern.-Rodr., Calvillo-Medina & Raymundo, with good statistical support (90% ML/0.92 PP).
The seven newly recorded strains and the known species in Rhytidhysteron clustered together in the phylogenetic tree with significant statistical support. Rhytidhysteron bruguierae (KUMCC 21-0484) grouped within five strains of R. bruguierae, with moderate statistical support (65% ML). Rhytidhysteron camporesii (KUMCC 21-0488) grouped with R. camporesii (KUN-HKAS 104277) with good statistical support (100% ML/1.00 PP). Rhytidhysteron hongheense (KUMCC 21-0487) grouped with R. hongheense (KUMCC 20-0222, H KAS112348, HKAS112349) with moderate statistical support (79% ML). Rhytidhysteron magnoliae (KUMCC 21-0478) grouped with R. magnoliae (MFLUCC 18-0719) with good statistical support (100% ML/1.00 PP). Rhytidhysteron neorufulum (KUMCC 21-0480) grouped with ten strains of R. neorufulum with low statistical support. Rhytidhysteron tectonae (KUMCC 21-0479) grouped with R. tectonae (MFLUCC 13-0710, MFLUCC 21-0037, MFLUCC 21-0034) with good statistical support (89% ML/0.98 PP). Rhytidhysteron thailandicum (KUMCC 21-0493) grouped within four strains of R. thailandicum with moderate statistical support (72% ML).
3.2. Taxonomy
Rhytidhysteron bannaense T.Y. Du and Tibpromma sp. nov. (Figure 2)
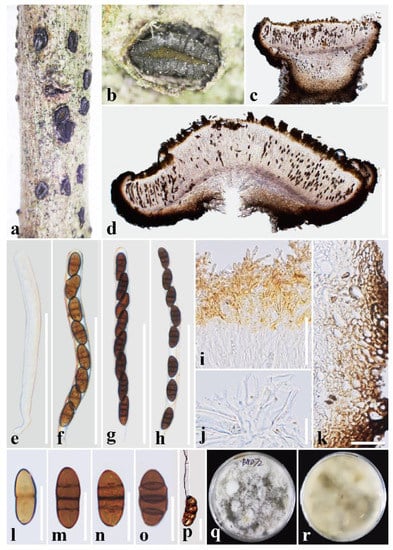
Figure 2.
Rhytidhysteron bannaense (HKAS 122695, holotype). (a,b) Appearance of hysterothecia on the host; (c,d) Vertical section through hysterothecium; (e–h) Asci; (i) Epithecium mounted in water; (j) Pseudoparaphyses; (k) Exciple; (l–o) Ascospores; (p) A germinating ascospore; (q,r) Colony on PDA medium (after four weeks). Scale bars: (c,d) = 500 μm; (e–h) = 100 μm; (i,j,l–p) = 20 μm; (k) = 50 μm.
MycoBank number: MB 845999. Facesoffungi number: FoF 12957
Etymology: Named after the region Xishuangbanna where the type specimen of the new species was collected.
Holotype: HKAS 122695
Saprobic on decaying wood of Buddleja officinalis (Loganiaceae). Sexual morph: Ascomata. 1000–1500 μm long × 550–1100 μm wide × 350–850 μm high (x = 1350 × 750 × 670 μm, n = 5), hysterothecial, solitary to aggregated, semi-immersed to superficial, black, apothecioid, navicular, rough, perpendicular striae, elongate and depressed, compressed at apex, opening through a longitudinal slit, green at the center. Exciple: 40–150 μm wide, composed of dark brown, thick-walled cells of textura angularis, outer layer brown to dark brown, inner layer pale brown to hyaline. Hamathecium: 1–2 μm wide, dense, hyaline, septate, branched, cellular pseudoparaphyses, forming an orange epithecium above asci when mounted in water, becoming a purple epithecium above the asci when mounted in 10% KOH and turns hyaline after 5 s. Asci: 140–189(–199) × 12–15(–16) μm (x = 166 × 14 μm, n = 20), 8-spored, bitunicate, cylindrical, with short pedicel, rounded at the apex, with an ocular chamber, J- apical ring. Ascospores: 22–27 × 10–12.8 μm (x = 25 × 11.5 μm, n = 30), uni-seriate, slightly overlapping, hyaline, 1-septate when immature, becoming brown to dark brown, 3-septate when mature, ellipsoidal, rounded to slightly pointed at both ends, smooth-walled, without guttules or mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h and germ tubes produced from one or both ends. Colonies on PDA reached a 6 cm diameter after two weeks at 28 °C. The colony is soft, circular, irregularly raised, with an undulated edge, white to gray on the forward, and grayish yellow in reverse.
Material examined: China, Yunnan Province, Xishuangbanna Prefecture, Jinghong City, Manlie Village, 101°0′1″ E, 21°55′15″ N, on a decaying wood piece of Buddleja officinalis Maxim. (Loganiaceae), 12 September 2021, T.Y. Du, BND72 (holotype, HKAS 122695, ex-type living culture, KUMCC 21-0482 = KUMCC 21-0483).
Notes: In phylogenetic analyses, Rhytidhysteron bannaense was well separated from R. thailandicum with relatively good statistical support (100% ML/1.00 PP). In morphology, R. bannaense is distinct from R. thailandicum, having 8-spored asci, and ascospores 3-septate when mature, without guttules, while R. thailandicum has (3–)6–8-spored asci, (1–)3-septate ascospores, and guttulate [6,16,54]. In addition, the asci and ascospore size in R. bannaense are larger than those in R. thailandicum (asci: 166 × 14 μm vs. 145 × 12.8 μm; ascospores: 25 × 11.5 μm vs. 24.5 × 9.5 µm) [16]. Moreover, according to the comparison results of different gene fragments (Table 2), R. bannaense is different from R. thailandicum in ITS and TEF genes (≥1.5%, without gaps). Therefore, in this study, R. bannaense is introduced as a new species.

Table 2.
Comparison of LSU, ITS, LSU, and TEF gene fragments of R. bannaense and four strains of R. thailandicum (without gaps).
Rhytidhysteron bruguierae Dayarathne, Mycosphere 11(1): 20 (2020) (Figure S1)
MycoBank number: MB 556574. Facesoffungi number: FoF 06154
Description: See Dayarathne et al. [47], Supplementary Notes 1
Distribution: China (this study), Thailand [47,48].
Host: Alnus nepalensis (this study), Bruguiera sp. [47], Chromolaena odorata [48].
Material examined: China, Yunnan Province, Xishuangbanna Prefecture, Jinghong City, Manlie Village, 101°1′1″ E, 21°54′0″ N, on decaying wood of Alnus nepalensis D. Don (Betulaceae), 12 September 2021, T.Y. Du, BND77 (HKAS 122690, living culture, KUMCC 21-0484).
Notes: Rhytidhysteron bruguierae was introduced by Dayarathne et al. (2020) based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. bruguierae (MFLUCC 18-0398, MFLUCC 17-1515, MFLUCC 17 1511, MFLUCC 17-1502, MFLUCC 17-1509). The morphological characteristics of our collection resemble R. bruguierae in having superficial, perpendicular striae, orange at the center hysterothecia, hamathecium comprising septate, branched pseudoparaphyses, forming a red epithecium above the asci, cylindrical, short pedicellate asci, and ellipsoidal to fusiform, brown ascospores [47,48]. Our collection and R. bruguierae are extremely similar in molecular data analyses and morphological characteristics. Previously, this species was only recorded in Thailand from Bruguiera sp. and Chromolaena odorata [47,48]. Therefore, our collection was introduced as a new geographical and host record of R. bruguierae from the decaying wood of Alnus nepalensis (Betulaceae) in the Yunnan Province of China. This is the first record of R. bruguierae on Alnus nepalensis.
Rhytidhysteron camporesii Ekanayaka & K.D. Hyde, Fungal Diversity 100: 5–277 (2020) (Figure S2)
MycoBank number: MB 556783. Facesoffungi number: FoF 06459
Description: See Hyde et al. [49], Supplementary Notes 2
Distribution: China [49] (this study).
Host: Cotoneaster franchetii (this study), unidentified wood [49].
Material examined: China, Yunnan Province, Kunming City, Panlong District, Changchong Mountain, on decaying wood of Cotoneaster franchetii Bois (Rosaceae), 5 September 2021, T.Y. Du, KMD93 (HKAS 122698, living culture, KUMCC 21-0488).
Notes: Rhytidhysteron camporesii was introduced by Hyde et al. [49] based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. camporesii (KUN-HKAS 104277). In addition, our collection shows similar morphological characteristics with R. camporesii, having black, striated hysterothecia, branched pseudoparaphyses, 8-spored, cylindrical, short pedicellate asci, and 3-septate, uni-seriate, ellipsoidal to fusiform, brown ascospores [49]. Therefore, we report our collection as a new host record of R. camporesii from decaying wood of Cotoneaster franchetii (Rosaceae) in the Yunnan Province of China. This is the first record of R. camporesii on Cotoneaster franchetii.
Rhytidhysteron coffeae T.Y. Du and Tibpromma sp. nov. (Figure 3)
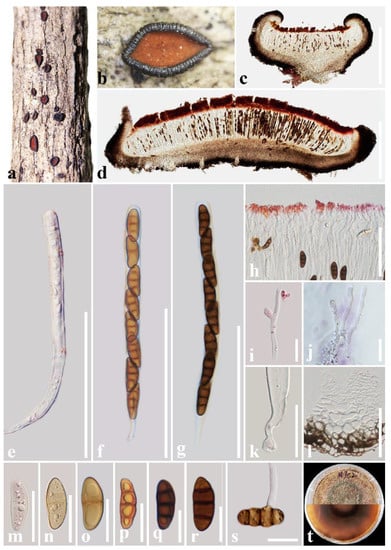
Figure 3.
Rhytidhysteron coffeae (HKAS 122701, holotype). (a,b) Appearance of hysterothecia on the host; (c,d) Vertical section through hysterothecium; (e–g) Asci; (h) Epithecium mounted in water; (i,j) Pseudoparaphyses; (k) Pedicel of asci; (l) Exciple; (m–r) Ascospores; (s) A germinating ascospore; (t) Colony on PDA medium (after four weeks). Scale bars: (c,d) = 500 μm; (e–g) = 100 μm; (h,l) = 50 μm; (i–k,m–s) = 20 μm.
MycoBank number: MB 846000. Facesoffungi number: FoF 12958
Etymology: Named after the host name, Coffea sp.
Holotype: HKAS 122700
Saprobic on decaying wood of Coffea sp. (Rubiaceae). Sexual morph: Ascomata 1000–1700 μm long × 1000–1200 μm wide × 300–600 μm high (x = 1520 × 1120 × 450 μm, n = 5), hysterothecial, solitary to aggregated, mostly solitary, superficial, base is embedded in the plant tissue, navicular, black, apothecioid, rough, perpendicular striae, elongate and depressed, compressed at apex, and opening through a nearly circular longitudinal slit, reddish brown at the center. Exciple: 70–160 μm wide (x = 95 μm, n = 10), composed of dark brown, thick-walled cells of textura angularis, outer layer brown to dark brown, inner layer pale brown to hyaline. Hamathecium: 2–3 μm wide, dense, hyaline, septate, branched, cellular pseudoparaphyses, forming a red to purple epithecium above asci when mounted in water, becoming dark purple epithecium above the asci when mounted in 10% KOH, and turns hyaline after 5 s. Asci: (162–)170–197 μm × (9–)10–14(–16) μm (x = 179.5 × 13 μm, n = 20), 8-spored, bitunicate, cylindrical, with short pedicel, rounded at the apex, with an ocular chamber, and J- apical ring. Ascospores: 23–28.5 μm × 8.5–11.5 μm (x = 26 × 10 μm, n = 30), uni-seriate, slightly overlapping, hyaline, 1-septate when immature, becoming reddish brown to brown, 3-septate when mature, ellipsoidal to fusoid, straight or curved, rounded to slightly pointed at both ends, guttulate, smooth-walled, without a mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h and germ tubes produced from one or both ends. Colonies on PDA reached a 6 cm diameter after two weeks at 28 °C. The colony is flossy, velvety, circular, slightly raised, with an entire edge, reddish brown on the forward and in reverse, with a green circle in the middle.
Material examined: China, Yunnan Province, Pu’er City, Mojiang County, Jinggong coffee plantation, 101°44′20″ E, 23°15′15″ N, decaying wood of Coffea sp. L. (Rubiaceae), 23 December 2020, L. Lu, MJC2 (holotype, HKAS 122700, ex-type living culture, KUMCC 21-0489); Yunnan Province, Pu’er City, Qixiang coffee plantation, 101°20′47″ E, 22°42′15″ N, on decaying wood of Coffea sp. (Rubiaceae), 25 December 2020, L. Lu, QXC8 (HKAS 122701 paratype, ex-paratype culture, KUMCC 21-0492).
Notes: In the phylogenetic analyses, Rhytidhysteron coffeae clearly separated from R. mangrovei with good statistical support (99% ML/1.00 PP). With respect to morphology, R. coffeae is distinct from R. mangrovei in having branched pseudoparaphyses, 8-spored asci, and 3-septate ascospores when mature, while R. mangrovei has unbranched pseudoparaphyses, (2–6–)8-spored asci, and (1–)3-septate ascospores [12]. In addition, the ascomata, asci, and ascospore size of R. coffeae are larger than those of R. mangrovei (ascomata:1520 × 1120 × 450 μm vs. 940 × 800 × 500 μm, asci: 179.5 × 13 μm vs. 146 × 9.5 μm, ascospores: 26 × 10 μm vs. 23 × 8.3 μm) [12]. Moreover, according to the comparison results of different gene fragments, R. coffeae is different from R. mangrovei in ITS (77/651 bp, 11.83%, without gaps) and TEF (21/960 bp, 2.19%, without gaps) genes (≥1.5%). Therefore, in this study, R. coffeae is introduced as a new species.
Rhytidhysteron hongheense Wanas. J. Fungi 7, 180 (2021) (Figure S3)
MycoBank number: MB 837992
Description: See Wanasinghe et al. [25], Supplementary Notes 3
Distribution: China [25] (this study).
Host: Dodonaea sp. [25], Phyllanthus emblica (this study).
Material examined: China, Yunnan Province, Kunming City, Panlong District, Kunming Institute of Botany, 102°45′5″ E, 25°8′30″ N, on decaying wood of Phyllanthus emblica (Euphorbiaceae), 1 February 2021, T.Y. Du, KMD24 (HKAS 122697, living culture, KUMCC 21-0487).
Notes: Rhytidhysteron hongheense was introduced by Wanasinghe et al. [25] based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. hongheense (KUMCC 20-0222, HKAS112348, and HKAS112349). In addition, our collection shows similar morphological characteristics with R. hongheense in having solitary to aggregated, slightly striated hysterothecia, branched pseudoparaphyses forming a red epithecium above asci, 8-spored, cylindrical, short pedicellate asci, and uni-seriate ascospores, brown when mature [25]. Therefore, we report our collection as a new host record of R. hongheense from decaying wood of Phyllanthus emblica (Euphorbiaceae) in the Yunnan Province of China. This is the first record of R. hongheense on Phyllanthus emblica.
Rhytidhysteron magnoliae N.I. de Silva, Lumyong S & K.D. Hyde, Asian Journal of Mycology 3(1): 295–306 (2019) (Figure S4)
MycoBank number: MB 557220. Facesoffungi number: FoF 07369
Description: See de Silva et al. [6], Supplementary Notes 4
Distribution: China [6] (this study).
Host: Hevea brasiliensis (this study), Magnolia grandiflora [6].
Material examined: China, Yunnan Province, Xishuangbanna Prefecture, Mengla County, Xishuangbanna Tropical Botanical Garden, 101°15′40″ E, 21°55′57″ N, on decaying wood of Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. (Euphorbiaceae), 24 November 2020, T.Y. Du, BND10 (HKAS 122693, living culture, KUMCC 21-0478).
Notes: Rhytidhysteron magnoliae was introduced by de Silva et al. [6] based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. magnoliae (MFLUCC 18-0719). In addition, our collection shows similar morphological characteristics to R. magnoliae in having solitary to aggregated, semi-immersed to superficial, coriaceous, striated hysterothecia, septate pseudoparaphyses slightly swollen at the apex and enclosed in a gelatinous matrix, 8-spored, cylindrical, short pedicellate asci, and ellipsoidal to fusoid, 1–3-septate, guttulate, brown to dark brown ascospores [6]. There is a small difference between our collection and R. magnoliae in that the perpendicular striae of ascomata in our collection is not as obvious as that in R. magnoliae. However, in the phylogenetic tree, our collection (KUMCC 21-0478) grouped with R. magnoliae (MFLUCC 18-0719) (100% ML/1.00 PP). Therefore, we report our collection as a new host record of R. magnoliae from decaying wood of Hevea brasiliensis (Euphorbiaceae) in the Yunnan Province of China. This is the first record of R. magnoliae on Hevea brasiliensis.
Rhytidhysteron mengziense T.Y. Du and Tibpromma sp. nov. (Figure 4)
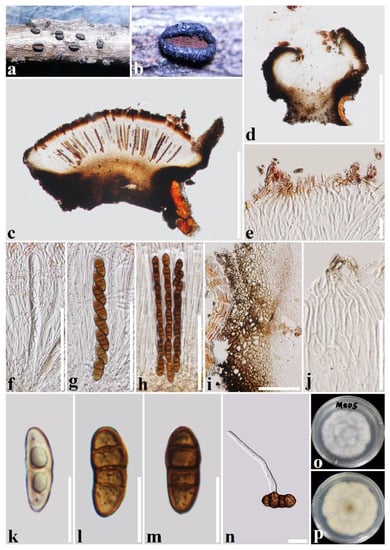
Figure 4.
Rhytidhysteron mengziense(HKAS 122699, holotype). (a,b) Appearance of hysterothecia on the host; (c,d) Vertical section through hysterothecium; (e) Epithecium mounted in water; (f–h) Asci; (i) Exciple; (j) Pseudoparaphyses; (k–m) Ascospores; (n) A germinating ascospore; (o,p) Colony on PDA medium (after one week). Scale bars: (c,d) = 500 μm; (e,k–n) = 20 μm; (f–i) = 100 μm; (j) = 50 μm.
MycoBank number: MB 846001. Facesoffungi number: FoF 12959
Etymology: Named after the region Mengzi where the type specimen was collected.
Holotype: HKAS 122699
Saprobic on decaying twigs of Crataegus scabrifolia (Rosaceae). Sexual morph: Ascomata: 1000–1600 μm long × 800–1000 μm wide × 400–700 μm high (x = 1400 × 910 × 640 μm, n = 5), hysterothecial, solitary to aggregated, mostly solitary, semi-immersed to superficial, navicular, black, apothecioid, smooth, perpendicular striae, elongate and depressed, compressed at apex, opening through a longitudinal slit, reddish brown at the center. Exciple: 60–135 μm wide, composed of outer layer brown to black, thick-walled cells of textura angularis, and inner layer light brown, thin-walled cells of textura prismatica. Hamathecium: 1–2.5 μm wide, dense, hyaline, septate, branched, cellular pseudoparaphyses, forming a reddish brown to brown epithecium above asci when mounted in water, becoming purple epithecium above the asci when mounted in 10% KOH, and turns hyaline after 30 s, while appendages turn dark brown. Asci: 150–176(–182) μm × 10–14(–16.3) μm (x = 164.5 × 13 μm, n = 20), 8-spored, bitunicate, cylindrical, with short pedicel, rounded at the apex, with an ocular chamber, J- apical ring, always fused with hamathecium. Ascospores: (22.5–)24.5–27.5(–29) μm × 10.5–12.5 μm (x = 27 × 12 μm, n = 30), slightly overlapping, uni-seriate, slightly overlapping, hyaline, 1-septate when immature, becoming reddish brown to brown, (1–)3-septate when mature, ellipsoidal to fusoid, straight or curved, rounded to slightly pointed at both ends, guttulate, rough-walled, without the mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h and germ tubes produced from one or both ends. Colonies on PDA reached a 6 cm diameter after one week at 28 °C. The colony is flossy, velvety, circular, slightly raised, with an undulated edge, white aerial hyphae on the forward and cream white in reverse.
Material examined: China, Yunnan Province, Honghe Prefecture, Mengzi City, on a decaying piece of wood of Crataegus scabrifolia (Rosaceae), 21 May 2020, S. Tibpromma, MZD5 (holotype, HKAS 122699, ex-type living culture, KUMCC 21-0490 = KUMCC 21-0491).
Notes: In phylogenetic analyses, Rhytidhysteron mengziense was well separated from R. camporesii with low statistical support. However, R. mengziense is distinct from R. camporesii in having exciple cells of textura angularis to prismatica, and rough-walled ascospores, while R. camporesii has exciple cells of textura globulosa to angularis, and smooth-walled of ascospores [49]. In addition, the ascomata and ascospore size of R. mengziense are larger than those of R. camporesii (ascomata: 1400 × 640 μm vs. 1002.4 × 570.1 µm, ascospores: 27 × 12 μm vs. 26.1 × 10.4 µm) [49]. Moreover, according to the comparison results of different gene fragments, R. mengziense is different from R. camporesii in ITS gene (10/651 bp, 1.54%, without gaps). Therefore, in this study, R. mengziense is introduced as a new species.
Rhytidhysteron neorufulum Thambug. & K.D. Hyde, Cryptog. Mycol. 37(1): 110 (2016) (Figure S5)
MycoBank number: MB 551865. Facesoffungi number: FoF 01840
Description: See Thambugala et al. [16], Supplementary Notes 5
Distribution: China (this study), Mexico [55], Thailand [16,18].
Host: Bursera sp. [55], Elaeagnus sarmentosa (this study), Hevea brasiliensis [16], Tectona grandis [18].
Material examined: China, Yunnan Province, Xishuangbanna Prefecture, Mengla County, Xishuangbanna Tropical Botanical Garden, 101°15′45″ E, 21°55′50″ N, on decaying wood of Elaeagnus sarmentosa Rehd. (Elaeagnaceae), 24 November 2020, T.Y. Du, BND49 (HKAS 122691, living culture, KUMCC 21-0480).
Notes: Rhytidhysteron neorufulum was introduced by Thambugala et al. [16] based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. neorufulum (MFLUCC 13-0216, GKM 361A, HUEFS 192194, MFLUCC 12-0528, CBS 306.38, MFLUCC 12-0011, MFLUCC 12-0567, MFLUCC 12-0569, EB 0381, MFLUCC 21-0035). In addition, our collection shows similar morphological characteristics to R. neorufulum, having solitary to aggregated, superficial, non-striated hysterothecia, septate pseudoparaphyses, 8-spored, cylindrical, short pedicellate asci, and uni-seriate, ellipsoidal to fusiform, ascospores, brown when mature [16,18]. This species was previously only recorded in Thailand and Mexico. Therefore, we report our collection as new geographical and host record of R. neorufulum from the decaying wood of Elaeagnus sarmentosa (Elaeagnaceae) in the Yunnan Province of China. This is the first record of R. neorufulum on Elaeagnus sarmentosa.
Rhytidhysteron tectonae Doilom & K.D. Hyde, Fungal Diversity. 82: 107–182 (2017) (Figure S6)
MycoBank number: MB 551964. Facesoffungi number: FoF 01849
Description: See Doilom et al. [53], Supplementary Notes 6
Distribution: China (this study), Thailand [18,53].
Host: Betula sp., an unidentified member of the Fabaceae [18], Magnolia delavayi (this study), and Tectona grandis [53].
Material examined: China, Yunnan Province, Xishuangbanna Prefecture, Mengla County, Xishuangbanna Tropical Botanical Garden, 101°15′25″ E, 21°55′37″ N, on decaying wood of Magnolia delavayi Franch. (Magnoliaceae), 24 November 2020, T.Y. Du, BND33 (HKAS 122692, living culture, KUMCC 21-0479).
Notes: Rhytidhysteron tectonae was introduced by Doilom et al. [53] based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. tectonae (MFLUCC 13-0710, MFLUCC 21-0034, and MFLUCC 21-0037). In addition, our collection shows similar morphological characteristics to R. tectonae, having solitary to aggregated, semi-immersed to superficial, non-striated, yellow at the center hysterothecia, septate, branched pseudoparaphyses, 8-spored, cylindrical, short pedicellate asci, and uni-seriate, slightly overlapping, 1–3-septate ascospores, dark brown when mature [18,53]. This species was previously only recorded in Thailand. Therefore, we report our collection as a new geographical and host record of R. tectonae from decaying wood of Magnolia delavayi (Magnoliaceae) in the Yunnan Province of China. This is the first record of R. tectonae on Magnolia delavayi.
Rhytidhysteron thailandicum Thambug. & K.D. Hyde, Cryptog. Mycol. 37(1): 110 (2016) (Figure S7)
MycoBank number: MB 551866. Facesoffungi number: FoF 01841
Description: See Thambugala et al. [16], Supplementary Notes 7
Distribution: China [6] (this study), Mexico [55], Thailand [16,54].
Host: Acacia sp. [55], Afzelia xylocarpa [54], Aquilaria sinensis (this study), Morus australis [6], and unidentified wood [16].
Material examined: China, Yunnan Province, Xishuangbanna Prefecture, Menghai County, agarwood plantation, on decaying wood of Aquilaria sinensis (Lour.) Spreng. (Thymelaeaceae), 15 September 2021, T.Y. Du, YNA62 (HKAS 122689, living culture, KUMCC 21-0493).
Notes: Rhytidhysteron thailandicum was introduced by Thambugala et al. [16] based on both morphology and phylogenetic analyses. According to phylogenetic analyses based on combined multi-gene (LSU, ITS, SSU, and TEF), our collection grouped together with R. thailandicum (MFLUCC 14-0503, MFLUCC 12-0530, MFLU17-0788, and MFLUCC 13-0051). In addition, our collection shows similar morphological characteristics to R. thailandicum, having solitary to aggregated, globose to subglobose, striated hysterothecia, exciple cells of textura angularis, septate, branched pseudoparaphyses, cylindrical, short pedicellate asci, and uni-seriate, brown ascospores [6,16,54]. There is a small difference between our collection and R. thailandicum in that the surface of the ascomata of our collection is covered with green. However, in the phylogenetic tree, our collection (KUMCC 21-0493) grouped with the four strains of R. thailandicum. Therefore, we report our collection as a new host record of R. thailandicum from decaying wood of Aquilaria sinensis (Thymelaeaceae) in the Yunnan Province of China. This is the first record of R. thailandicum on Aquilaria sinensis.
Rhytidhysteron yunnanense T.Y. Du and Tibpromma, sp. nov. (Figure 5)
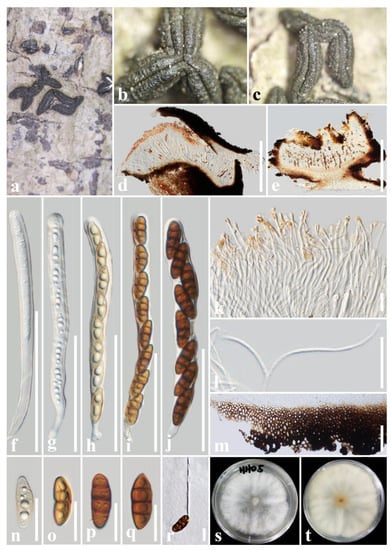
Figure 5.
Rhytidhysteron yunnanense (HKAS 122696, holotype). (a–c) Appearance of hysterothecia on the host; (d,e) Vertical section through hysterothecium; (f–j) Asci; (k) Epithecium mounted in water; (l) Pseudoparaphyses; (m) Exciple; (n–q) Ascospores; (r) A germinating ascospore; (s,t) Colony on PDA medium (after one week); Scale bars: (d,e) = 500 μm; (f–j) = 100 μm; (k,l,n–r) = 20 μm; (m) = 50 μm.
MycoBank number: MB 846002. Facesoffungi number: FoF 12960
Etymology: Named after the region Yunnan where the type specimen was collected.
Holotype: HKAS 122696
Saprobic on decaying wood of Rhus chinensis (Anacardiaceae). Sexual morph: Ascomata: 1900–3000 μm long × 400–800 μm wide × 300–600 μm high (x = 2510 × 625 × 455 μm, n = 5), hysterothecial, solitary to aggregated, mostly aggregated, semi-immersed, navicular to irregular, black, apothecioid, rough, each hysterothecia has two parallel striae parallel to the longitudinal slit, and slight perpendicular striae, elongate and depressed, compressed at apex, longitudinal slit, no opening. Exciple: 60–180 μm wide, composed of dark brown, thick-walled cells of textura globulosa, outer layer brown to dark brown, inner layer pale brown to hyaline. Hamathecium: 1–2.5 μm wide, dense, hyaline, septate, branched, cellular pseudoparaphyses, forming a yellow epithecium above asci when mounted in water, and becoming hyaline epithecium above the asci when mounted in 10% KOH, while appendages turn dark. Asci: (205–)215–250(–265) μm × 12–16(–17) μm (x = 230 × 14 μm, n = 20), 8-spored, bitunicate, cylindrical, short with club-like perdicel, rounded at the apex, with an ocular chamber, J- apical ring. Ascospores: 28.5–36 μm × 11–14.5 μm (x = 32.5 × 13 μm, n = 30), uni-seriate when mature, hyaline, 1-septate when immature, becoming reddish brown to brown, 3-septate when mature, ellipsoidal to fusoid, straight or curved, rounded to slightly pointed at both ends, guttulate, without the mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h and germ tubes produced from one or both ends. Colonies on PDA reached a 6 cm diameter after two weeks at 28 °C. The colony is velvety, circular, slightly raised, with a filiform edge, white on the forward and white in reverse.
Material examined: China, Yunnan Province, Honghe Prefecture, Honghe County, 102°14′24″ E, 23°25′30″ N, on a decaying wood piece of Rhus chinensis Mill. (Anacardiaceae), 8 December 2020, T.Y. Du, HHD5 (holotype, HKAS 122696, ex-type living culture, KUMCC 21-048 = KUMCC 21-0486).
Notes: In phylogenetic analyses, Rhytidhysteron yunnanense was well separated from R. mesophilum with good statistical support (90% ML/0.92 PP). In morphology, R. yunnanense is distinct from R. mesophilum, having navicular to irregular ascomata, each hysterothecia has two parallel striae parallel to the longitudinal slit, and slight perpendicular striae, and a longitudinal slit with no opening, while R. mesophilum has boat-shaped ascomata, with perpendicular striae with a perpendicular to longitudinal slit, and a longitudinal slit opening [24]. In addition, the asci and ascospore size of R. yunnanense are smaller than those of R. mesophilum (asci: 230 × 14 μm vs. 267–282 × 15.5–16 µm, ascospores: 32.5 × 13 μm vs. 44.2 × 13.6 μm) [24]. Moreover, according to the comparison results of different gene fragments, R. yunnanense is different from R. mesophilum in the ITS (28/651 bp, 4.30%, without gaps) gene. Therefore, in this study, R. yunnanense is introduced as a new species.
Moreover, in the previous study of the genus Rhytidhysteron, ascomata have a transverse-striae, perpendicular to the longitudinal slit, and this study is the first to find parallel striae that are parallel to the longitudinal slit.
4. Discussion
Based on the morphological study and phylogenetic analyses, four new species and seven new records of Rhytidhysteron are introduced in this paper. Rhytidhysteron bannaense sp. nov., R. coffeae sp. nov., R. mengziense sp. nov., and R. yunnanense sp. nov. are proposed as new to science based on their unique morphological characteristics and moderate to good statistical support. Seven collections of R. bruguierae, R. camporesii, R. hongheense, R. magnoliae, R. neorufulum, R. tectonae, and R. thailandicum are identified as new records because of their identical morphological characteristics with the type species of the same species and high statistical support.
In this study, we found that the pseudoparaphyses of all 11 species are branched and septate. Interestingly, the epithecia of ten species all turn purple in 10% KOH, but the purple color of the epithecium fades and becomes hyaline in a short period of time (5–30 s). On the contrary, the epithecium of R. yunnanense sp. nov. (HKAS 122696) turns hyaline in 10% KOH, and the appendages become dark. In addition, R. yunnanense is unique in Rhytidhysteron because it has both parallel and perpendicular striae, relative to the longitudinal slit. This is also the first discovery of parallel striae in this genus, while other species of Rhytidhysteron have perpendicular striae or are non-striated (Table 3). The presence or absence of striae on the margin of ascomata is one of the important characteristics of this genus and is used to identify different species [16].

Table 3.
Morphological characteristics, hosts, and location information of species of Rhytidhysterons. Host and location information of taxa are from the following references and Farr and Rossman [26].
The results of the phylogenetic tree generated in this study are consistent with those reported by Ren et al. [18], and R. erioi Ekanayaka & K.D. Hyde is grouped as a sister to R. bruguierae. Therefore, to find out the correct taxonomic placement of R. erioi, it is necessary to recollect more samples and confirm their placement. Boehm et al. [13] suggested that R. opuntiae (J.G. Br.) M.E. Barr should be removed from Rhytidhysteron based on morphological and molecular data. Subsequently, Almeida et al. [15] suggested that R. opuntiae should be accommodated by a new genus in future studies. The main reason is that R. opuntiae grouped with Hysterodifractum partisporum D.A.C. Almeida, Gusmão & A.N. Mill. [15,56], but morpho-molecular differences exist between Hysterodifractum D.A.C. Almeida, Gusmão & A.N. Mill. and R. opuntiae [12]. This study agrees with Boehm et al. [13] and Almeida et al. [15] in that R. opuntiae should be included in a new genus, due to the fact that morpho-molecular data of R. opuntiae are different from those of Rhytidhysteron and Hysterodifractum. Therefore, more studies on R. opuntiae are needed.
Rhytidhysteron neorufulum and R. rufulum are the most common and most reported species in this genus [51]. Rhytidhysteron rufulum is considered a complex species based on studies on molecular and chemical data [16,22,27,51,57]. Thambugala et al. [16] indicated that some fungal specimens were incorrectly classified as R. rufulum, which needs to be reviewed exhaustively, as they might represent new species [51]. This action is meaningful, and can clarify the taxonomic placement of unclear species and enrich the diversity of Rhytidhysteron. Unfortunately, in this study, no strains with very similar morpho-molecular data to R. rufulum were found.
In previous studies, six species were reported in China. These are R. camporesii [49], R. hongheense [25], R. magnoliae [6], R. rufulum [15], R. thailandicum [6], and R. xiaokongense [18]. This study provides seven additional species from China—R. bannaense sp. nov., R. bruguierae, R. coffeae sp. nov., R. mengziense sp. nov., R. neorufulum, R. tectonae, and R. yunnanense sp. nov. With these additions, the number of species of Rhytidhysteron in China increases from six to thirteen. At the same time, the four new species added in this study expand the species of Rhytidhysteron from thirty-three to thirty-seven, and the known hosts for Rhytidhysteron expand from 52 to 62 records. However, only 22 species of Rhytidhysteron have sequence data (including this study), so more research needs to be carried out, and more samples need to be collected, isolated, and sequenced. This study also summarizes the morphological characteristics, hosts, and countries of the species of this genus for the first time (Table 3), which provides references for future research on Rhytidhysteron.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9020148/s1, Supplementary Notes S1–S7: Description of seven new records; Supplementary Figures S1–S7: Photo plates of seven new records.
Author Contributions
Conceptualization, S.C.K. and S.T.; Data curation, T.-Y.D. and L.L.; Formal analysis, T.-Y.D.; Funding acquisition, S.C.K., D.-Q.D., N.S., A.M.E., S.A.-R. and S.T.; Methodology, T.-Y.D. and L.L.; Software, T.-Y.D.; Supervision, S.C.K. and S.T.; Writing—original draft, T.-Y.D.; Writing—review & editing, S.C.K., D.-Q.D., A.M., L.L., N.S., S.L.S., A.M.E., S.A.-R. and S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number NSFC 31760013, 31950410558, 32260004, High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program), and the Researchers Supporting Project, number (RSP2023R120), King Saud University, Riyadh, Saudi Arabia. The authors extend their appreciation to Chiang Mai University for financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Gui-Qing Zhang for her help. Tian-Ye Du thanks Mae Fah Luang University for the award of fee-less scholarship. Nakarin Suwannarach thanks Chiang Mai University, Thailand for financial support. Shaun Pennycook is thanked for his assistance in selecting a species epithet for the new species.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Chethana, K.W.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Pem, D.; Bhat, J.D.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- De Silva, N.I.; Tennakoon, D.S.; Thambugala, K.M.; Karunarathna, S.C.; Lumyong, S.; Hyde, K.D. Morphology and multigene phylogeny reveal a new species and a new record of Rhytidhysteron (Dothideomycetes, Ascomycota) from China. Asian J. Mycol. 2020, 3, 295–306. [Google Scholar] [CrossRef]
- Zogg, H. Die Hysteriaceae s. str. und Lophiaceae, unter besonderer berücksichtigung der mitteleuropäischen formen. Ber. Zur Kryptogamenflora Der Schweiz. 1962, 11, 1–190. [Google Scholar]
- Müller, E.; von Arx, J.A. Einige aspekte zur systematik pseudopharilaler ascomyceten. Ber. Schweiz. Bot. Ges. 1950, 60, 329–397. [Google Scholar]
- Luttrell, E.S. The ascostromatic Ascomycetes. Mycologia 1955, 47, 511–532. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; David, J.C.; Stalpers, J.A. Ainsworth & Bisby’s Dictionary of the Fungi, 9th ed.; CABI: Wallingford, UK, 2001. [Google Scholar]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J.A. Dictionary of the fungi, 10th edn. CAB International, Wallingford fermentations. Appl. Microbiol. Biotechnol. 2008, 39, 36–41. [Google Scholar]
- Kumar, V.; Cheewangkoon, R.; Thambugala, K.M.; Jones, G.E.; Brahmanage, R.S.; Doilom, M.; Jeewon, R.; Hyde, K.D. Rhytidhysteron mangrovei (Hysteriaceae), a new species from mangroves in Phetchaburi Province, Thailand. Phytotaxa 2019, 401, 166–178. [Google Scholar] [CrossRef]
- Boehm, E.W.; Schoch, C.L.; Spatafora, J.W. On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycol. Res. 2009, 113, 461–479. [Google Scholar] [CrossRef] [PubMed]
- Boehm, E.W.A.; Mugambi, G.; Miller, A.N.; Huhndorf, S.; Marincowitz, S.; Schoch, C.L.; Spatafora, J.W. A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Stud. Mycol. 2009, 64, 49–83. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.A.C. A new genus and three new species of hysteriaceous Ascomycetes from the semiarid region of Brazil. Phytotaxa 2014, 176, 298–308. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Hyde, K.D.; Eungwanichayapant, P.D.; Romero, A.I.; Liu, Z.Y. Additions to the genus Rhytidhysteron in Hysteriaceae. Cryptogam. Mycol. 2016, 37, 99–116. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Jeewon, R.; Maharachchikumbura, S.S.N.; Liu, J.K.; Bhat, D.J.; Jones, E.B.G.; McKenzie, E.H.C.; Camporesi, E.; Bulgakov, T.S.; et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017, 83, 1–261. [Google Scholar] [CrossRef]
- Ren, G.C.; Wanasinghe, D.N.; Jeewon, R.; Monkai, J.; Mortimer, P.E.; Hyde, K.D.; Xu, J.C.; Gui, H. Taxonomy and phylogeny of the novel Rhytidhysteron-like collections in the Greater Mekong Subregion. MycoKeys 2022, 86, 65. [Google Scholar] [CrossRef]
- Spegazzini, C. Fungi argentini additis nonnullis brasiliensibus montevideensibusque. Pugillus quartus (Continuacion). An. Soc. Cient. Argent. 1881, 12, 174–189. [Google Scholar]
- Clements, F.E.; Shear, C.L. The Genera of Fungi; Hafner Publishing Co.: New York, NY, USA, 1931; p. 632. [Google Scholar]
- Samuels, G.J.; Müller, E. Life-history studies of Brazilian Ascomycetes. 7. Rhytidhysteron rufulum and the genus Eutryblidiella. Sydowia 1979, 32, 277–292. [Google Scholar]
- Yacharoen, S.; Tian, Q.; Chomnunti, P.; Boonmee, S.; Chukeatirote, E.; Bhat, J.D.; Hyde, K.D. Patellariaceae revisited. Mycosphere 2015, 6, 290–326. [Google Scholar] [CrossRef]
- Index Fungorum. 2022. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 7 October 2022).
- Cobos-Villagrán, A.; Valenzuela, R.; Hernández-Rodríguez, C.; Calvillo-Medina, R.P.; Villa-Tanaca, L.; Mateo-Cid, L.E.; Pérez-Valdespino, A.; Martínez-González, C.R.; Raymundo, T. Three new species of Rhytidhysteron (Dothideomycetes, Ascomycota) from Mexico. MycoKeys 2021, 83, 123. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Mortimer, P.E.; Xu, J. Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaea viscosa in Honghe (China). J. Fungi 2021, 7, e180. [Google Scholar] [CrossRef] [PubMed]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. 2022. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 12 November 2022).
- Soto-Medina, E.; Lücking, R. A new species of Rhytidhysteron (Ascomycota: Patellariaceae) from Colombia, with a provisional working key to known species in the world Artículo original. Rev. De La Acad. Colomb. De Cienc. Exactas Físicas Y Nat. 2017, 41, 59–63. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi, collection examination isolation sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S. Primers for Elongation Factor 1-Alpha (EF1-Alpha); Insect Biocontrol Laboratory, USDA, ARS, PSI: Beltsville, MD, USA, 2001. [Google Scholar]
- Du, T.Y.; Hyde, K.D.; Mapook, A.; Mortimer, P.E.; Xu, J.C.; Karunarathna, S.C.; Tibpromma, S. Morphology and phylogenetic analyses reveal Montagnula puerensis sp. nov. (Didymosphaeriaceae, Pleosporales) from southwest China. Phytotaxa 2021, 514, 001–025. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER, program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE); IEEE Computer Society: New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8, a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef]
- Rambaut, A. 2012—FigTree version 1. 4. University of Edinburgh: Edinburgh, Scotland, 2012. [Google Scholar]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 175. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.P.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2018, 91, 23–36. [Google Scholar] [CrossRef]
- Cobos-Villagrán, A.; Raymundo, T.; Calvillo-Medina, R.P.; Valenzuela, R. Rhytidhysteron mexicanum sp. nov. (Dothideomycetes, Ascomycota) from the Sierra of Guadalupe, Trans Mexican Volcanic Belt. Phytotaxa 2021, 479, 275–286. [Google Scholar] [CrossRef]
- Mugambi, G.K.; Huhndorf, S.M. Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Syst. Biodivers. 2009, 7, 453–464. [Google Scholar] [CrossRef]
- Doilom, M.; Dissanayake, A.J.; Wanasinghe, D.N.; Boonmee, S.; Liu, J.K.; Bhat, D.J.; Taylor, J.E.; Bahkali, A.H.; McKenzie, E.H.C.; Hyde, K.D. Microfungi on Tectona grandis (teak) in northern Thailand. Fungal Divers. 2016, 82, 107–182. [Google Scholar] [CrossRef]
- Hyde, K.D.; de Silva, N.I.; Jeewon, R.; Bhat, D.J.; Phookamsak, R.; Doilom, M.; Boonmee, S.; Jayawardena, R.S.; Maharachchikumbura, S.S.N.; Senanayake, I.C.; et al. AJOM new records and collections of fungi: 1–100. Asian J. Mycol. 2020, 3, 22–294. [Google Scholar] [CrossRef]
- Cobos-Villagrán, A.; Hernández-Rodríguez, C.; Valenzuela, R.; Villa-Tanaca, L.; Calvillo-Medina, R.P.; Mateo-Cid, L.E.; Martínez-Pineda, M.; Raymundo, T. El género Rhytidhysteron (Dothideomycetes, Ascomycota) en México. Acta Bot. Mex. 2020, 127, e1675. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; Perŝoh, D.D.; Camporesi, E.; Kang, J.C. Taxonomic novelties of hysteriform Dothideomycetes. Mycosphere 2018, 9, 803–837. [Google Scholar] [CrossRef]
- Murillo, C.; Federico, J.A.; Julieta, C.; Thorsten, L.; Giselle, T. Molecular data indicate that Rhytidhysteron rufulum (Ascomycetes, Patellariales) in Costa Rica consists of four distinct lineages corroborated by morphological and chemical characters. Mycol. Res. 2009, 113, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Petrak, F. Ein beitrag zur pilzflora Floridas. Sydowia 1953, 7, 103–116. [Google Scholar]
- Sarbhoy, A.K.; Lal, G.; Varshney, J.L. Fungi of India (1967–71); Navyug Traders: New Delhi, India, 1971; p. 148. [Google Scholar]
- Dingley, J.M.; Fullerton, R.A.; McKenzie, E.H.C. Survey of agricultural pests and diseases. Records of fungi, bacteria, algae, and angiosperms pathogenic on plants in Cook Islands, Fiji, Kiribati, Niue, Tonga, Tuvalu, and Western Samoa. Tech. Rep. 1981, 2, 485. [Google Scholar]
- Yuan, Z.Q. Fungi and associated tree diseases in Melville Island, Northern Territory, Australia. Austral. Syst. Bot. 1996, 9, 337–360. [Google Scholar] [CrossRef]
- Pande, A.; Rao, V.G. A Compendium of Fungi on Legumes from India; Scientific Publishers: Rajasthan, India, 1998; p. 188. [Google Scholar]
- McKenzie, E.H.C.; Buchanan, P.K.; Johnston, P.R. Checklist of fungi on Nothofagus species in New Zealand. N. Z. J. Bot. 2000, 38, 635–720. [Google Scholar] [CrossRef]
- Minter, D.W.; Rodriguez Hernandez, M.; Mena Portales, J. Fungi of the Caribbean: An Annotated Checklist; PDMS Publishing: Surrey, UK, 2001; p. 946. [Google Scholar]
- Urtiaga, R. Host index of plant diseases and disorders from Venezuela—addendum. Unknown journal or publisher. 2004; 268. [Google Scholar]
- Urtiaga, R. Indice de enfermedades en plantas de Venezuela y Cuba, second edition. Unknown journal or publisher. 2004; 301. [Google Scholar]
- Sierra-López, D. Contribución al estudio de los ascomicetes bitunicados de Cataluña. Acta Bot. Barcinon. 2006, 50, 5–434. [Google Scholar]
- Pande, A. Ascomycetes of Peninsular India; Scientific Publishers: Rajasthan, India, 2008; p. 584. [Google Scholar]
- Niranjan, M.; Tiwari, S.; Baghela, A.; Sarma, V.V. New records of ascomycetous fungi from Andaman Islands, India and their molecular sequence data. Curr. Res. Environ. Appl. Mycol. 2018, 8, 331–350. [Google Scholar] [CrossRef]
- Camino-Vilaro, M.; Castro-Hernandez, L.; Abreu-Herrera, Y.; Mena-Portales, J.; Cantillo-Perez, T. Fungi associated with invasive plant species in Cuba. Phytotaxa 2019, 419, 239–267. [Google Scholar] [CrossRef]
- Reale Accademia delle scienze fisiche e matematiche di NapoliSocietà reale di Napoli. Atti della Reale accademia delle scienze fisiche e matematiche; Tip. della R. Accademia delle scienze fisiche et matematiche Napoli 1863–1935; 1879; Volume 8. [Google Scholar]
- Sociedad Científica Argentina and Congreso Científico Latino Americano and Congreso Científico Internacional Americano. Anales de la Sociedad Científica Argentina; Buenos Aires, [Sociedad Científica Argentina]: Buenos Aires, Argentina, 1879; t.8-9.
- Sociedad Científica Argentina and Congreso Científico Latino Americano and Congreso Científico Internacional Americano. Anales de la Sociedad Científica Argentina; Buenos Aires, [Sociedad Científica Argentina]: Buenos Aires, Argentina, 1888; Volume 25.
- Anahosur, K.H. Ascomycetes of Coorg (India) II. M. A. C. S. Research Institute, Poona-4 (India). Sydowia 1970, 24, 177–182. [Google Scholar]
- New York State Museum and University of the State of New York. Annual Report of the Regents; University of the State of New York: Albany, NY, USA, 1893. [Google Scholar]
- Desai, B.G.; Pathak, V.N. A new Tryblidiella from India. Sydowia 1970, 24, 198–200. [Google Scholar]
- Sociedad Científica Argentina and Congreso Científico Latino Americano and Congreso Científico Internacional Americano. Anales de la Sociedad Científica Argentina; Buenos Aires, [Sociedad Científica Argentina]: Buenos Aires, Argentina, 1881; t.12.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).