Abstract
Diagnosis of endemic mycoses is still challenging. The moderated availability of reliable diagnostic methods, the lack of clinical suspicion out of endemic areas and the limitations of conventional techniques result in a late diagnosis that, in turn, delays the implementation of the correct antifungal therapy. In recent years, molecular methods have emerged as promising tools for the rapid diagnosis of endemic mycoses. However, the absence of a consensus among laboratories and the reduced availability of commercial tests compromises the diagnostic effectiveness of these methods. In this review, we summarize the advantages and limitations of molecular methods for the diagnosis of endemic mycoses.
1. Introduction
The common term “endemic fungi” usually refers to fungal species within the Onygenales order sharing, among others, four distinctive features: (i) thermal dimorphism, (ii) geographical distribution restricted to specific regions of the world, (iii) ability to cause a disease in otherwise healthy humans, although illness tends to be more severe in immunocompromised individuals, and (iv) high mortality rates if the illness fails to be timely diagnosed and treated [1]. Recently, the WHO has released the fungal priority pathogen list to strengthen the global response to fungal infections. Several endemic fungi are listed within the high and medium priority groups [2].
1.1. Epidemiology of Endemic Mycoses
Endemic mycoses (EM) are caused by species of the genera Histoplasma, Blastomyces, Coccidioides, Paracoccidioides, Talaromyces, Sporothrix, Lacazia, and the recently described Emergomyces. The distribution area of endemic cases encompasses countries across the five continents. Coccidiomycosis, paracoccidiomycosis and lobomycosis are restricted to the American continent, whereas sporothrichosis and histoplasmosis have a cosmopolite distribution with high presence in the Americas and Africa. Blastomycosis extends mainly across Africa, the western basins of United States of America (USA), and the south-western Canadian border. Talaromycosis cases are typically found in south-eastern Asia, while emergomycosis is frequently diagnosed in South Africa, but cases have also been reported in North America, Europe, Asia and India [3,4]. Certain species within endemic genera can be found only in specific areas of the world, usually associated to particular environmental conditions of heat, moisture, pH or nutrients, among others [5].
The true epidemiology of endemic fungal infections is unknown. Many primary infections are asymptomatic or present with mild self-resolving symptoms not requiring the search of medical care, and frequently the etiological agent of the infection fails to be identified due to lack of awareness and limited access to the appropriate diagnostic tools. This is particularly concerning outside hyperendemic territories, where the vast majority of EM cases are imported and associated to immigration and travels from endemic areas [6]. Despite EM global burden is increasing, clinical infections are not subjected to mandatory notification to Public Health systems with exceptions restricted to specific areas [7,8].
The incidence of histoplasmosis has been estimated to range between 0.1–100 cases/100,000 inhabitants, with lowest rates observed in areas with temperate climates, and the highest incidence in humid tropical territories [9]. Serologic studies indicate that up to 40% of the population living in highly endemic areas may have been exposed to the fungus, with seropositivity reaching up to 87% in specific populations [7]. The real number of coccidioidomycosis cases has been estimated to exceed 350,000 per year in the USA, with an increasing trend observed over the last years [10,11,12]. In Brazil, paracoccidioidomycosis is estimated to affect 3–4 new patients/100,000 inhabitants/year, with an incidence that may reach up to 40 patients/100,000 inhabitants depending on the location. In this country, paracoccidioidomycosis represents the main cause of hospitalization and death among the overall systemic mycoses [3]. Talaromycosis is one of the most neglected and underrecognized EM as its prevalence is largely unknown. This disease is strongly associated to poverty and uncontrolled advanced HIV disease, especially in areas where the access to healthcare is limited. Some reports describe that the burden of this disease could exceed 17,000 cases/year, being lethal in as much as 1 in 3 cases [13]. With approximately 40,000 new cases every year sporotrichosis is considered the most prevalent EM in South America, and the most frequent EM in regions of Southern Brazil [14]. It is also endemic in Mexico and Northern China, and has been responsible of large outbreaks in North-America, Australia, and South Africa [15]. Emergomycosis has been described as an HIV-associated infection in South Africa, where it ranked the second most frequent EM only after sporotrichosis in a recent review of contemporary cases spanning 10 years [16]. Scattered reports locate Emergomyces spp. also in Europe, North America, and Asia. The true incidence of emergomycosis is unknown, but after the introduction of molecular techniques, many cases initially classified as histoplasmosis on the basis of histopathology have been demonstrated to be emergomycosis, indicating that its prevalence may be more frequent than previously thought [17]. Lobomycosis prevalence is unknown but an increase in new cases has been observed in recent years [18].
EM incidence has been reported to be on the rise in recent years [3,7,19,20]. This has been mainly attributed to environmental changes, travels, and expansion of at-risk populations, along with increased awareness and wider access to improved diagnostic techniques. As more research and educational actions are undertaken, new areas devoted to combatting these diseases will be uncovered [7,13,21,22]. Thus, a wide availability of sensitive, specific, rapid, and versatile diagnostic techniques will become an immediate necessity.
1.2. Diagnosis of Endemic Mycoses
To date, the laboratory diagnosis of EM is an unsolved issue [23]. The diagnostic yield of currently available microbiological techniques has been extensively reviewed by an international team of experts in a joint ECMM-ISHAM initiative, resulting in evidence-based recommendations of use recently published [4]. Conventional techniques, such as histopathology and culture, are not difficult to implement, but exhibit a number of limitations that should be taken into consideration. Firstly, these techniques require a high level of expertise and special caution is needed when handling specimens and cultures, as some species are classified as BSL-3 microorganisms; depending on the specimen and phase of the illness [24]. Moreover, cultures may delay the diagnosis up to four-to-six weeks, as these fungal species are slow-growing, and the confirmation of the dimorphism may be required for the final identification. In addition, their diagnostic yield is hampered by lack of sensitivity, particularly in non-invasive chronic forms.
Culture independent commercial assays, which rely on the detection of antigens or antibodies in clinical samples, are only available for the most prevalent EM. Antibody-based diagnosis is determined by the immune status of the host, as immunosuppresed patients fail to produce high antibody titers and seropositivity remains long time after the infection [25]. Moreover, these tests exhibit cross-reactivity among EM-causing species and with other human fungal pathogens. Antigen tests have been proved to be useful for rapid diagnosis in populations generally affected by severe immunosuppresion and disseminated forms of disease [26], but little information is available on their applications in other contexts. Specificity of antigen tests is reduced by cross-reactivity issues with other fungi [27]. During the diagnostic process, the possibility of cross-reactivity of antigens and antibodies shall be considered in areas where endemic genera co-exist. Point-of-care methods have been developed for the detection of Coccidioides spp. and H. capsulatum, although studies to date are limited, results seem to be promising [28,29].
Molecular techniques have been key for taxonomic placement and to uncover cryptic species withing the EM-causing species [30], but their application to clinical diagnosis is far from being of routine use. Most specific PCR techniques have been developed by reference laboratories without a consensus about the technology used (conventional PCR, quantitative PCR, LAMP etc.) or the genomic regions targeted by the assays. Despite several techniques for the detection of EM have been reported to be useful for diagnosis, only a Coccidioides-specific PCR is commercially available ((accessed on 28 December 2022)).
Despite the efforts on validating molecular assays in diverse types of patients and samples, the limited presence of molecular techniques in international diagnostic guidelines is due to the need of further standardization, and the lack of solid multicentered studies involving large populations. Recently, an European initiative has established a working group devoted to perform intercomparison multicenter diagnostic studies with the objective of improving EM diagnosis and acquiring a better knowledge about the epidemiology of these neglected fungal infections (https://www.ecmm.info/working-groups/working-group-on-the-diagnosis-and-the-epidemiology-of-endemic-mycoses (accessed on 28 December 2022)). Notwithstanding these limitations, molecular techniques currently seem to represent a good immediate alternative for a fast and specific diagnosis of such infections, as well as a feasible tool to go deeper into the knowledge of their epidemiology [31].
This review is intended to summarize the techniques, targets, applications of molecular techniques to the diagnosis of endemic mycoses, covering the full spectrum of techniques, from the most traditional PCR protocols to the most advanced sequencing methods (Figure 1).
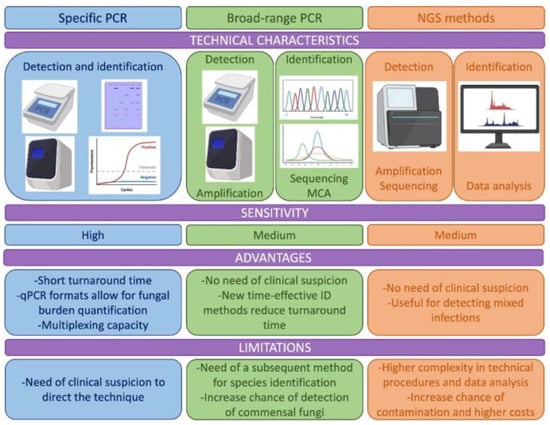
Figure 1.
Description of the technical characteristics, advantages and limitations of the molecular methods used for the diagnosis of endemic mycoses including specific PCR methods, methods based on broad range PCR and new methods as those based on Next Generation Sequencing (NGS). MCA: melting curve analysis.
2. Specific PCR Assays
Specific PCR assays have been developed last years in reference laboratories mainly focused on the detection of H. capsulatum and Coccidioides spp. For the remaining EM species, there are considerably fewer studies. In general, commercial tests and inter-comparison studies are lacking. The global SARS-CoV-2 pandemic has allowed the implementation of conventional and real-time PCR (qPCR) technology in several laboratories worldwide, including endemic regions, which offers an excellent opportunity to expand the application of molecular techniques for the detection of these neglected pathogens in a near future.
2.1. Histoplasmosis
Methods based on PCR (conventional or real time) for the detection of H. capsulatum target different genomic regions: (i) ribosomal DNA (rDNA) multicopy regions as 18S [32], ITS1 and ITS2 regions [33,34,35,36] and the ribosomal small subunit RNA [37], or (ii) unicopy targets as genes coding the 100-kDa-like protein or the M antigen [38,39,40,41,42] and, more recently, PPK and CFP4 genes [43].
DNA from clinical and reference isolates or/and clinical samples has been used for validation of these specific PCR assays. The type of clinical samples varies including respiratory secretions, biopsies, bone marrow, blood, or sera. In general, better sensitivity values were reported using clinical specimens sampled at the site of the infection, such as respiratory and biopsy samples. However, less invasive samples, such as sera and blood, were often preferred in disseminated infections [33,37,41]. Methods for the nucleic acid extraction from clinical samples also differed depending on the assay with some including sample pretreatment and others using total nucleic acids, the latter introducing a reverse transcription step before the amplification of the target to improve sensitivity [37]. Although the number of clinical samples in some publications was very limited, sensitivity values reported in these studies ranged from 70–100% [35,38]. A recent meta-analysis focused on HIV+ patients with progressive disseminated histoplasmosis reported an overall sensitivity and specificity of (95% CI) of 95.4% (88.8–101.9) and 98.7% (95.7–101.7), respectively, in different type of samples including respiratory, biopsies, blood and bone marrow [44].
LAMP methods described for the diagnosis of histoplamosis are scarce. These assays have been designed to target the ITS region [45] or the 100-kDa-like protein [46] showing variable sensitivity results.
Regarding the establishment of a consensus about histoplasmosis PCR diagnostic methods, to date, only one multicenter study involving laboratories from four Latin American countries and Spain has been published [47]. In this work, seven different PCR protocols were compared using the same DNA panel for testing the assays. Although the overall sensitivity and specificity was 86 and 100%, respectively, PCR real-time based protocols were demonstrated to be the most sensitive and reproducible approaches compared to conventional PCR assays. Methods targeting unicopy genes showed the poorest sensitivity.
2.2. Coccidiomycosis
Molecular techniques for the detection of Coccidioides spp. have been developed to be used on both clinical [48,49] and environmental settings, such as endemic regions from USA, where a steady rise in coccidioidomycosis infections has been reported [50,51]. These assays were designed to target the ITS region of the ribosomal DNA and genes encoding both Antigen 2 and Proline rich Antigen, with sensitivity ranging from 74 to 100%. Clinical samples used to test these assays were mainly respiratory, fresh and paraffin embedded biopsies and cerebrospinal fluid. When comparing different clinical samples, the best performance was obtained when using respiratory samples, fresh tissues reached 93% sensitivity, and paraffin-embedded tissues sensitivity was reported to be around 73% [48]. In 2018, the FDA authorized a commercial assay for the rapid detection of coccidioidomycosis, the Genestat MDX Coccidioides (https://www.aacc.org/cln/articles/2018/march/fda-clears-first-molecular-test-for-valley-fever (accessed on 28 December 2022)). In a multicenter study, this method reached a 100% sensitivity, with a specificity that ranged between 93.8% and 100% depending on the sample tested [52].
Of interest, Coccidioides spp. is the only fungal genus included in the international lists of potential bioterrorism agents [53], making essential to be able to face this contingency with the aid of a rapid detection method. In this line, molecular techniques represent an excellent option to be included in preparedness and response protocols due to their short turnaround response time and remarkable sensitivity and specificity scores. However, further standardization and consensus are needed.
2.3. Paracoccidioidomycosis
Several “in house” molecular techniques have been described for the detection of Paracoccidioides spp., especially in laboratories from Brazil and other non-endemic regions (Table 1). Most of these assays were based on conventional PCR methodologies [54,55,56,57]. On the other hand, two methods based on qPCR for their use in paracoccidioidomycosis diagnosis have been published [31,58]. Targets selected for amplification were the multicopy ITS rDNA region and the genes encoding the proteins Gp43 or Pb27. The clinical samples tested in theses assays were mainly respiratory, biopsies, blood and sera. Of note, sera samples were not recommended in two of these studies [31,56] as authors never detected DNA in these kinds of samples. The overall sensitivity ranges reported were 91–100%, showing a great potential of these techniques for clinical use.
Only one LAMP method has been described to date targeting the gene encoding the Gp43 protein; however, the sensitivity reported on sputum samples was moderate (61%) [59].
Paracoccidioides spp. are considered fastidious microorganisms as recovering these pathogens from culture is hard and time-consuming, commercial antigen tests are still not available and serological methods have strong limitations. Considering all the above, the inclusion of these molecular diagnostic techniques in the routine of clinical microbiology laboratories is substantially justified. This is even more imperative in non-endemic regions where the delay in diagnosis has fatal consequences in paracoccidioidomycosis patients [60,61,62].
2.4. Blastomycosis
Fewer assays have been described for the diagnosis of blastomycosis. The BAD1 gene, an important conserved adhesion-promoting protein and virulence factor of Blastomyces spp. has been chosen as target in several assays developed for the detection of the fungus in soil [63] or in clinical samples [64]. Other targets as DRK1 gene have also been used [65]. Although there is little evidence of the usefulness of theses assays in a clinical setting, the results obtained were very promising with high specificity and sensitivity values reported.
2.5. Talaromycosis
A recent meta-analysis has reviewed the methods based on PCR developed for the rapid diagnosis of talaromycosis [66]. Most of them have been published by authors from endemic regions (China, Vietnam, Thailand) which used conventional nested PCR [67] or real-time PCR [68,69] targeting the ribosomal DNA or other gene encoding regions [70]. Samples tested included plasma, blood, serum and bone marrow reporting an overall sensitivity and specificity of 84% and 99%, respectively. A LAMP assay has been published recently showing a suitable sensitivity and detecting all the biopsy samples tested [71].
2.6. Conclusions
Although data are very heterogeneous among works, specific PCR assays are rapid sensitive and specifics. Some studies used a limited number of samples for the validation of the assays, and studies focus on blastomycosis and talaromycosis are scarce. Reaching consensus about targets and kind of samples should be a priority (Table 1).

Table 1.
Details of the studies where specific PCR assays were used to diagnose endemic mycoses.
Table 1.
Details of the studies where specific PCR assays were used to diagnose endemic mycoses.
| PCR Technology | Target | Sample | Sensitivity (Cases)/Specificity | Specificity | Ref |
|---|---|---|---|---|---|
| Histoplasmosis | |||||
| Conventional (nested) | 18S rDNA | Blood, spleen, lung (mice) | 83.1% | ND | [32] |
| Conventional (nested) | 100-kDa-like protein gene | Biopsy | 70% | 100% | [72] |
| Conventional | M antigen gene | ND | 100% | 100% | [39] |
| Conventional (semi-nested) | M antigen gene | Biopsy, blood, mucose, BM | ND (30) | ND | [38] |
| Real-time | ITS rDNA | BAL, lung biopsy, BM | 100% (3) | 100% | [35] |
| Conventional (nested) | 100-kDa-like protein gene | Blood, serum, BAL, BAS, biopsy, CSF, others | 100% (40) | 100% | [41] |
| Real-time | ITS rDNA | Blood, serum, BM, sputum, BAS, BAL, biopsy, CSF, others | 89% Proven H (54) 60% Probable H (13) | 100% | [31] |
| Real-time | ITS rDNA | BAL, biopsy, BM, CSF | 95.4% (348) | 96% | [36] |
| Real-time (multiplex) | ITS rDNA | BAL, biopsy, serum, BM | 92.5% (72) | 100% | [34] |
| Real-time | mtSSU gene | Blood, serum, BAL, BAS, biopsy, CSF, others | 97.7% (44) | ND | [37] |
| Conventional Real-time | PPK, CFP4 | FFPE tissue | 100% (2) | ND | [43] |
| Paracoccidioidomycosis | |||||
| Conventional (nested) | Gp43 | Biopsy (mice) | 91% (23) | ND | [57] |
| LAMP | Gp43 | Sputum | 60% (18) | ND | [59] |
| Conventional (semi-nested) | ITS rDNA | Biopsy (mice) | 100% (4) | 100% | [54] |
| Real-time | ITS rDNA | Serum, blood, sputum | 100% (6) | ND | [73] |
| Conventional | ITS rDNA | Serum, biopsy | ND | ND | [56] |
| Conventional (semi-nested) | ITS rDNA | Sputum | 100% (14) | ND | [74] |
| Conventional (nested) | GP43 gene | BAL, biopsy, sputum | 100% (25) | 100% | [55] |
| Real-time | Pb27 gene | Blood, serum, biopsy and others | 94% (78) | 100% | [58] |
| Coccidioidomycosis | |||||
| Conventional (nested)/real-time | Antigen2/Proline-Rich Antigen, | FFPE- biopsy | 100% (3) | ND | [75] |
| Real-time | ITS rDNA | Respiratory, biopsy, FFPE-biopsy | 89% (480) | 98% | [48] |
| Real-time | ITS rDNA | Mice samples | 98% (44) | 100% | [49] |
| Real-time | GeneSTAT Coccidioides assay | BAL/BW | 100% (332) | 93.85–100% | [52] |
| Blastomycosis | |||||
| Conventional (nested) | WI-1 (BAD 1) | PE-biopsy (dogs) | ND (73) | ND | [76] |
| Real-time | DRK-1 | Respiratory, biopsy and others | 86% (14) | 99.4% | [65] |
| Real-time | BAD-1 | FFPE-biopsy | 83% (12) | 100% | [64] |
| Real-time (duplex) | BAD-1 | FFPE-biopsy, respiratory and others | ND (33) | ND | [77] |
| Talaromycosis | |||||
| Real-time | 5.8S rDNA | Blood | 60% (20) | 100% | [78] |
| Conventional (nested) | 18S rDNA | Serum | 68.6% (35) | 100% | [67] |
| LAMP | ITS rDNA | Biopsy | 100% (12) | 100% | [71] |
| Conventional (nested)/ real-time | ITS rDNA | Blood, serum | 82% (22)/91% (22) | 75%/63% | [68] |
| Real-time | ITS rDNA | Serum | 86.11% (36) | ND | [69] |
ND: no data; FFPE-biopsy: formalin-fixed paraffin-embedded biopsy; BAL: brochoalveolar lavage; BAS: brochoaspirate; BW: bronchial wash; CSF: cerebrospinal fluid; BM: bone marrow.
3. Broad-Range PCRs
Broad-range or panfungal PCR assays are especially useful for EM diagnosis, generally used when there is not a clear suspicion of the fungal agent causing the disease, which is one of the hallmarks of EM, or when the infection is not frequent in the setting of the diagnostic laboratory, as it is in non-endemic areas. This approach relies on the use of fungal (or fungal group)-specific primers to amplify fungal DNA directly from clinical samples followed by an identification method, mainly Sanger sequencing, to confirm the causative agent [79]. With the aim of improving sensitivity, classic multi-copy targets as the ribosomal operon [37] are often selected for panfungal amplification, while fresh tissue samples are preferred over formalin-fixed, paraffin-embedded samples [80].
Sample contamination, detection of commensal fungi, PCR bias due to primer mismatches and, the lack of adequate reference databases for fungi identification are the main limitations of panfungal PCR assays. However, the limitation of delay in response time associated to species determination has been addressed by replacing Sanger sequencing identification with other time-saving post-PCR methods such as melting curve analysis, DNA microarray, electrospray-ionization mass spectrometry analysis and T2 magnetic resonance [81].
In conclusion, although proper studies directed to EM diagnosis by using broad-range PCRs are still missing, there are plenty reports in the literature showing the ability of this technique to provide a definite diagnosis when paired with other reference methods. This technique has the advantage of being cost-effective and can be an alternative to specific PCR considering their limitations (Table 2).

Table 2.
Details of the studies where broad-range PCR was used to diagnose endemic mycoses.
4. Next Generation Sequencing (NGS)
NGS has revolutionized the diagnosis of fungal and other microbial infections and it is already considered the future replacement for the current broad-range PCR methods. The most used NGS approach for diagnosis nowadays is targeted amplicon sequencing or metabarcoding. By using fungal-specific primers, thousands of copies of different DNA templates are amplified and sequenced simultaneously, reducing turnaround time and costs [95]. However, shotgun metagenomic sequencing can be also used to target most parts of the genomes of the microorganisms present in the sample. This approach is more expensive and computationally demanding, but allows for further characterization of the infecting agent as other features, such as identifying the subtype or the antimicrobial resistance profile, could be retrieved from the sequenced data [96]. In general, NGS methods face the same limitations as broad-range PCR assays but with the additional requirement of expertise in data analysis and increasing complexity in the technical procedures [97].
NGS technologies were originally standardized as an exploratory tool to study the fungal community profile (mycobiome) of human specimens. As an example, McTaggart LR and colleagues developed an NGS-based method for the analysis of the lung mycobiome during Blastomyces dermatitidis/gilchristii infection [98]. The successful detection of the causative agent as well as other fungal pathogens indicated the potential of this method for the diagnosis of EM. However, proper standardization and retrospective studies including a substantial number of clinical isolates are still missing, it not being currently possible to recommend or suggest a method or to consider NGS as a suitable tool for EM diagnosis. Most studies reported in the literature describe brief case reports or anecdotical presence of EM samples in bigger specimen sets. Nevertheless, NGS methods have already been employed successfully in the differential diagnosis of infections with similar clinical symptoms and the identification of the biological source of an outbreak (Table 3). Recently, the assessment of the clinical performance of NGS for the rapid diagnosis of talaromycosis in HIV patients has been evaluated [99]. The sensitivity of the new method was significantly higher than culture and serum galactomannan determination (98.3% vs. 66.7% and 83.3%, respectively) underlining the potential use of NGS for EM diagnosis. In conclusion, although the NGS-based method seems to be promising, more studies need to be able of consider it as a tool for the diagnosis of EM (Table 3).

Table 3.
Details of the studies where NGS was used to diagnose endemic mycoses.
5. Conclusions and Perspectives
Diagnosis of EM is still difficult in endemic regions and even more complicated out of these regions, where the lack of suspicion and expertise are the major shortcomings. Molecular techniques have shown their great potential for the rapid diagnosis of EM in several studies performed in reference laboratories in the last years. The recent COVID-19 pandemic has not only increased the awareness on how critical a rapid diagnosis is but paved the way to the generalized implementation of the molecular diagnosis of infectious diseases. As summarized in this review, several molecular techniques developed in recent years show a great potential for the rapid diagnosis of EM. In non-endemic countries, where the availability of some other useful techniques, as antigen detection, is limited, qPCR-based molecular assays have been developed to this purpose, extending their usefulness to difficult-to-diagnose forms of infection [34,37]. The introduction of multiplex formats also allows for performing a differential diagnosis with other pathogens causing similar clinical patterns reducing costs [118]. In endemic areas, especially in resource-limited settings, cost-effective molecular methods such as LAMP could be a promising alternative. However, in general terms, there is still great variability in published methods to date and commercial kits are practically non-existent. An effort to standardize and achieve a consensus should be performed among the different laboratories. Technical issues such as the selection of genomic targets or nucleic acid extraction methods, coupled with the implementation of inter-comparison studies should be prioritized to include these techniques in the future guidelines for patient management. Panfungal assays stand for an interesting alternative to specific assays as these techniques are easy to implement and more cost-effective; however, limitations of these tests should be considered when performing a final diagnosis. Recently, NGS has emerged as an alternative to overcome some of these limitations soon. As a conclusion, the implementation of molecular techniques in clinical settings will revolutionize the rapid diagnosis of EM, especially in countries where laboratories use diagnostic PCR routinely.
Author Contributions
C.V. review the panfugal and NGS methods, elaborated the figures and Tables, organized the bibliography and participated in the redactions. M.J.B. and M.T.M.-G. designed the structure of the paper and reviewed the final version. M.T.M.-G. elaborated the introduction and participated in the redaction. M.J.B. elaborated the specific PCR section and the Table associate. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by research project PI21CIII/00007 from Spanish Fondo de Investigaciones Sanitarias of the Instituto de Salud Carlos III.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
M.J.B. is a founding partner and holds shares of Micologia Molecular S.L. She has received grant support from the Instituto de Salud Carlos III and has been paid for talks on behalf of United Medical LTDA.
References
- Gnat, S.; Lagowski, D.; Nowakiewicz, A.; Dylag, M. A global view on fungal infections in humans and animals: Infections caused by dimorphic fungi and dermatophytoses. J. Appl. Microbiol. 2021, 131, 2688–2704. [Google Scholar] [CrossRef] [PubMed]
- WHO. Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Shikanai-Yasuda, M.A.; Mendes, R.P.; Colombo, A.L.; Queiroz-Telles, F.; Kono, A.S.G.; Paniago, A.M.M.; Nathan, A.; Valle, A.; Bagagli, E.; Benard, G.; et al. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 2017, 50, 715–740. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.R., 3rd; Le, T.; Chindamporn, A.; Kauffman, C.A.; Alastruey-Izquierdo, A.; Ampel, N.M.; Andes, D.R.; Armstrong-James, D.; Ayanlowo, O.; Baddley, J.W.; et al. Global guideline for the diagnosis and management of the endemic mycoses: An initiative of the European Confederation of Medical Mycology in cooperation with the International Society for Human and Animal Mycology. Lancet Infect. Dis. 2021, 21, e364–e374. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.E.; Cat, L.A.; Zender, C.S.; Treseder, K.K.; Randerson, J.T. Coccidioidomycosis Dynamics in Relation to Climate in the Southwestern United States. Geohealth 2018, 2, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Salzer, H.J.F.; Stoney, R.J.; Angelo, K.M.; Rolling, T.; Grobusch, M.P.; Libman, M.; Lopez-Velez, R.; Duvignaud, A.; Asgeirsson, H.; Crespillo-Andujar, C.; et al. Epidemiological aspects of travel-related systemic endemic mycoses: A GeoSentinel analysis, 1997-2017. J. Travel Med. 2018, 25, tay055. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, N.; Kubat, R.C.; Poplin, V.; Adenis, A.A.; Denning, D.W.; Wright, L.; McCotter, O.; Schwartz, I.S.; Jackson, B.R.; Chiller, T.; et al. Re-drawing the Maps for Endemic Mycoses. Mycopathologia 2020, 185, 843–865. [Google Scholar] [CrossRef]
- Vallabhaneni, S.; Mody, R.K.; Walker, T.; Chiller, T. The Global Burden of Fungal Diseases. Infect. Dis. Clin. N. Am. 2016, 30, 1–11. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Beale, M.A.; Hagen, F.; Fisher, M.C.; Terra, P.P.D.; de Hoog, S.; Brilhante, R.S.N.; de Aguiar Cordeiro, R.; de Souza Collares Maia Castelo-Branco, D.; Rocha, M.F.G.; et al. The global epidemiology of emerging Histoplasma species in recent years. Stud. Mycol. 2020, 97, 100095. [Google Scholar] [CrossRef]
- Benedict, K.; McCotter, O.Z.; Brady, S.; Komatsu, K.; Sondermeyer Cooksey, G.L.; Nguyen, A.; Jain, S.; Vugia, D.J.; Jackson, B.R. Surveillance for Coccidioidomycosis—United States, 2011-2017. MMWR Surveill Summ. 2019, 68, 1–15. [Google Scholar] [CrossRef]
- Sondermeyer Cooksey, G.L.; Nguyen, A.; Vugia, D.; Jain, S. Regional Analysis of Coccidioidomycosis Incidence—California, 2000-2018. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1817–1821. [Google Scholar] [CrossRef]
- Van Dyke, M.C.C.; Thompson, G.R.; Galgiani, J.N.; Barker, B.M. The Rise of Coccidioides: Forces Against the Dust Devil Unleashed. Front. Immunol. 2019, 10, 2188. [Google Scholar] [CrossRef] [PubMed]
- Narayanasamy, S.; Dat, V.Q.; Thanh, N.T.; Ly, V.T.; Chan, J.F.; Yuen, K.Y.; Ning, C.; Liang, H.; Li, L.; Chowdhary, A.; et al. A global call for talaromycosis to be recognised as a neglected tropical disease. Lancet Glob. Health 2021, 9, e1618–e1622. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Bonifaz, A.; Gutierrez-Galhardo, M.C.; Mochizuki, T.; Li, S. Global epidemiology of sporotrichosis. Med. Mycol. 2015, 53, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Mapengo, R.E.; Maphanga, T.G.; Grayson, W.; Govender, N.P. Endemic mycoses in South Africa, 2010-2020: A decade-long description of laboratory-diagnosed cases and prospects for the future. PLoS Negl. Trop. Dis. 2022, 16, e0010737. [Google Scholar] [CrossRef]
- Schwartz, I.S.; Kenyon, C.; Feng, P.; Govender, N.P.; Dukik, K.; Sigler, L.; Jiang, Y.; Stielow, J.B.; Munoz, J.F.; Cuomo, C.A.; et al. 50 Years of Emmonsia Disease in Humans: The Dramatic Emergence of a Cluster of Novel Fungal Pathogens. PLoS Pathog. 2015, 11, e1005198. [Google Scholar] [CrossRef]
- Goncalves, F.G.; Rosa, P.S.; Belone, A.F.F.; Carneiro, L.B.; de Barros, V.L.Q.; Bispo, R.F.; Sbardelott, Y.; Neves, S.; Vittor, A.Y.; Woods, W.J.; et al. Lobomycosis Epidemiology and Management: The Quest for a Cure for the Most Neglected of Neglected Tropical Diseases. J. Fungi 2022, 8, 494. [Google Scholar] [CrossRef]
- Baker, J.; Setianingrum, F.; Wahyuningsih, R.; Denning, D.W. Mapping histoplasmosis in South East Asia—Implications for diagnosis in AIDS. Emerg. Microbes Infect. 2019, 8, 1139–1145. [Google Scholar] [CrossRef]
- McCotter, O.Z.; Benedict, K.; Engelthaler, D.M.; Komatsu, K.; Lucas, K.D.; Mohle-Boetani, J.C.; Oltean, H.; Vugia, D.; Chiller, T.M.; Sondermeyer Cooksey, G.L.; et al. Update on the Epidemiology of coccidioidomycosis in the United States. Med. Mycol. 2019, 57, S30–S40. [Google Scholar] [CrossRef]
- Amona, F.M.; Denning, D.W.; Moukassa, D.; Develoux, M.; Hennequin, C. Histoplasmosis in the Republic of Congo dominated by African histoplasmosis, Histoplasma capsulatum var. duboisii. PLoS Negl. Trop. Dis. 2021, 15, e0009318. [Google Scholar] [CrossRef]
- Rakislova, N.; Hurtado, J.C.; Palhares, A.E.M.; Ferreira, L.; Freire, M.; Lacerda, M.; Monteiro, W.; Navarro, M.; Casas, I.; Teixeira, M.M.; et al. High prevalence and mortality due to Histoplasma capsulatum in the Brazilian Amazon: An autopsy study. PLoS Negl. Trop. Dis. 2021, 15, e0009286. [Google Scholar] [CrossRef]
- Caceres, D.H.; Echeverri Tirado, L.C.; Bonifaz, A.; Adenis, A.; Gomez, B.L.; Flores, C.L.B.; Canteros, C.E.; Santos, D.W.; Arathoon, E.; Soto, E.R.; et al. Current situation of endemic mycosis in the Americas and the Caribbean: Proceedings of the first international meeting on endemic mycoses of the Americas (IMEMA). Mycoses 2022, 65, 1179–1187. [Google Scholar] [CrossRef]
- Wheat, L.J. Approach to the diagnosis of the endemic mycoses. Clin. Chest Med. 2009, 30, 379–389, viii. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.; Page, I. Role of Serological Tests in the Diagnosis of Mold Infections. Curr. Fungal Infect. Rep. 2018, 12, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Kassis, C.; Durkin, M.; Holbrook, E.; Myers, R.; Wheat, L. Advances in Diagnosis of Progressive Pulmonary and Disseminated Coccidioidomycosis. Clin. Infect. Dis. 2021, 72, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Govender, N.P.; Chakrabarti, A.; Robert-Gangneux, F.; Boulware, D.R.; Zafar, A.; Oladele, R.O.; Richardson, M.D.; Gangneux, J.P.; Alastruey-Izquierdo, A.; et al. Essential in vitro diagnostics for advanced HIV and serious fungal diseases: International experts’ consensus recommendations. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1581–1584. [Google Scholar] [CrossRef]
- Cáceres, D.H.; Gómez, B.L.; Tobon, A.M.; Minderman, M.; Bridges, N.; Chiller, T.; Lindsley, M.D. Validation and Concordance Analysis of a New Lateral Flow Assay for Detection of Histoplasma Antigen in Urine. J. Fungi 2021, 7, 799. [Google Scholar] [CrossRef]
- Donovan, F.M.; Ramadan, F.A.; Khan, S.A.; Bhaskara, A.; Lainhart, W.D.; Narang, A.T.; Mosier, J.M.; Ellingson, K.D.; Bedrick, E.J.; Saubolle, M.A.; et al. Comparison of a Novel Rapid Lateral Flow Assay to Enzyme Immunoassay Results for Early Diagnosis of Coccidioidomycosis. Clin. Infect. Dis. 2021, 73, e2746–e2753. [Google Scholar] [CrossRef]
- Van Dyke, M.C.C.; Teixeira, M.M.; Barker, B.M. Fantastic yeasts and where to find them: The hidden diversity of dimorphic fungal pathogens. Curr. Opin. Microbiol. 2019, 52, 55–63. [Google Scholar] [CrossRef]
- Buitrago, M.J.; Bernal-Martinez, L.; Castelli, M.V.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Histoplasmosis and paracoccidioidomycosis in a non-endemic area: A review of cases and diagnosis. J. Travel Med. 2011, 18, 26–33. [Google Scholar] [CrossRef]
- Bialek, R.; Fischer, J.; Feucht, A.; Najvar, L.K.; Dietz, K.; Knobloch, J.; Graybill, J.R. Diagnosis and monitoring of murine histoplasmosis by a nested PCR assay. J. Clin. Microbiol. 2001, 39, 1506–1509. [Google Scholar] [CrossRef] [PubMed]
- Buitrago, M.J.; Berenguer, J.; Mellado, E.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Detection of imported histoplasmosis in serum of HIV-infected patients using a real-time PCR-based assay. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Gago, S.; Esteban, C.; Valero, C.; Zaragoza, O.; Puig de la Bellacasa, J.; Buitrago, M.J. A multiplex real-time PCR assay for identification of Pneumocystis jirovecii, Histoplasma capsulatum, and Cryptococcus neoformans/Cryptococcus gattii in samples from AIDS patients with opportunistic pneumonia. J. Clin. Microbiol. 2014, 52, 1168–1176. [Google Scholar] [CrossRef]
- Martagon-Villamil, J.; Shrestha, N.; Sholtis, M.; Isada, C.M.; Hall, G.S.; Bryne, T.; Lodge, B.A.; Reller, L.B.; Procop, G.W. Identification of Histoplasma capsulatum from culture extracts by real-time PCR. J. Clin. Microbiol. 2003, 41, 1295–1298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simon, S.; Veron, V.; Boukhari, R.; Blanchet, D.; Aznar, C. Detection of Histoplasma capsulatum DNA in human samples by real-time polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2010, 66, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Gits-Muselli, M.; Lanternier, F.; Sturny-Leclère, A.; Benazra, M.; Hamane, S.; Rodrigues, A.M.; García-Hermoso, D.; Lortholary, O.; Dromer, F.; et al. Evaluation of a New Histoplasma spp. Quantitative RT-PCR Assay. J. Mol. Diagn. 2021, 23, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Bracca, A.; Tosello, M.E.; Girardini, J.E.; Amigot, S.L.; Gomez, C.; Serra, E. Molecular detection of Histoplasma capsulatum var. capsulatum in human clinical samples. J. Clin. Microbiol. 2003, 41, 1753–1755. [Google Scholar] [CrossRef]
- Guedes, H.L.; Guimaraes, A.J.; Muniz Mde, M.; Pizzini, C.V.; Hamilton, A.J.; Peralta, J.M.; Deepe, G.S., Jr.; Zancopé-Oliveira, R.M. PCR assay for identification of Histoplasma capsulatum based on the nucleotide sequence of the M antigen. J. Clin. Microbiol. 2003, 41, 535–539. [Google Scholar] [CrossRef][Green Version]
- López, L.F.; Munoz, C.O.; Cáceres, D.H.; Tobon, A.M.; Loparev, V.; Clay, O.; Chiller, T.; Litvintseva, A.; Gade, L.; Gonzalez, A.; et al. Standardization and validation of real time PCR assays for the diagnosis of histoplasmosis using three molecular targets in an animal model. PLoS ONE 2017, 12, e0190311. [Google Scholar] [CrossRef]
- Maubon, D.; Simon, S.; Aznar, C. Histoplasmosis diagnosis using a polymerase chain reaction method. Application on human samples in French Guiana, South America. Diagn. Microbiol. Infect. Dis. 2007, 58, 441–444. [Google Scholar] [CrossRef]
- Rickerts, V.; Bialek, R.; Tintelnot, K.; Jacobi, V.; Just-Nubling, G. Rapid PCR-based diagnosis of disseminated histoplasmosis in an AIDS patient. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 821–823. [Google Scholar] [CrossRef]
- Gallo, J.E.; Torres, I.; Gomez, O.M.; Rishishwar, L.; Vannberg, F.; Jordan, I.K.; McEwen, J.G.; Clay, O.K. New Histoplasma Diagnostic Assays Designed via Whole Genome Comparisons. J. Fungi 2021, 7, 544. [Google Scholar] [CrossRef] [PubMed]
- Caceres, D.H.; Knuth, M.; Derado, G.; Lindsley, M.D. Diagnosis of Progressive Disseminated Histoplasmosis in Advanced HIV: A Meta-Analysis of Assay Analytical Performance. J. Fungi 2019, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Zatti, M.D.S.; Arantes, T.D.; Fernandes, J.A.L.; Bay, M.B.; Milan, E.P.; Naliato, G.F.S.; Theodoro, R.C. Loop-mediated Isothermal Amplification and nested PCR of the Internal Transcribed Spacer (ITS) for Histoplasma capsulatum detection. PLoS Negl. Trop. Dis. 2019, 13, e0007692. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.M.; Zhou, Y.; Theodoro, R.C.; Abrams, B.; Balajee, S.A.; Litvintseva, A.P. Development of a loop-mediated isothermal amplification method for detection of Histoplasma capsulatum DNA in clinical samples. J. Clin. Microbiol. 2014, 52, 483–488. [Google Scholar] [CrossRef]
- Buitrago, M.J.; Canteros, C.E.; Frias De Leon, G.; Gonzalez, A.; Marques-Evangelista De Oliveira, M.; Munoz, C.O.; Ramirez, J.A.; Toranzo, A.I.; Zancope-Oliveira, R.; Cuenca-Estrella, M. Comparison of PCR protocols for detecting Histoplasma capsulatum DNA through a multicenter study. Rev. Iberoam Micol. 2013, 30, 256–260. [Google Scholar] [CrossRef]
- Binnicker, M.J.; Buckwalter, S.P.; Eisberner, J.J.; Stewart, R.A.; McCullough, A.E.; Wohlfiel, S.L.; Wengenack, N.L. Detection of Coccidioides species in clinical specimens by real-time PCR. J. Clin. Microbiol. 2007, 45, 173–178. [Google Scholar] [CrossRef]
- Gago, S.; Buitrago, M.J.; Clemons, K.V.; Cuenca-Estrella, M.; Mirels, L.F.; Stevens, D.A. Development and validation of a quantitative real-time PCR assay for the early diagnosis of coccidioidomycosis. Diagn. Microbiol. Infect. Dis. 2014, 79, 214–221. [Google Scholar] [CrossRef]
- Bowers, J.R.; Parise, K.L.; Kelley, E.J.; Lemmer, D.; Schupp, J.M.; Driebe, E.M.; Engelthaler, D.M.; Keim, P.; Barker, B.M. Direct detection of Coccidioides from Arizona soils using CocciENV, a highly sensitive and specific real-time PCR assay. Med. Mycol. 2019, 57, 246–255. [Google Scholar] [CrossRef]
- Lauer, A.; Baal, J.D.; Baal, J.C.; Verma, M.; Chen, J.M. Detection of Coccidioides immitis in Kern County, California, by multiplex PCR. Mycologia 2012, 104, 62–69. [Google Scholar] [CrossRef]
- Saubolle, M.A.; Wojack, B.R.; Wertheimer, A.M.; Fuayagem, A.Z.; Young, S.; Koeneman, B.A. Multicenter Clinical Validation of a Cartridge-Based Real-Time PCR System for Detection of Coccidioides spp. in Lower Respiratory Specimens. J. Clin. Microbiol. 2018, 56, e01277-17. [Google Scholar] [CrossRef] [PubMed]
- Warnock, D.W. Coccidioides species as potential agents of bioterrorism. Future Microbiol. 2007, 2, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Koishi, T.; Yasuoka, K.; Zeng, X.C.; Fujikawa, S. Molecular dynamics simulations of urea-water binary droplets on flat and pillared hydrophobic surfaces. Faraday Discuss 2010, 146, 185–193; discussion 195–215, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Gaviria, M.; Rivera, V.; Munoz-Cadavid, C.; Cano, L.E.; Naranjo, T.W. Validation and clinical application of a nested PCR for paracoccidioidomycosis diagnosis in clinical samples from Colombian patients. Braz. J. Infect. Dis. 2015, 19, 376–383. [Google Scholar] [CrossRef]
- Dias, L.; de Carvalho, L.F.; Romano, C.C. Application of PCR in serum samples for diagnosis of paracoccidioidomycosis in the southern Bahia-Brazil. PLoS Negl. Trop. Dis. 2012, 6, e1909. [Google Scholar] [CrossRef][Green Version]
- Bialek, R.; Ibricevic, A.; Aepinus, C.; Najvar, L.K.; Fothergill, A.W.; Knobloch, J.; Graybill, J.R. Detection of Paracoccidioides brasiliensis in tissue samples by a nested PCR assay. J. Clin. Microbiol. 2000, 38, 2940–2942. [Google Scholar] [CrossRef]
- Rocha-Silva, F.; Maria de Figueiredo, S.; Rutren La Santrer, E.F.; Machado, A.S.; Fernandes, B.; Assuncao, C.B.; Goes, A.M.; Caligiorne, R.B. Paracoccidioidomycosis: Detection of Paracoccidioides brasiliensis genome in biological samples by quantitative chain reaction polymerase (qPCR). Microb. Pathog. 2018, 121, 359–362. [Google Scholar] [CrossRef]
- Tatibana, B.T.; Sano, A.; Uno, J.; Kamei, K.; Igarashi, T.; Mikami, Y.; Miyaji, M.; Nishimura, K.; Itano, E.N. Detection of Paracoccidioides brasiliensis gp43 gene in sputa by loop-mediated isothermal amplification method. J. Clin. Lab. Anal. 2009, 23, 139–143. [Google Scholar] [CrossRef]
- Onda, H.; Komine, M.; Murata, S.; Ohtsuki, M. Letter: Imported paracoccidioidomycosis in Japan. Dermatol. Online J. 2011, 17, 11. [Google Scholar] [CrossRef]
- Ginarte, M.; Pereiro, M., Jr.; Toribio, J. Imported paracoccidioidomycosis in Spain. Mycoses 2003, 46, 407–411. [Google Scholar] [CrossRef]
- Botas-Velasco, M.; Jover-Diaz, F.; Ortiz de la Tabla-Duccase, V.; Martinez-Garcia, C. [Imported paracoccidioidomycosis in Spain]. Enferm. Infecc. Microbiol. Clin. 2010, 28, 259–260. [Google Scholar] [CrossRef] [PubMed]
- Burgess, J.W.; Schwan, W.R.; Volk, T.J. PCR-based detection of DNA from the human pathogen Blastomyces dermatitidis from natural soil samples. Med. Mycol. 2006, 44, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Sidamonidze, K.; Peck, M.K.; Perez, M.; Baumgardner, D.; Smith, G.; Chaturvedi, V.; Chaturvedi, S. Real-time PCR assay for identification of Blastomyces dermatitidis in culture and in tissue. J. Clin. Microbiol. 2012, 50, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Babady, N.E.; Buckwalter, S.P.; Hall, L.; Le Febre, K.M.; Binnicker, M.J.; Wengenack, N.L. Detection of Blastomyces dermatitidis and Histoplasma capsulatum from culture isolates and clinical specimens by use of real-time PCR. J. Clin. Microbiol. 2011, 49, 3204–3208. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Lai, J.; Wei, W.; Zhou, B.; Huang, J.; Jiang, J.; Liang, B.; Liao, Y.; Zang, N.; Cao, C.; et al. Accuracy of rapid diagnosis of Talaromyces marneffei: A systematic review and meta-analysis. PLoS ONE 2018, 13, e0195569. [Google Scholar] [CrossRef]
- Pongpom, M.; Sirisanthana, T.; Vanittanakom, N. Application of nested PCR to detect Penicillium marneffei in serum samples. Med. Mycol. 2009, 47, 549–553. [Google Scholar] [CrossRef]
- Lu, S.; Li, X.; Calderone, R.; Zhang, J.; Ma, J.; Cai, W.; Xi, L. Whole blood Nested PCR and Real-time PCR amplification of Talaromyces marneffei specific DNA for diagnosis. Med. Mycol. 2016, 54, 162–168. [Google Scholar] [CrossRef]
- Li, X.; Zheng, Y.; Wu, F.; Mo, D.; Liang, G.; Yan, R.; Khader, J.A.; Wu, N.; Cao, C. Evaluation of quantitative real-time PCR and Platelia galactomannan assays for the diagnosis of disseminated Talaromyces marneffei infection. Med. Mycol. 2020, 58, 181–186. [Google Scholar] [CrossRef]
- Hien, H.T.A.; Thanh, T.T.; Thu, N.T.M.; Nguyen, A.; Thanh, N.T.; Lan, N.P.H.; Simmons, C.; Shikuma, C.; Chau, N.V.V.; Thwaites, G.; et al. Development and evaluation of a real-time polymerase chain reaction assay for the rapid detection of Talaromyces marneffei MP1 gene in human plasma. Mycoses 2016, 59, 773–780. [Google Scholar] [CrossRef]
- Sun, J.; Li, X.; Zeng, H.; Xie, Z.; Lu, C.; Xi, L.; de Hoog, G.S. Development and evaluation of loop-mediated isothermal amplification (LAMP) for the rapid diagnosis of Penicillium marneffei in archived tissue samples. FEMS Immunol. Med. Microbiol. 2010, 58, 381–388. [Google Scholar] [CrossRef]
- Bialek, R.; Feucht, A.; Aepinus, C.; Just-Nubling, G.; Robertson, V.J.; Knobloch, J.; Hohle, R. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 2002, 40, 1644–1647. [Google Scholar] [CrossRef] [PubMed]
- Buitrago, M.J.; Merino, P.; Puente, S.; Gomez-Lopez, A.; Arribi, A.; Zancope-Oliveira, R.M.; Gutierrez, M.C.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Utility of real-time PCR for the detection of Paracoccidioides brasiliensis DNA in the diagnosis of imported paracoccidioidomycosis. Med. Mycol. 2009, 47, 879–882. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pitz Ade, F.; Koishi, A.C.; Tavares, E.R.; Andrade, F.G.; Loth, E.A.; Gandra, R.F.; Venancio, E.J. An optimized one-tube, semi-nested PCR assay for Paracoccidioides brasiliensis detection. Rev. Soc. Bras. Med. Trop. 2013, 46, 783–785. [Google Scholar] [CrossRef] [PubMed]
- Bialek, R.; Kern, J.; Herrmann, T.; Tijerina, R.; Cecenas, L.; Reischl, U.; Gonzalez, G.M. PCR assays for identification of Coccidioides posadasii based on the nucleotide sequence of the antigen 2/proline-rich antigen. J. Clin. Microbiol. 2004, 42, 778–783. [Google Scholar] [CrossRef]
- Bialek, R.; Cirera, A.C.; Herrmann, T.; Aepinus, C.; Shearn-Bochsler, V.I.; Legendre, A.M. Nested PCR assays for detection of Blastomyces dermatitidis DNA in paraffin-embedded canine tissue. J. Clin. Microbiol. 2003, 41, 205–208. [Google Scholar] [CrossRef]
- Kaplan, M.; Zhu, Y.; Kus, J.V.; McTaggart, L.; Chaturvedi, V.; Chaturvedi, S. Development of a Duplex Real-Time PCR Assay for the Differentiation of Blastomyces dermatitidis and Blastomyces gilchristii and a Retrospective Analysis of Culture and Primary Specimens from Blastomycosis Cases from New York (2005 to 2019). J. Clin. Microbiol. 2021, 59, e02078-20. [Google Scholar] [CrossRef]
- Pornprasert, S.; Praparattanapan, J.; Khamwan, C.; Pawichai, S.; Pimsarn, P.; Samleerat, T.; Leechanachai, P.; Supparatpinyo, K. Development of TaqMan real-time polymerase chain reaction for the detection and identification of Penicillium marneffei. Mycoses 2009, 52, 487–492. [Google Scholar] [CrossRef]
- Kidd, S.E.; Chen, S.C.; Meyer, W.; Halliday, C.L. A New Age in Molecular Diagnostics for Invasive Fungal Disease: Are We Ready? Front. Microbiol. 2019, 10, 2903. [Google Scholar] [CrossRef]
- White, P.L.; Alanio, A.; Brown, L.; Cruciani, M.; Hagen, F.; Gorton, R.; Lackner, M.; Millon, L.; Morton, C.O.; Rautemaa-Richardson, R.; et al. An overview of using fungal DNA for the diagnosis of invasive mycoses. Expert. Rev. Mol. Diagn. 2022, 22, 169–184. [Google Scholar] [CrossRef]
- Buitrago, M.J.; Valero, C. Diagnosis of Fungal Infections; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Imhof, A.; Schaer, C.; Schoedon, G.; Schaer, D.J.; Walter, R.B.; Schaffner, A.; Schneemann, M. Rapid detection of pathogenic fungi from clinical specimens using LightCycler real-time fluorescence PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2003, 22, 558–560. [Google Scholar] [CrossRef][Green Version]
- Trubiano, J.A.; Dennison, A.M.; Morrissey, C.O.; Chua, K.Y.; Halliday, C.L.; Chen, S.C.; Spelman, D. Clinical utility of panfungal polymerase chain reaction for the diagnosis of invasive fungal disease: A single center experience. Med. Mycol. 2016, 54, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Ala-Houhala, M.; Koukila-Kahkola, P.; Antikainen, J.; Valve, J.; Kirveskari, J.; Anttila, V.J. Clinical use of fungal PCR from deep tissue samples in the diagnosis of invasive fungal diseases: A retrospective observational study. Clin. Microbiol. Infect. 2018, 24, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Lindner, A.K.; Rickerts, V.; Kurth, F.; Wilmes, D.; Richter, J. Chronic oral ulceration and lip swelling after a long term stay in Guatemala: A diagnostic challenge. Travel. Med. Infect. Dis. 2018, 23, 103–104. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, D.; McCormick-Smith, I.; Lempp, C.; Mayer, U.; Schulze, A.B.; Theegarten, D.; Hartmann, S.; Rickerts, V. Detection of Histoplasma DNA from Tissue Blocks by a Specific and a Broad-Range Real-Time PCR: Tools to Elucidate the Epidemiology of Histoplasmosis. J. Fungi 2020, 6, 319. [Google Scholar] [CrossRef]
- Buitrago, M.J.; Bernal-Martinez, L.; Castelli, M.V.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Performance of panfungal--and specific-PCR-based procedures for etiological diagnosis of invasive fungal diseases on tissue biopsy specimens with proven infection: A 7-year retrospective analysis from a reference laboratory. J. Clin. Microbiol. 2014, 52, 1737–1740. [Google Scholar] [CrossRef][Green Version]
- Morjaria, S.; Otto, C.; Moreira, A.; Chung, R.; Hatzoglou, V.; Pillai, M.; Banaei, N.; Tang, Y.W.; Figueroa, C.J. Ribosomal RNA gene sequencing for early diagnosis of Blastomyces dermatitidis infection. Int. J. Infect. Dis. 2015, 37, 122–124. [Google Scholar] [CrossRef]
- Rooms, I.; Mugisha, P.; Gambichler, T.; Hadaschik, E.; Esser, S.; Rath, P.M.; Haase, G.; Wilmes, D.; McCormick-Smith, I.; Rickerts, V. Disseminated Emergomycosis in a Person with HIV Infection, Uganda. Emerg. Infect. Dis. 2019, 25, 1750–1751. [Google Scholar] [CrossRef]
- Beltrame, A.; Danesi, P.; Farina, C.; Orza, P.; Perandin, F.; Zanardello, C.; Rodari, P.; Staffolani, S.; Bisoffi, Z. Case Report: Molecular Confirmation of Lobomycosis in an Italian Traveler Acquired in the Amazon Region of Venezuela. Am. J. Trop. Med. Hyg. 2017, 97, 1757–1760. [Google Scholar] [CrossRef]
- Valero, C.; de la Cruz-Villar, L.; Zaragoza, O.; Buitrago, M.J. New Panfungal Real-Time PCR Assay for Diagnosis of Invasive Fungal Infections. J. Clin. Microbiol. 2016, 54, 2910–2918. [Google Scholar] [CrossRef]
- Gade, L.; Hurst, S.; Balajee, S.A.; Lockhart, S.R.; Litvintseva, A.P. Detection of mucormycetes and other pathogenic fungi in formalin fixed paraffin embedded and fresh tissues using the extended region of 28S rDNA. Med. Mycol. 2017, 55, 385–395. [Google Scholar] [CrossRef][Green Version]
- Gomez, C.A.; Budvytiene, I.; Zemek, A.J.; Banaei, N. Performance of Targeted Fungal Sequencing for Culture-Independent Diagnosis of Invasive Fungal Disease. Clin. Infect. Dis. 2017, 65, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Sabino, R.; Simoes, H.; Verissimo, C. Detection of deep fungal infections: A polyphasic approach. J. Med. Microbiol. 2019, 68, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Suarez, C.J.; Banaei, N.; Pinsky, B.A. Next-Generation Sequencing for Infectious Disease Diagnosis and Management: A Report of the Association for Molecular Pathology. J. Mol. Diagn. 2015, 17, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Knox, N.C.; Ronholm, J.; Pagotto, F.; Reimer, A. Metagenomics: The Next Culture-Independent Game Changer. Front. Microbiol. 2017, 8, 1069. [Google Scholar] [CrossRef] [PubMed]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef]
- McTaggart, L.R.; Copeland, J.K.; Surendra, A.; Wang, P.W.; Husain, S.; Coburn, B.; Guttman, D.S.; Kus, J.V. Mycobiome Sequencing and Analysis Applied to Fungal Community Profiling of the Lower Respiratory Tract During Fungal Pathogenesis. Front. Microbiol. 2019, 10, 512. [Google Scholar] [CrossRef]
- Mao, Y.; Shen, H.; Yang, C.; Jia, Q.; Li, J.; Chen, Y.; Hu, J.; Huang, W. Clinical performance of metagenomic next-generation sequencing for the rapid diagnosis of talaromycosis in HIV-infected patients. Front. Cell Infect Microbiol. 2022, 12, 962441. [Google Scholar] [CrossRef]
- Zhu, Y.M.; Ai, J.W.; Xu, B.; Cui, P.; Cheng, Q.; Wu, H.; Qian, Y.Y.; Zhang, H.C.; Zhou, X.; Xing, L.; et al. Rapid and precise diagnosis of disseminated T. marneffei infection assisted by high-throughput sequencing of multifarious specimens in a HIV-negative patient: A case report. BMC Infect. Dis. 2018, 18, 379. [Google Scholar] [CrossRef]
- Wang, D.M.; Ma, H.L.; Tan, M.Q.; Wu, Y.M.; Wang, S.N. Next-generation sequencing confirmed the diagnosis of isolated central nervous system infection caused by Talaromyces marneffei in an immunocompetent patient. Chin. Med. J. 2020, 133, 374–376. [Google Scholar] [CrossRef]
- Zhang, W.; Ye, J.; Qiu, C.; Wang, L.; Jin, W.; Jiang, C.; Xu, L.; Xu, J.; Li, Y.; Wang, L.; et al. Rapid and precise diagnosis of T. marneffei pulmonary infection in a HIV-negative patient with autosomal-dominant STAT3 mutation: A case report. Ther. Adv. Respir. Dis. 2020, 14, 1753466620929225. [Google Scholar] [CrossRef]
- Du, R.; Feng, Y.; Liu, L.N.; Liu, Y.B.; Ye, H.; Lu, X.J.; Wang, X.H.; Zong, Z.Y. [Pathogen Diagnosis of a Febrile HIV Case by the Metagenomic Next-generation Sequencing]. Sichuan Da Xue Xue Bao Yi Xue Ban 2020, 51, 257–260. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, D.; Du, J.; Zhou, Y.; Cai, Y.; Sun, R.; Zhou, J.; Tian, J.; Wu, H.; Lu, M.; et al. Rapid diagnosis of Talaromyces marneffei infection assisted by metagenomic next-generation sequencing in a HIV-negative patient. IDCases 2021, 23, e01055. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Wu, L.; Cai, J.; Chen, H. An Iris Tumor Secondary to Talaromyces marneffei Infection in a Patient with AIDS and Syphilis. Ocul. Immunol. Inflamm. 2022, 30, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, N.; Qian, G. Case Report: Metagenomic Next-Generation Sequencing in Diagnosis of Talaromycosis of an Immunocompetent Patient. Front. Med. 2021, 8, 656194. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, Y.; Wen, Y. Gastrointestinal manifestations of Talaromyces marneffei infection in an HIV-infected patient rapidly verified by metagenomic next-generation sequencing: A case report. BMC Infect. Dis. 2021, 21, 376. [Google Scholar] [CrossRef]
- Shen, Q.; Sheng, L.; Zhou, J. HIV-negative case of Talaromyces marneffei pulmonary infection with a TSC2 mutation. J. Int. Med. Res. 2021, 49, 3000605211016761. [Google Scholar] [CrossRef]
- Wilson, M.R.; O’Donovan, B.D.; Gelfand, J.M.; Sample, H.A.; Chow, F.C.; Betjemann, J.P.; Shah, M.P.; Richie, M.B.; Gorman, M.P.; Hajj-Ali, R.A.; et al. Chronic Meningitis Investigated via Metagenomic Next-Generation Sequencing. JAMA Neurol. 2018, 75, 947–955. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, W.; Ling, H.; Dong, X.; Zhang, Y.; Li, J.; Zhang, Y.; Song, J.; Liu, W.J.; Li, Y.; et al. Identification of Histoplasma causing an unexplained disease cluster in Matthews Ridge, Guyana. Biosaf. Health 2019, 1, 150–154. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Li, Z.; Chen, G.; Liu, X.; Ding, L. Metagenomic next-generation sequencing identified Histoplasma capsulatum in the lung and epiglottis of a Chinese patient: A case report. Int. J. Infect. Dis. 2020, 101, 33–37. [Google Scholar] [CrossRef]
- Muldoon, J.L.; Wysozan, T.R.; Toubin, Y.; Relich, R.F.; Davis, T.E.; Zhang, C.; Alomari, A.K. An unusual presentation of cutaneous histoplasmosis as a recurrent solitary and spontaneously healing lesion in an immunocompetent patient. Access Microbiol. 2020, 2, acmi000156. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, C.; Tang, C.; Wang, L. Case Report and Literature Review: Disseminated Histoplasmosis Infection Diagnosed by Metagenomic Next-Generation Sequencing. Infect. Drug. Resist. 2022, 15, 4507–4514. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Yadav, M.; Singhania, N.; Samal, S.; Singhania, G. Blastomycosis Detected by Microbial Cell-Free DNA in Renal Transplant Recipient. Am. J. Med. 2020, 133, e599–e600. [Google Scholar] [CrossRef]
- Wang, N.; Luo, Z.; Deng, S.; Li, Q. A young male with chronic nonproductive cough diagnosed with blastomycosis in China: A case report. BMC Pulm. Med. 2020, 20, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.C.; Zhang, Q.R.; Ai, J.W.; Cui, P.; Wu, H.L.; Zhang, W.H.; Wang, T. The role of bone marrow metagenomics next-generation sequencing to differential diagnosis among visceral leishmaniasis, histoplasmosis, and talaromycosis marneffei. Int. J. Lab. Hematol. 2020, 42, e52–e54. [Google Scholar] [CrossRef]
- Larkin, P.M.K.; Lawson, K.L.; Contreras, D.A.; Le, C.Q.; Trejo, M.; Realegeno, S.; Hilt, E.E.; Chandrasekaran, S.; Garner, O.B.; Fishbein, G.A.; et al. Amplicon-Based Next-Generation Sequencing for Detection of Fungi in Formalin-Fixed, Paraffin-Embedded Tissues: Correlation with Histopathology and Clinical Applications. J. Mol. Diagn. 2020, 22, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Martinez, L.; Herrera, L.; Valero, C.; de la Cruz, P.; Ghimpu, L.; Mesa-Arango, A.C.; Santoni, G.; Goterris, L.; Millan, R.; Buitrago, M.J. Differential Diagnosis of Fungal Pneumonias vs. Tuberculosis in AIDS Patients by Using Two New Molecular Methods. J. Fungi 2021, 7, 336. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).