Abstract
Alternaria is a ubiquitous fungal genus including saprobic, endophytic, and pathogenic species associated with a wide variety of substrates. It has been separated into 29 sections and seven monotypic lineages based on molecular and morphological data. Alternaria sect. Porri is the largest section, containing the majority of large-spored Alternaria species, most of which are important plant pathogens. Since 2015, of the investigations for large-spored Alternaria species in China, 13 species were found associated with Compositae plants based on morphological comparisons and phylogenetic analyses. There were eight known species and five new species (A. anhuiensis sp. nov., A. coreopsidis sp. nov., A. nanningensis sp. nov., A. neimengguensis sp. nov., and A. sulphureus sp. nov.) distributed in the four sections of Helianthiinficientes, Porri, Sonchi, and Teretispora, and one monotypic lineage (A. argyranthemi). The multi-locus sequence analyses encompassing the internal transcribed spacer region of rDNA (ITS), glyceraldehydes-3-phosphate dehydrogenase (GAPDH), Alternaria major allergen gene (Alt a 1), translation elongation factor 1-alpha (TEF1), and RNA polymerase second largest subunit (RPB2), revealed that the new species fell into sect. Porri. Morphologically, the new species were illustrated and compared with other relevant large-spored Alternaria species in the study. Furthermore, A. calendulae, A. leucanthemi, and A. tagetica were firstly detected in Brachyactis ciliate, Carthamus tinctorius, and Calendula officinalis in China, respectively.
1. Introduction
Alternaria is a cosmopolitan and widely distributed fungal genus described originally by Nees (1816), which is characterized by the dark-coloured phaeodictyospores in chains and a beak of tapering apical cells []. It is also associated with nearly every environmental substrate including animal, plant, agricultural product, soil, and the atmosphere. Species of Alternaria are known as serious plant pathogens, causing enormous losses on many crops [,]. The taxonomy is mainly based on sporulation patterns and their conidial shape, size, and septation [,]. Around 280 species are summarised and recognised on the basis of morphology [], comprising two groups, large-spored (60–100 μm long conidial body) and small-spored (below 60 μm conidial body) [,,].
Since the 20th century, molecular approaches, especially multi-locus phylogenetic analyses, have been used to identify Alternaria species [,,,]. Over ten gene regions are used in the classification, such as the internal transcribed spacer region of rDNA (ITS), large subunit ribosomal DNA (LSU), mitochondrial small subunit (mtSSU), glyceraldehydes-3-phosphate dehydrogenase (GAPDH), Alternaria major allergen gene (Alt a 1), translation elongation factor 1-alpha (TEF1), RNA polymerase second largest subunit (RPB2), and plasma membrane ATPase [,,,,,,,,,,,]. Alternaria has been separated into 29 sections and seven monotypic lineages [,,]. The introduction of a molecular phylogenetic approach has helped to clarify their taxonomy, combining many allied genera into one large genus of Alternaria complex [].
Due to the effects of Alternaria on humans and their surroundings, the identification is particularly important to agriculture, medicine, and science. The Compositae plants serve as food plant, oil seed, seed plant, ornamental, and sources of medicine and insecticide worldwide [], of which nearly 3000 species almost 240 genera have been found in China []. Most Alternaria are commonly plant pathogens leading to substantial economic losses caused by Alternaria leaf spots and defoliation [,,,]. Large-spored Alternaria species encompassing 148 species are almost phytopathogenic demonstrated [].
During the investigation of large-spored Alternaria in China, five new species were encountered from diseased leave samples of composite plants. The objectives of this study were to identify them on the basis of the cultural and conidial morphology incorporate with multi-loci phylogeny (ITS, GAPDH, Alt a 1, TEF1, and RPB2). The present multi-locus analysis supplemented with cultural and morphological data forms an example for Alternaria species recognition. The five new species described in this study add species diversity to large-spored Alternaria and provide theoretical and practical basis for the further identification and disease management.
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
Symptomatic samples of composite plants (14) have been randomly collected from different provinces in China since 2015. For fungal isolation, the samples were put into sterile plastic bags and taken to the laboratory. Small leaf segments (2 mm) with disease lesions were placed into petri dishes with moist filter papers and incubated at 25 °C in dark for conidial sporulation. Single spore of large-spored Alternaria was picked by a sterilized glass needle under the stereoscopic microscope and transferred to potato dextrose agar (PDA: Difco, Montreal, Canada). Over ten similar spores were randomly picked from a sample for sub-culturing to obtain the pure cultures, and two to three strains were selected for deposition when exhibiting similar cultural morphology on PDA. A total of 81 strains were kept in test-tube slants and deposited at 4 °C. Living ex-type strains were preserved in the Fungi Herbarium of Yangtze University (YZU), in Jingzhou, Hubei, China.
2.2. Morphological Observations
To determine cultural characteristics including growth rate, color and texture of colonies [], mycelial plugs (6 mm in diameter) were taken from the edge of colonies grown on PDA. Then, the plugs were put on fresh PDA plates (90 mm) at 25 °C for 7 days in darkness. To observe the conidial morphology (conidial sporulation patterns, shape, size, etc.), mycelia were grown on potato carrot agar (PCA) and V8 juice agar (V8A) inoculated at 22 °C with a light period of 8 h light/16 h dark []. After 7 days, conidia and sporulation patterns were observed. Conidiophores and conidia were mounted with lactophenol picric acid solution and photographed with a Nikon ECLIPSE Ni-U microscope (Nikon, Japan). Randomly selected conidia (n = 50) were separately measured for each characterization.
2.3. DNA Extraction and PCR Amplification
Genomic DNA extraction was performed using fresh mycelia collected from colonies grown on PDA []. Polymerase chain reaction (PCR) amplifications of the internal transcribed spacer region of rDNA (ITS), glyceraldehydes-3-phosphate dehydrogenase (GAPDH), Alternaria major allergen gene (Alt a 1), translation elongation factor 1-alpha (TEF1), and RNA polymerase second largest subunit (RPB2) gene regions were amplified with the primer pairs ITS5/ITS4 [], EF1-728F/EF1-986R [], gpd1/gpd2 [], Alt-for/Alt-rev [], and RPB2-5F2/RPB2-7cR [,], respectively. A 25 μL of the PCR reaction mixture comprising 21 μL of 1.1 × Taq PCR Star Mix (TSINGKE, Beijing, China), 2 μL template DNA and 1 μL of each primer was applied and performed in a BIORAD T100 thermocycler []. Successfully amplified PCR products were purified and sequenced by TSINGKE company (Beijing, China).
2.4. Phylogenetic Analyses
The resulted sequences were examined by BioEdit v.7.0.9 [] and assembled with PHYDIT 3.2 []. All newly generated sequences were deposited in GenBank (Table 1). Relevant sequences [] were retrieved from NCBI database based on the results of BLAST searches (Table 1). The concatenated sequence dataset of multiple loci was aligned using MEGA v.6.0 []. Phylogenetic analyses of each alignment were performed using maximum likelihood (ML) and Bayesian inference (BI) methods. ML analysis was conducted using RAxML v.7.2.8 []. Bootstrapping with 1000 replicates was performed using the model of nucleotide substitution obtained by MrModeltest. For the BI analysis, it was performed using parameters including 1,000,000 Markov chain Monte Carlo (MCMC) algorithm with Bayesian posterior probabilities []. MrModel test v.2.3 used the best-fit model (GTR+I+G) according to the Akaike Information Criterion (AIC). Two MCMC chains were run from random trees for 106 generations, and the trees were sampled every 100th generation. After discarding the first 25% of the samples, the 50% majority rule consensus tree and posterior probability values were calculated. Finally, the resulting trees were edited in FigTree v.1.3.1 []. Branch support of the groupings (>60%/0.6 for ML bootstrap value-BS/posterior probability-PP) were indicated in the phylogram. Alternaria gypsophilae CBS 107.41 in sect. Gypsophilae was used as an outgroup.

Table 1.
GenBank accession numbers of Alternaria species used in phylogenetic analyses.
3. Results
In the present study, large-spored Alternaria species associated with Compositae leaf spot in China since a survey from 2015 are summarized based on the phylogenetic analysis of GAPDH and RPB2 gene fragments (Figure S1 and Table S1). A total of 13 species including the present five new taxa revealed in four sections of Helianthiinficientes (A. helianthiinficiens), Porri (A. calendulae, A. tagetica and A. zinniae), Sonchi (A. cinerariae and A. sonchi), and Teretispora (A. leucanthemi), and one monotypic lineage (A. argyranthemi) (Figure S1). Meanwhile, a comprehensive description of the five new species in sect. Porri are described as A. anhuiensis sp. nov., A. coreopsidis sp. nov., A. nanningensis sp. nov., A. neimengguensis sp. nov., and A. sulphureus sp. nov..
3.1. Phylogenetic Analysis
The multi-gene phylogeny was constructed to determine the accurate positions of the new Alternaria based on five sequence loci (ITS + GAPDH + Alt a 1 + TEF1 + RPB2) (Table 1). The analysis comprised sequences of the ITS (504 characters), GAPDH (526 characters), Alt a 1 (457 characters), TEF1 (342 characters), and RPB2 (672 characters) gene regions with a total length of 2501 characters. The tree topologies (Figure 1) computed from the ML and BI analyses, were similar to each other, resulting in identical species-clades and the ML topology was presented as basal tree. The present strains fell into five separate branches in sect. Porri of Alternaria. Strain YZU 171206 was sister to A. alternariacida supported with a PP value of 1.0, which close to A. silybi with low BS and PP values surpport. Strains YZU 161159 and YZU 161160 formed an independent clade (BS/PP = 100%/1.0). Strain YZU 171523 fell into an individual branch close to A. obtecta and A. tillandsiae well-supported by 97%/0.99 (BS/PP). Strain YZU 171784 was clustered with A. cirsinoxia, A. centaureae, A. cichorii, and A. cantannaches supported by values of 79%/1.0 (BS/PP). Strain YZU 191448 was out group of strain YZU 171206, A. silybi and A. alternariacida with BS and PP values below 60% and 0.6. The results indicated that the five branches represent five new species from three different hosts (Coreopsis basalis, Cosmos sulphureus, and Lactuca seriola).
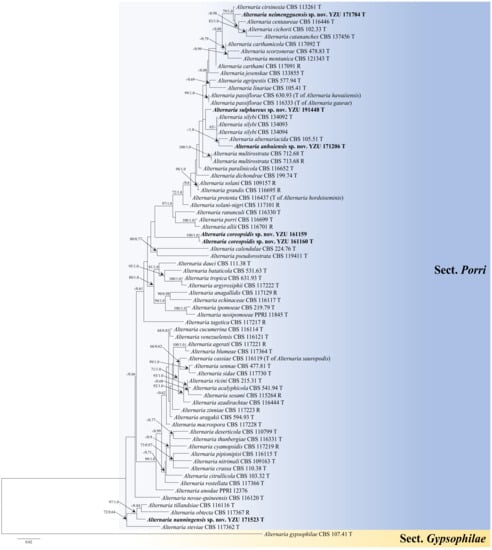
Figure 1.
Maximum likelihood (ML) phylogram of new five Alternaria species from the Compositae family based on a combined dataset of ITS, GAPDH, Alt a 1, TEF1, and RPB2 gene sequences. The RAxML bootstrap support values >60%(ML) and Bayesian posterior probabilities >0.6 (PP) are given at the nodes (ML/PP). The present strains are in bold.
3.2. Taxonomy
Alternaria anhuiensis H. Luo and J.X. Deng, sp. nov. (Figure 2).
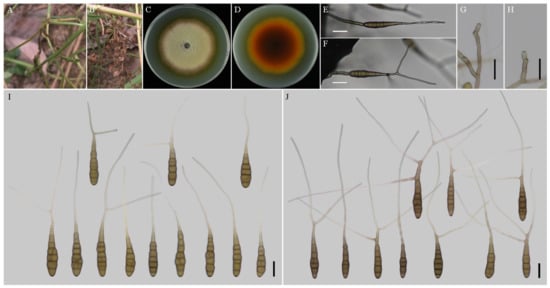
Figure 2.
Morphology of Alternaria anhuiensis sp. nov. (A,B) Natural symptoms of Coreopsis basalis; (C,D) Colony phenotypes (on PDA for 7 days at 25 °C); (E,F) Sporulation patterns (on V8A at 22 °C); (G,H) Conidiophores (on V8A at 22 °C); (I) Conidia (on V8A at 22 °C); (J) Conidia (on PCA at 22 °C). Bars: (E–J) = 25 μm.
MycoBank No: 844033.
Etymology: Named after the collecting locality, Anhui Province.
Typification: China, Anhui Province, Hefei City, from leaf spot of Coreopsis basalis. June, 2017, J.X Deng, ex-type culture YZU 171206.
Description: Colonies on PDA circular, buff in the centre, flocculent with brown halo at the edge; reverse crimson pigment at centers, light yellow at margins, 59–60 mm in diam, at 25 °C for 7 days. On V8A, conidiophores arising from substrate or lateral of aerial hyphae with geniculate conidiogenous loci at or near apex, straight or curved, smooth-walled, septate, pale to dark brown, (40–) 60–145 (–203) × (4.5–) 5–7.5 (–8) μm; conidia solitary, long-narrow ovoid or ellipsoid body, apex rounded, base narrow, smooth-walled, single to double beak, dark brown, 61–100 (–111.5) × (11.5–) 13–19.5 μm, 6–11 transverse septa, 0–1 (–2) longitudinal septa; beak long-narrowed filiform, 1-beak, (32–) 58–133 (–150.5) × 2.5–4 (–4.5) μm; 2-beak, (22–) 60.5–90.5 (–116.5) × 2.5–3.5 μm. On PCA, conidiophores straight or curved, smooth-walled, septate, (42.5–) 50–140 × 4.5–6.5 (–9) μm; conidia solitary, long-narrow ovoid or ellipsoid body, single to double beak, triple or quadruple beaks not common, black brown, (55–) 66–105 × 11–16 μm, 5–10 (–11) transverse septa, 0–1 longitudinal septum; beak long-narrowed filiform, 1-beak, 100–180 (–202) × 2.5–4 μm; 2-beak, 95–217 (–236) × 2.5–4 (–5.5) μm; 3-beak, 60–140 × 2.5–3.5 μm; 4-beak (n = 1), 82 × 3 μm.
Notes: Phylogenetic analysis of the species based on a combined dataset of ITS, GAPDH, Alt a 1, TEF1, and RPB2 gene fragments falls in an individual clade close to A. alternariacida and A. silybi in sect. Porri (Figure 1). Morphologically, its primary conidiophores can generate geniculate conidiogenous loci at or near apex which differed from those two species (Figure 2, Table 2). It can be easily distinguished from A. alternariacida by producing more transverse septa and shorter beaks. Moreover, its conidia are solitary while A. alternariacida forms solitary or in unbranched chains of 2 (–3) conidia.

Table 2.
Morphological comparisons of the five new Alternaria species and their closely related species.
Alternaria coreopsidis H. Luo and J.X. Deng, sp. nov. (Figure 3).
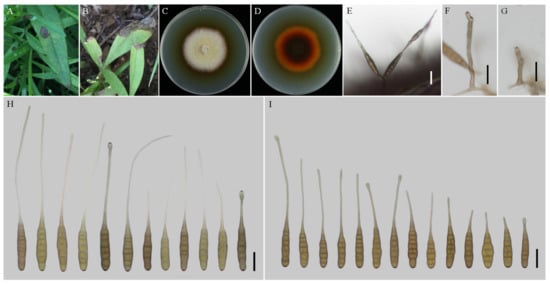
Figure 3.
Morphology of Alternaria coreopsidis sp. nov. (A,B) Natural symptoms of Coreopsis basalis; (C,D) Colony phenotypes (on PDA for 7 days at 25 °C); (E) Sporulation patterns (on V8A at 22 °C); (F,G) Conidiophores (on V8A at 22 °C); (H) Conidia (on V8A at 22 °C); (I) Conidia (on PCA at 22 °C). Bars: (E–I) = 25 μm.
MycoBank No: 844034.
Etymology: Named after the host genus name, Coreopsis.
Typification: China, Shaanxi Province, Xian City, from leaf spot of Coreopsis basalis. June, 2016, J.X Deng, ex-type culture YZU 161160.
Description: Colonies on PDA circular, buff halo in the centre, villiform with white at the edge; reverse dark brown at centers, vinaceous buff pigment at margins, 47–48 mm in diam, at 25 °C for 7 days. On V8A, conidiophores arising from substrate or lateral of aerial hyphae, solitary, simple, straight to slightly curved, septate, pale to dark brown, apical conidiogenous locus, pale brown, (34–) 50–86 (–115.5) × 5–7 (–9) μm; conidia solitary or in unbranched chains of 2 conidia, long-narrow ovoid or ellipsoid body, smooth-walled, single beak, yellow or brown, (48.5–) 55–80 (–85) × (9–) 10–15 μm, 6–9 transverse septa, 0–1 longitudinal septa; beak filamentous, 1-beak, (20–) 30–140 (–206) × (2–) 2.5–4 μm; normally, false beak swollen at the apex, around 8–10.5 (–14) × 4.5–5 (–6) μm. On PCA, conidiophores straight or curved, smooth-walled, septate, (24–) 50–90 (–135) × 5–7.5 (–9) μm; conidia long-narrow ovoid or ellipsoid body, apex rounded, single beak, pale brown, (40–) 45–70 × 9–13 μm, (5–) 6–8 (–9) transverse septa, 0–1 longitudinal septa; beak filamentous, 1-beak, (0–) 15–100 (–175) × (0–) 2–4 μm; swollen apex of false beak commonly 10–13 (–16.5) × 5–6 (–6.5) μm.
Materials examined: China, Shaanxi Province, Xian City, from leaf spot of Coreopsis basalis. June 2016, J.X Deng, living culture YZU 161159.
Notes: Phylogenetically, the species falls into an independent lineage outside of a clade comprising type species of A. porri of sect. Porri (Figure 1). It can be delimited based on either of GAPDH and RPB2 gene sequences (Figure S1). The species is characterized by producing conidia with false beak swollen at the apex up to 8–13 (–16.5) × 4.5–6.5 μm (Figure 3; Table 2).
Alternaria nanningensis H. Luo and J.X. Deng, sp. nov. (Figure 4).
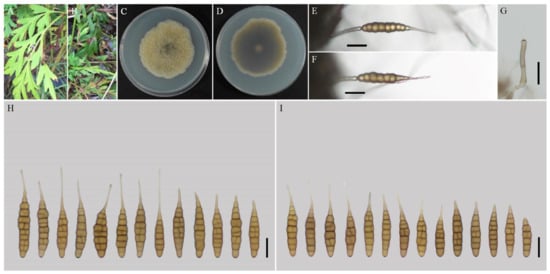
Figure 4.
Morphology of Alternaria nanningensis sp. nov. (A,B) Natural symptoms of Cosmos sulphureus; (C,D) Colony phenotypes (on PDA for 7 days at 25 °C); (E,F) Sporulation patterns (on V8A at 22 °C); (G) Conidiophores (on V8A at 22 °C); (H) Conidia (on V8A at 22 °C); (I) Conidia (on PCA at 22 °C). Bars: (E–J) = 25 μm.
MycoBank No: 844035.
Etymology: Named after the collecting locality, Nanning City.
Typification: China, Guangxi Province, Nanning City, from leaf spot of Cosmos sulphureus. July 2017, J.X Deng, ex-type culture YZU 171523.
Description: Colonies on PDA irregular, pistac, entire; reverse dark olive green, slightly protuberant with white at margins, 56–57 mm in diam, at 25 °C for 7 days. On V8A, conidiophores arising from substrate or lateral of aerial hyphae with geniculate conidiogenous loci at apex, straight or curved, smooth-walled, septate, pale brown, 38–59 (–64)× 4–5 (–6) μm; conidia solitary, ovoid or ellipsoid body, base narrow, smooth-walled, single beak, pale to yellow brown, (40.5–) 47–79 (–87) × 9–13.5 (–15) μm, 6–10 (–11) transverse septa, 0–1 longitudinal septa; beak long-narrowed filiform, 1-beak, 10–30 × (1–) 1.5–2 (–3) μm. On PCA, conidiophores straight or curved, smooth-walled, septate; 32–70 (–86) × 4–5.5 μm; conidia solitary, ovoid or ellipsoid body, single beak, pale to yellow brown, (49–) 55–77 (–82) × 10.5–13.5 (–15) μm, (5–) 6–9 (–10) transverse septa, 0–1 longitudinal septum; beak long-narrowed filiform, 1-beak, 13–26 (–44) × 1.5–2 (–2.5) μm.
Notes: The species is phylogenetically recognized as a distinct species in sect. Porri based on ITS, GAPDH, Alt a 1, TEF1, and RPB2 which displays a close relationship with A. obtecta, A. tillandsiae, and A. steviae (Figure 1). Compared with them, it is quite different by producing smaller conidia with short beaks (Figure 4; Table 2). Furthermore, its conidia are smooth-walled while some conidia of A. obtecta and A. steviae are minutely punctulate. Alternaria nanningensis forms simple conidiophores (solitary). But many conidiophores of A. steviae produce geniculate extensions and additional conidia, yielding tiny distal clumps of sporulation.
Alternaria neimengguensis H. Luo and J.X. Deng, sp. nov. (Figure 5).
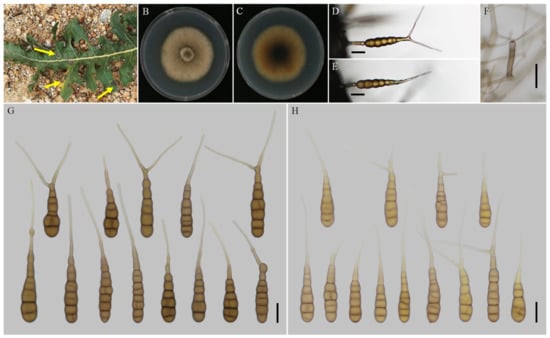
Figure 5.
Morphology of Alternaria neimengguensis sp. nov. (A) Natural symptoms of Lactuca seriola; (B,C) Colony phenotypes (on PDA for 7 days at 25 °C); (D,E) Sporulation patterns (on V8A at 22 °C); (F) Conidiophores (on V8A at 22 °C); (G) Conidia (on V8A at 22 °C); (H) Conidia (on PCA at 22 °C). Bars: (D–H) = 25 μm.
MycoBank No: 844036.
Etymology: Named after the collecting locality, Inner Mongolia Autonomous Region.
Typification: China, Inner Mongolia Autonomous Region, Inner Mongolia Agricultural University, IMAU, from leaf spot of Lactuca seriola. September 2017, J.X Deng, ex-type culture YZU 171784.
Description: Colonies on PDA circular, pale brown en masse, flocculent, reverse dark olive green at centers, pale brown at margins, 51–54 mm in diam, at 25 °C for 7 days. On V8A, conidiophores arising from substrate or lateral of aerial hyphae, straight or curved, smooth-walled, septate, brown, 26–45 (–51) × 5–7 (–8) μm; conidia solitary, ovoid or ellipsoid body, apex rounded, base wide, smooth-walled, single to double beak, brown, (70–) 77–130 (–143.5) × (13–) 15–20 (–23) μm, 6–11 (–12) transverse septa, 0–1 (–2) longitudinal septa; beak long-narrowed filiform, 1-beak, (24.5–) 35–65 (–76) × (1.5–) 2–3 (–4) μm; 2-beak, (33–) 45–65 (–92) × (2–) 2.5–3 (–3.5) μm. On PCA, conidiophores straight or curved, smooth-walled, septate; 35–70 (–75)× 5–6.5 (–7.5) μm; conidia solitary, ovoid or ellipsoid body, apex rounded, single to double beak, pale to yellow brown, (59–) 66–104 (–120.5) × 13–18 (–20) μm, (5–) 6–10 (–11) transverse septa, 0–1 (–2) longitudinal septa; beak long-narrowed filiform, 1-beak, (13–) 31.5–60 (–93) × 1.5–3 μm; 2-beak, (12–) 26–53 (–80) × 1.5–2.5 (–3) μm.
Notes: In the phylogeny, the species is sister to A. cirsinoxia, A. centaureae, A. cichorii, and A. catananches (Figure 1). The conidiophores are distinct to A. cirsinoxia whose are 2–3 arm branches near a conidiophore tip and progressively geniculate, yielding tufts of several conidia. They are different from A. cichorii whose are frequently branch or proliferate in a geniculate manner near the apex, yielding terminal clumps of 4–5 conidia. In conidial morphology, it is obviously different from those four species by producing larger conidia (Table 2).
Alternaria sulphureus L. Zhao and J.X. Deng, sp. nov. (Figure 6).

Figure 6.
Morphology of Alternaria sulphureus sp. nov. (A,B) Natural symptoms of Cosmos sulphureus; (C,D) Colony phenotypes (on PDA for 7 days at 25 °C); (E,F) Sporulation patterns (on PCA at 22 °C); (G) Conidiophores (on PCA at 22 °C); (H) Conidia (on V8A at 22 °C); (I) Conidia (on PCA at 22 °C). Bars: (E–I) = 25 μm.
MycoBank No: 844037.
Etymology: Named after the host species name, Cosmos sulphureus.
Typification: China, Shanxi Province, from leaf spot of Cosmos sulphureus. September 2019, J.X Deng, ex-type culture YZU 191448.
Description: Colonies on PDA circular, light brown in the centre, buff texture velutinous at the edge, reverse black brown at centers, 62–63 mm in diam, at 25 °C for 7 days. On V8A, conidiophores arising from substrate, solitary, simple, straight to slightly curved, septate, apical conidiogenous locus, pale brown; (50–) 63–100 (–108) × 6–8 (–9) μm; conidia solitary, sometimes in chains of two conidia, ovoid, ellipsoid or obovoid body, smooth-walled, pale to yellow, (64–) 74–116 × (12.5–) 14–20 (–25.5) μm, (5–) 7–11 transverse septa, 0–1 (–2) longitudinal septa; beak long-narrowed filiform, 1-beak, (25.5–) 34–151 (–159.5) × 2.5–4.5 (–5.5) μm; 2-beak (n = 1), 129 × 4 μm. On PCA, conidiophores straight or curved, smooth-walled, septate, (34.5–) 40.5–56 (–85) × 5–7.5 μm; conidia ovoid, ellipsoid, or obovoid body, apex rounded, single to double beak, triple beaks not common, pale brown, 80–110 × 16–24 μm, 6–10 transverse septa, 0–1 longitudinal septum; beak long-narrowed filiform, 1-beak, (73–) 110–195 × 3–5 μm; 2-beak, (74–) 96–170 × 3–4 μm; 3-beak (n = 1), 109.5 × 3.5 μm.
Notes: This species is phylogenetically related to A. silybi, A. alternariacida and A. anhuiensis sp. nov. in sect. Porri (Figure 1). It could be distinguished from A. silybi and A. alternariacida by forming larger conidia (Figure 6; Table 2) and is quite different from A. alternariacida by producing multiple and shorter beaks.
4. Discussion
Thirteen large-spored Alternaria species associated with Compositae leaf spot in China were assigned to four sections and one monotypic lineage in this study. Among theses species, five new species (A. anhuiensis sp. nov., A. coreopsidis sp. nov., A. nanningensis sp. nov., A. neimengguensis sp. nov., and A. sulphureus sp. nov.) were clearly recognized in section Porri. The section is speciose assessing encompassing 117 large-spored Alternaria []. In 2014, the section is reduced 82 morphospecies in to 63 phylogenetic species []. They are commonly pathogenic and could induce typical black necrotic lesions surrounded by chlorotic areas. There are some important famous plant pathogens, such as A. porri on Allium plants (Liliaceae), A. solani for potato (Solanaceae), A. sesami for sesame (Pedaliaceae) and A. dauci for carrot (Umbelliferae) []. Twenty-one species are comprised in sect. Porri associated with the Compositae family []. This study provides new data supplements for the Alternaria taxonomy of sect. Porri.
Morphologically, large-spored Alternaria species in sect. Porri are characterised by broadly ovoid, obclavate, ellipsoid, subcylindrical or obovoid, medium to large conidia containing multiple transverse and longitudinal septa, solitary or in short chains with a simple or branched, long to filamentous beak []. Among these characteristics, sporulation patterns, conidial body, transverse septa, and beak type provide useful information for the preliminary separation into sections []. Morphology is quite important for new fungal species identification, which can be defined based on unique morphological characters when the molecular data is not well-supported []. Morphological comparisons of the present new species and their relevant species in sect. Porri were conducted (Table 2). For the sporulation patterns, the conidia of A. anhuiensis, A. nanningensis, A. neimengguensis, and A. sulphureus are solitary produced except A. coreopsidis, which similar to A. alternariacida, A. cichorii, A. cirsinoxia, and A. steviae forming chain of 2 (–3) units [,]. In conidial morphology, A. anhuiensis, A. nanningensis, A. neimengguensis, and A. sulphureus are distinguishable from their closely related species based on the size of conidial bodies (Table 2) and also the wall ornamentations [,]. On the other hand, A. anhuiensis, A. neimengguensis, and A. sulphureus are readily be distinguished by producing multiple beaks. By the way, there are no significant differences on conidial morphology of PCA and V8A medium for all species.
In addition, morphological variation and fundamental pleomorphism complicate the Alternaria species recognition, and host plants reflect some evidences for the identification []. With the discovery of Alternaria species, it has been found from various plants of Compositae [,,,,]. Alternaria calendulae has been reported from Calendula officinalis in Czech Republic [], Germany [], Japan [], and Korea []. It also is found on C. officinalis in China and firstly on Brachyactis ciliata in the study. Alternaria leucanthemi has previously been found on Chrysanthemum maximum from Netherlands [] and Helianthus annuus from China []. It is firstly isolated from Carthamus tinctorius in this study. In addition, A. tagetica is commonly associated with Tagetes plants (Tagetes erecta and Tagetes patula) [,,,,], which firstly encountered from Calendula officinalis in this study. Interestedly, the five new species are isolated from three different composite hosts (Coreopsis basalis, Cosmos sulphureus, and Lactuca seriola) and A. cinerariae are found on five different composite plants in China (Figure S1; Table S1). The results suggest that an Alternaria species may associated with several host plants.
5. Conclusions
The present data indeed revealed a diversity of large-spored Alternaria associated with Compositae plants in China. A total of 13 large-spored Alternaria species were obtained and circumscribed as eight known species and five new species belonging to the four sections of Helianthiinficientes, Porri, Sonchi, and Teretispora, and one monotypic lineage (A. argyranthemi) based on the morphological characteristics and molecular properties of multiple DNA sequences (ITS, GAPDH, Alt a 1, TEF1, and RPB2). Alternaria calendulae, A. leucanthemi, and A. tagetica were firstly isolated from Brachyactis ciliate, Carthamus tinctorius, and Calendula officinalis in China, respectively. Since large-spored Alternaria species are almost demonstrated phytopathogens, further study on the pathogenicity is needed to verify in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8060607/s1, Figure S1: Phylogenetic tree of large-spored Alternaria from the Compositae family in China using a maximum likelihood (ML) analysis based on combined GAPDH and RPB2 gene sequences. The RAxML bootstrap support values > 60% (ML) and Bayesian posterior probabilities >0.6 (PP) are given at the nodes (ML/PP); Table S1: The other Altenraria species associated with the Compositae plants from China analyzed by phylogeny.
Author Contributions
The contributions of L.Z. and H.L. are consistent. Conceptualization, L.Z. and J.-X.D.; methodology, L.Z. and J.-X.D.; software, H.L.; validation, H.L., H.C. and Y.-N.G.; formal analysis, H.C. and Y.-N.G.; data curation, L.Z. and H.L.; writing—original draft preparation, L.Z. and H.L.; writing—review and editing, L.Z., J.-X.D. and Z.-H.Y.; visualization, L.Z.; supervision, J.-X.D.; project administration, J.-X.D. All authors have read and agreed to the published version of the manuscript.
Funding
The financial support was given by the National Natural Science Foundation of China (No. 31400014 and No. 31570022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences newly generated in this study have been submitted to the GenBank database.
Acknowledgments
The authors would like to thank Xue-Feng Wei for providing the leaf samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Woudenberg, J.H.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Zhang, T.Y. Flora Fungorum Sinicorum, Alternaria; Science Press: Beijing, China, 2003; Volume 16. [Google Scholar]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef] [Green Version]
- Gannibal, P.B. Distribution of Alternaria species among sections. 1. Section Porri. Mycotaxon 2015, 130, 207–213. [Google Scholar] [CrossRef]
- Pinto, V.E.F.; Patriarca, A. Alternaria Species and Their Associated Mycotoxins; Mycotoxigenic Fungi Humana Press: New York, NY, USA, 2017; pp. 13–32. [Google Scholar] [CrossRef]
- Pryor, B.M.; Bigelow, D.M. Molecular characterization of Embellisia and Nimbya species and their relationship to Alternaria, Ulocladium and Stemphylium. Mycologia 2003, 95, 1141–1154. [Google Scholar] [CrossRef]
- Park, M.S.; Romanoski, C.E.; Pryor, B.M. A re-examination of the phylogenetic relationship between the causal agents of carrot black rot, Alternaria radicina and A. carotiincultae. Mycologia 2008, 100, 511–527. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.F.; Liao, J.; Chen, X.Y.; Liu, Q.K.; Yu, Z.H.; Deng, J.X. A novel species and a new record of Alternaria isolated from two Solanaceae plants in China. Mycol. Prog. 2019, 18, 1005–1012. [Google Scholar] [CrossRef]
- Htun, A.A.; Liu, H.F.; He, L.; Zhou, X.Z.; Aung, S.L.L.; Deng, J.X. New species and new record of Alternaria from onion leaf blight in Myanmar. Mycol. Prog. 2022, 21, 59–69. [Google Scholar] [CrossRef]
- Pryor, B.M.; Gilbertson, R.L. Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mt SSU rDNA sequences. Mycol. Res. 2000, 104, 1312–1321. [Google Scholar] [CrossRef] [Green Version]
- Hong, G.S.; Cramer, R.A.; Lawrence, C.B.; Pryor, B.M. Alt a 1 allergen homologs from Alternaria and related taxa: Analysis of phylogenetic content and secondary structure. Fungal Genet. Biol. 2005, 42, 119–129. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Park, M.S.; Pryor, B.M. Nimbya and embellisia revisited, with nov.comb. for Alternaria celosiae and A. perpunctulata. Mycol. Prog. 2012, 11, 799–815. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, D.P.; Gannibal, P.B.; Dugan, F.M.; Pryor, B.M. Characterization of Alternaria isolates from the infectoria species-group and a new taxon from Arrhenatherum, Pseudoalternaria arrhenatheria sp. nov. Mycol. Prog. 2014, 13, 257–276. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; Vries, M.D.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Pei, D.F.; Aung, S.L.L.; Liu, H.F.; Liu, Q.K.; Yu, Z.H.; Deng, J.X. Alternaria hydrangeae sp. nov. (Ascomycota: Pleosporaceae) from Hydrangea paniculata in China. Phytotaxa 2019, 401, 287–295. [Google Scholar] [CrossRef]
- He, L.; Cheng, H.; Htun, A.A.; Ge, H.; Xia, Z.Z.; Guo, J.J.; Deng, J.X.; Du, T. Phylogeny and taxonomy of two new Alternaria (Ascomycota: Pleosporaceae) species in section Gypsophilae from China. Mycol. Prog. 2021, 20, 355–363. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Rotondo, F.; Gannibal, P.B. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol. Prog. 2016, 15, 3. [Google Scholar] [CrossRef]
- Ghafri, A.A.; Maharachchikunbura, S.S.; Hyde, K.D.; Nadiya, A.A.S.; Abdullah, M.A.S. A new section and a new species of Alternaria encountered from Oman. Phytotaxa 2019, 405, 279–289. [Google Scholar] [CrossRef]
- Gannibal, P.B.; Orina, A.S.; Gasich, E.L. A new section for Alternaria helianthiinficiens found on sunflower and new asteraceous hosts in Russia. Mycol. Prog. 2022, 21, 34. [Google Scholar] [CrossRef]
- Hind, N. Introduction to the compositae, the largest family of flowering plants. Curtis’s Bot. Mag. 2018, 35, 332–338. [Google Scholar] [CrossRef]
- Luo, H.; Yu, Z.; Lu, H.; Zhao, M.; Zhou, Y.; Deng, J. Taxonomy of Alternaria from Compositae: Research progress. Chin. Agric. Sci. Bull. 2018, 34, 63–70. [Google Scholar]
- Mackinnon, S.L.; Keifer, P.; Ayer, W.A. Components from the phytotoxic extract of Alternaria brassicicola, a black spot pathogen of canola. Phytochemistry 1999, 51, 215–221. [Google Scholar] [CrossRef]
- Andersen, B.; Krøger, E.; Roberts, R.G. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol. Res. 2001, 105, 291–299. [Google Scholar] [CrossRef]
- Thomma, B.P. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.X.; Li, M.J.; Paul, N.C.; Oo, M.M.; Lee, H.B.; Oh, S.K.; Yu, S.H. Alternaria brassicifolii sp. nov. isolated from Brassica rapa subsp. pekinensis in Korea. Mycobiology 2018, 46, 172–176. [Google Scholar] [CrossRef] [Green Version]
- Cenis, J.L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992, 20, 2380. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi–gene phylogeny of Clavicipitaceae (Ascomycota, fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenetics Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Chun, J. Computer Assisted Classification and Identification of Actinomycetes. Ph.D. Thesis, Unversity of Newcastle, Newcastle upon Tyne, UK, 1995. Available online: http://theses.ncl.ac.uk/jspui/handle/10443/410 (accessed on 1 May 2022).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A. FigTree v.1.3.1. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, UK, 2010. [Google Scholar]
- Gannibal, P.B. Taxonomic studies of Alternaria from Russia: New species on Asteraceae. Mycotaxon 2010, 114, 109–114. [Google Scholar] [CrossRef]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Luo, H.; Xia, Z.Z.; Chen, Y.Y.; Zhou, Y.; Deng, J.X. Morphology and Molecular Characterization of Alternaria argyranthemi on Chrysanthemum coronarium in China. Mycobiology 2018, 46, 278–282. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Liu, H.F.; Htun, A.A.; Ge, H.; Deng, J.X.; Du, T. First Report of Alternaria cinerariae Causing Leaf Spot on Tussilago farfara in China. Plant Dis. 2020, 104, 3264. [Google Scholar] [CrossRef]
- Lee, Y.H.; Cho, W.D.; Kim, W.K.; Jin, K.S.; Lee, E.J. Report on host-unrecorded diseases identified from economical crops in Korea. Res. Rep. Rural Developm. Admin. 1991, 33, 15–19. [Google Scholar]
- Zhao, G.; Zhang, T.Y.; Cao, A.X.; Wang, H.K. Phylogenetic relationships of Alternaria and related genera and taxonomic status of A. leucanthemi inferred from ITS rDNA sequence analysis. Mycosystema 2006, 25, 184–191. [Google Scholar] [CrossRef]
- Tomioka, K.; Sato, T.; Koganezawa, H. Marigold leaf spot caused by Alternaria tagetica new to Japan. J. Gen. Plant Pathol. 2000, 664, 294–298. [Google Scholar] [CrossRef]
- Li, M.J.; Deng, J.X.; Paul, N.C.; Lee, H.B.; Yu, S.H. Characterization and pathogenicity of Alternaria vanuatuensis, a new record from Allium plants in Korea and China. Mycobiology 2014, 42, 412–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, N.C.; Deng, J.X.; Lee, H.B.; Yu, S.H. Characterization and pathogenicity of Alternaria burnsii from seeds of Cucurbita maxima (Cucurbitaceae) in Bangladesh. Mycobiology 2015, 43, 384–391. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).