Abstract
Fungi of genus Epichloë (Ascomycota, Clavicipitaceae) are common endophytic symbionts of Poaceae, including wild and agronomically important cool-season grass species (subfam. Poöideae). Here, we examined the genetic diversity of Epichloë from three European species of Brachypodium (B. sylvaticum, B. pinnatum and B. phoenicoides) and three species of Calamagrostis (C. arundinacea, C. purpurea and C. villosa), using DNA sequences of tubB and tefA genes. In addition, microsatellite markers were obtained from a larger set of isolates from B. sylvaticum sampled across Europe. Based on phylogenetic analyses the isolates from Brachypodium hosts were placed in three different subclades within the Epichloë typhina complex (ETC) but did not strictly group according to host grass species, suggesting that the host does not always select for particular endophyte genotypes. Analysis of microsatellite markers confirmed the presence of genetically distinct lineages of Epichloë sylvatica on B. sylvaticum, which appeared to be tied to different modes of reproduction (sexual or asexual). Among isolates from Calamagrostis hosts, two subclades were detected which were placed outside ETC. These endophyte lineages are recognized as distinct species for which we propose the names E. calamagrostidis Leuchtm. & Schardl, sp. nov. and E. ftanensis Leuchtm. & A.D. Treindl, sp. nov. This study extends knowledge of the phylogeny and evolutionary diversification of Epichloë endophytes that are symbionts of wild Brachypodium and Calamagrostis host grasses.
1. Introduction
In plant parasitic fungi, host association is expected to be a major driver for the emergence of new species given their dependence on the host [1,2]. Underlying mechanisms of speciation may include cospeciating between parasite and host, host-range expansion and host jumps [3,4]. Although most well-studied cases of speciation either concern model systems or come from studies in agronomic settings, fewer examples have been investigated from natural ecosystems.
Epichloë species (Ascomycota, Clavicipitaceae) are common endophytic symbionts of grasses of the subfamily Poöideae [5,6]. They systemically infect above-ground parts of the host plant, and for reproduction may exhibit two different life history strategies. Sexually reproducing species form fruiting structures (called stromata) producing ascospores around developing grass inflorescences, thereby preventing host flowering and seed set (choke disease). Ascospores are released into the air and mediate contagious spread to new hosts. Most asexual species cause no symptoms in their hosts, grow into host ovules and seeds and thus are clonally propagated. [7,8]. In some grass/endophyte associations intermediate levels of choking occur, which allow for both strategies of fungal reproduction [9]. The type of reproduction is usually characteristic of a species or strain in association with a particular host and appears to be controlled mostly by the genotype of the fungus [10,11,12]. However, in some species environmental factors such as high soil nutrient level following fertilization can reduce or prevent disease expression [13,14].
The genus Epichloë currently encompasses 39 species, of which 13 are stroma-forming and mostly sexual, and 26 are asexual [6,15,16,17,18,19]. Furthermore, among the asexual species the majority are allopolyploid, interspecific hybrids that presumably resulted from hybridizations between two or more ancestral sexual species [20]. Most sexual Epichloë species were originally identified as distinct mating populations (MP) based on their interfertility, and a biological species concept has generally been applied in their descriptions [21,22,23,24,25]. The largest MP is represented by several taxa of the Epichloë typhina complex (ETC), which includes E. typhina (Pres.) Brockm. With ssp. Clarkii and spp. Poae, and E. sylvatica Leuchtm. & Schardl with ssp. Pollinensis, which together may infect at least 15 different host grass species [6]. More recent investigations, however, suggest that subspecies status of E. clarkii J.F. White and E. poae Tadych, K.V. Ambrose, F.C. Belanger & J.F. White may no longer be tenable, and these taxa should be recognized as species [25,26].
In recent years, numerous investigations have been made on the diversity of Epichloë species symbiotic with particular grass hosts. These hosts included agronomically important species of genus Festuca and Lolium [27,28,29], as well as genera of grasses growing naturally in woodlands and prairies [16,30,31,32,33]. A well-studied host of natural habitats in woodlands is the grass species Brachypodium sylvaticum (Huds.) Beauv. (false brome), which is widely distributed in temperate Eurasia and North Africa [34,35] and appears to be almost ubiquitously infected by Epichloë sylvatica Leuchtm. & Schardl in its native Eurasian range [10,36,37,38,39]. Epichloë sylvatica is mainly associated with B. sylvaticum, but may rarely be found on Hordelymus europaeus (L.) Harz [21,31]. The endophyte can reproduce sexually, which involves outcrossing between individuals with different mating types within a distinct MP. However, expression of stromata is rarely observed in infected populations of B. sylvaticum, but when it is, either stroma formation is restricted to a subset of infected plants, or individuals have both choked and healthy flowering tillers [10]. Moreover, plants with mixed symptoms are typically infected by more than one endophyte genotype, which may be responsible for the different disease expression [12,40].
Two other species of Brachypodium, B. pinnatum (L.) B. Beauv. and B. phoenicoides (L.) Roem. & Schult., are occasionally found to be infected with stroma-forming Epichloë [21,39,41]. Isolates from these hosts have been assigned to E. typhina based on their mating compatibility and genetic relatedness. However, isolates infecting B. pinnatum are not uniform and some may have a closer phylogenetic relationship to E. sylvatica than to E. typhina [6].
A second host genus of Epichloë examined in the present study is Calamagrostis (reed grass) which is distributed around the globe in temperate zones and comprises approximately 250 species that are particularly rich in South America [42]. However, a minority of the species have been examined so far and only approximately 18 species are reported to be infected. These included host species with symptomless infections mainly in South America [43,44] and species that had stromata. A stroma-forming Epichloë species, E. amarillans J.F. White, was identified from North American C. canadensis (Michx.) P. Beauv. [45], whereas collections from European Calamagrostis species have been assigned to E. baconii J.F. White [46]. A third stroma-forming species, E. stromatolonga (Y.L. Ji, L.H. Zhan & Z.W. Wang) Leuchtm., was more recently described from C. epigejos (L.) Roth in China [47].
In this study we report on the genetic diversity of Epichloë species symbiotic with Brachypodium and Calamagrostis species in Europe. Particular emphasis is given to endophytes infecting B. sylvaticum, which were obtained from a large sample across Europe. To evaluate diversity we use microsatellite markers and DNA sequence data from tubB and tefA genes. Two endophyte lineages infecting Calamagrostis hosts are recognized as new species. This study extends our knowledge on the phylogeny and evolutionary diversification of Epichloë endophytes in wild Brachypodium and Calamagrostis host grass species.
2. Materials and Methods
2.1. Host Grass Species
The genus Brachypodium has approximately 20 species classified in its own tribe Brachypodieae [48]. Among these, three species have been found to be infected by Epichloë endophytes. (1) Brachypodium sylvaticum is a perennial, caespitose (tussock forming) grass that is native to Europe, temperate Asia and north-western Africa, and introduced to other parts of the world, e.g., Pacific North West of the United States [49]. It is most commonly found in forests and woodlands, preferring the moist, shaded canopy, but may also grow in open areas. In contrast to the other species of the genus that are predominantly wind pollinated, a very high level of selfing is observed in B. sylvaticum [35]. This host is commonly infected by E. sylvatica [21]. (2) Brachypodium pinnatum is a perennial, stolon-forming grass with similar distribution as B. sylvaticum in temperate Eurasia. It prefers dry, nutrient poor grassland or light deciduous woods and may be a pioneer in open soil or clearings. It can be infected by choke-forming E. typhina which is not seed-transmitted on this host [6]. (3) Brachypodium phoenicoides is also stolon-forming and grows in light oak woods in the south-western part of the Mediterranean region. This species may be infected by stroma-forming E. typhina [41].
The genus Calamagrostis of tribe Agrostideae comprises approximately 250 species worldwide. In our study, we examined endophytes from four Calamagrostis hosts. (1) C. villosa (Chaix) J.F. Gmel., a stolon-forming species occurring on acid soils in the understory of pine and larch forest in central and southern European mountains, (2) C. varia (Schrad.) Host that grows on lime rich soils preferably in open scree or other pioneer sites and has a similar distribution, and (3) C. purpurea (Trin.) Trin. that grows on humid to damp sites in woodlands or aside standing water with a native range from subalpine to subarctic. C. purpurea has been repeatedly reported to be infected by stroma-forming Epichloë mainly in Nordic European countries but without assigning a distinct species name to it [36,50]. (4) C. arundinacea (L.) Roth has a Euro-Siberian distribution and occurs on mostly lime-free, dry to humid soils, preferentially in mountain woodlands.
2.2. Endophyte Sampling and Isolation
Sampling was conducted in different years from host grasses growing at natural sites. Grass shoots (with or without stromata) or seeds were collected and taken to the laboratory. One isolate from B. sylvaticum was obtained from Japan as a cultured strain. Geographic origin, source of isolation and mode of reproduction of all isolates used in this study are listed in Table 1. In addition, location of collection sites of Epichloë-infected Brachypodium hosts are indicated in Figure 1.

Table 1.
Epichloë isolates from Brachypodium and Calamagrostis hosts used in this study with mode of reproduction (sexual or asexual) and source of isolation (culm, culm with stroma, seed, stroma tissue or ascopores). In addition, use for microsatellite analysis (+) and GenBank accession numbers are indicated (new sequences of this study are in bold).
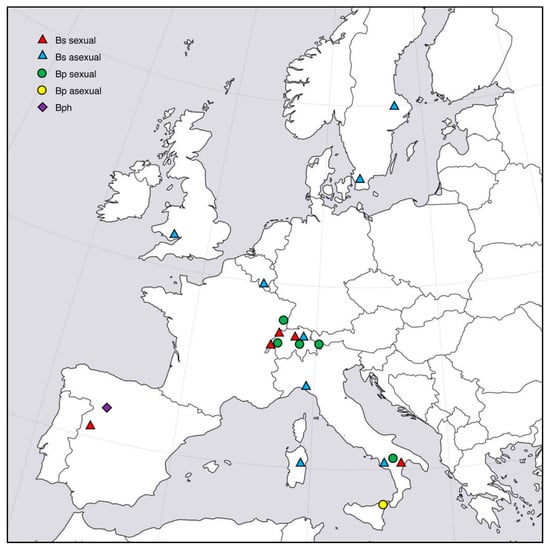
Figure 1.
Location of collection sites in Europe of Epichloë-infected Brachypodium species: B. sylvaticum (Bs), B. pinnatum (Bp) and B. phoenicoides (Bph). Symbols distinguish between host, and stroma-forming (sexual) or symptomless (asexual) infections.
Endophytes were isolated from surface-disinfected plant tissues (culm) or from seeds that were placed on standard nutrient agar medium supplemented with 50 mg/L oxytetracycline (Pfizer, New York, NY, USA) in Petri dishes as previously described [51]. Isolates from stroma-forming plants were either obtained from ascospores or in some cases from mycelium taken from the innermost part of young, clean stromata after splitting them open under sterile condition. Colonies growing from plant tissues, seeds or mycelium were checked under the microscope for purity, and Epichloë identity was confirmed based on the characteristic sporulation [52]. For DNA extraction isolates were grown on V-8 liquid medium on a rotary shaker for 12–14 days as previously described [31].
2.3. Microsatellite Analyses
Genomic DNA was extracted from freeze-dried mycelia with the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions.
Microsatellite analyses followed the protocol developed by Schirrmann et al. [26] using a multiplex PCR approach. The 16 markers were arranged in four multiplex sets (Table 2) and each set was amplified using approximately 1 ng of genomic DNA in a PCR volume of 10 µL. Amplification conditions were as follows: initial denaturation of dsDNA at 94 °C for 3 min, 30 cycles of 30 s denaturation at 94 °C with 1 min annealing at 56 °C and 30 s extension at 72 °C, and final elongation at 72 °C for 5 min. Signals of the PCR products were detected on a 3130xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) with GeneScan-500 LIZ as size standard. Electropherograms were analyzed using Geneious 9.1.8 (Biomatters, Auckland, New Zealand).

Table 2.
Microsatellite loci from Epichloë spp. with repeat motifs, primer sequences and size ranges used in four fluorescent labelled multiplexes (M1–M4).
Principal coordinate analysis (PCoA) was applied to microsatellite data to visualize proximity of isolates based on genetic distances using GenAlEx 6.5 [53].
2.4. DNA Sequencing and Phylogenetic Analyses
PCR amplification and sequencing was performed as previously described [31]. Amplification of the translation elongation factor 1-alpha gene (tefA) segment including introns 1–4 employed primer pair 5′-GGG TAA GGA CGA AAA GAC TCA-3′ and 5′-CGG CAG CGA TAA TCA GGA TAG-3′. Amplifications of the β-tubulin gene (tubB) segment including introns 1–3 employed primer pair 5′-TGG TCA ACC AGC TCA GCA CC-3′ and 5′-TGG TCA ACC AGC TCA GCA CC-3′ [46]. Reactions were performed in a final volume of 25 µL containing about 10 ng of genomic DNA in a standard touchdown cycle. Labelling reactions involved BigDye® Terminator v3.1 chemistry (Applied Biosystems™, Foster City, CA, USA) and sequences were obtained on a 3130xl Genetic Analyzer (Applied Biosystems™, Foster City, CA, USA). New sequences of this study have been submitted to GenBank (www.ncbi.nlm.nih.gov) and their accession numbers are indicated in bold in Table 1.
After manually editing, sequences were aligned with MUSCLE implemented in Geneious 9.1.8 (Biomatters, Auckland, New Zealand) including previously published sequences and selected reference sequences. Phylogenetic trees were inferred by maximum likelihood (ML) with likelihood settings of best-fit models selected by automated AICc model selection using PAUP* 4.0a [54]. The ML trees were generated in a heuristic search with gaps treated as missing information and random sequence additions. Bootstrap support values were estimated from 100 ML replications with random number seed and stepwise sequence addition.
2.5. Morphological Examinations
Colony growth was examined from cultures on potato dextrose agar (PDA; BD Comp., Sparks, MA, USA). Petri dishes were inoculated with 2 mm agar blocks, sealed and incubated at 24 °C in the dark. Colony diameter was measured after 21 days from three replicates per strain, and cultures were characterized and photographed. Microscopic observations of asci and ascospores, conidiogenous cells and conidia were made with an Olympus BX40 microscope (Olympus Corp., Tokyo, Japan) from stromata of herbarium specimens and from mycelium grown on PDA mounted in lactic acid. Measurements were taken with an ocular micrometer at 400× or 1000× using phase contrast optics. Of each fungal structure 20 measurements were taken and range with average in parenthesis is given. All specimens examined are deposited at the Fungarium of ETH Zürich (ZT).
3. Results
3.1. Microsatellite Data
Microsatellite data were obtained from 30 endophyte isolates from B. sylvaticum, 11 isolates from B. pinnatum and one isolate from B. phoenicoides (Table 1, Supplementary Materials File S1). Principal coordinate analysis (PCoA) of haplotypes based on 16 microsatellite loci grouped isolates in three main clusters (Figure 2). One cluster contained all but one asexual isolate from B. sylvaticum (Bsa), a second cluster included the majority of the sexual isolates from B. sylvaticum (Bss) and one asexual isolate (Bs 9301), and a third cluster included the remaining Bss isolates and isolates from B. pinnatum (Bp) and B. phoenicoides (Bph). In this cluster, Bss and Bp isolates tended to be separated from each other forming two subclusters. A separate PCoA made with only asexual isolates from B. sylvaticum revealed some subclustering, but clusters did not reflect geographic origin (Supplementary Materials File S2). Likewise, PCoA of isolates from B. pinnatum showed subclusters that in part were related to the site of origin (population) but not to the overall geography (Supplementary Materials File S3).
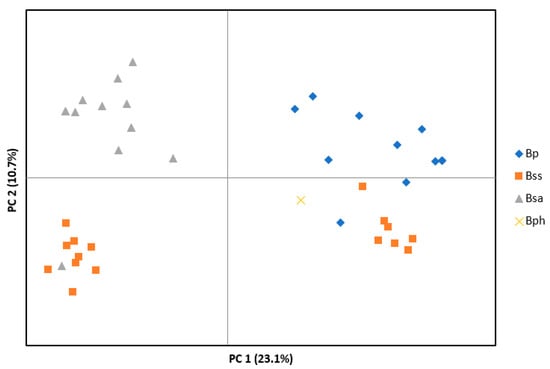
Figure 2.
Principal coordinate analysis (PCoA) based on 16 microsatellite loci of Epichloë isolates from Brachypodium hosts: B. sylvaticum (Bss, sexual isolates; Bsa, asexual isolates), B. pinnatum (Bp) and B. phoenicoides (Bph).
3.2. tubB Phylogeny
Isolates from B. sylvaticum (Bs), B. pinnatum (Bp) and B. phoenicoides (Bph) grouped within the E. typhina complex but did not form separate subclades according to different host plant species (Figure 3). For example, subclade I and IV of the tubB phylogeny included both Bp and Bs isolates. Subclade I comprised most Bs isolates together with two Bp isolates from Perroudaz, Switzerland and Basilicata, Italy. Subclade IV was placed at the base of the E. typhina main clade next to the isolates of E. poae and included the remaining Bp isolates together with a Bs isolate from Salamanca, Spain, and E. sylvatica ssp. pollinensis, a taxon that has been described from Hordelymus europaeus in Southern Italy. The isolate from B. phoenicoides (Bph) grouped in subclade III with E. poae, but without significant bootstrap support.

Figure 3.
Phylogenetic tree inferred from maximum likelihood (ML) analysis of partial tubB gene sequences including introns 1–3 of Epichloë isolates obtained from Brachypodium and Calamagrostis host grasses: B. sylvaticum (Bs), B. pinnatum (Bp), B. phoenicoides (Bph), C. arundinacea (Ca), C. purpurea (Cp) and C. villosa (Cv). Asexual isolates are indicated with filled squares. representative sequences of all other sexual Epichloë species or subspecies described to date are included. Dashed lines with Roman numerals denote distinct subclades. The tree is midpoint rooted at the left edge. Branch support values were estimated by 100 ML bootstrap replicates and are indicated if above 50%.
The tubB phylogeny placed the new isolates from Calamagrostis species in subclades of a main clade that encompassed five haploid Epichloë species described to date. The three isolates from C. arundinacea (Ca) had identical tubB sequences, which grouped them in a well-supported subclade with sequence relationship to E. stromatolonga, an Asian species described from Calamagrostis epigejos. The isolate from C. purpurea (Cp) and three isolates from C. villosa (Cv) grouped in a well-supported subclade that formed a polytomy with the E. baconii subclade and another subclade that included E. festucae Leuchtm., Schardl & M.R. Siegel, E. mollis (Morgan-Jones & W. Gams) Leuchtm. & Schardl, E. stromatolonga, and the Ca isolates.
3.3. tefA Phylogeny
In the tefA phylogeny all isolates from Brachypodium hosts grouped in a moderately supported clade in the E. typhina complex (Figure 4). As in the tubB tree, Bs and Bp isolates were not resolved by host species in the tefA tree. A moderately supported subclade (Ia) of the tefA tree encompassed all asexual Bs isolates together with some of the sexual Bs isolates. Otherwise, the clade exhibited no resolution based on host or geography.
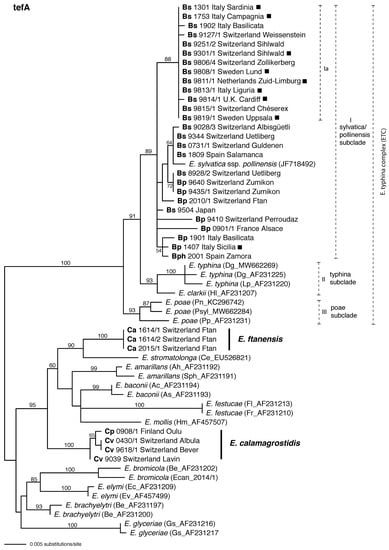
Figure 4.
Phylogenetic tree inferred from maximum likelihood (ML) analysis of partial tefA gene sequences including introns 1–4 of Epichloë isolates obtained from Brachypodium and Calamagrostis host grasses: B. sylvaticum (Bs), B. pinnatum (Bp), B. phoenicoides (Bph), C. arundinacea (Ca), C. purpurea (Cp) and C. villosa (Cv). Asexual isolates are indicated with filled squares. Included are representative sequences of all other sexual Epichloë species or subspecies described to date. Dashed lines with Roman numerals denote distinct subclades. The tree is midpoint rooted at the left edge. Branch support values were estimated by 100 ML bootstrap replicates and are indicated if above 50%.
The tefA phylogeny indicated the same relationships of isolates from Calamagrostis as did the tubB phylogeny. Considering only well-supported branches, the phylogenetic relationships to other members of that clade were consistent with the tubB phylogeny. One strongly supported clade included the three Ca isolates, and another strongly supported clade included the one Cp and three Cv isolates.
4. Discussion
Evolutionary diversification of parasites often depends on the association of infecting organisms with a particular host. Here, we examined the Epichloë endophytes found on different host grass species of genus Brachypodium and Calamagrostis. Following the concept of previously defined mating populations that were based on interfertility tests, B. sylvaticum is the host of a distinct species recognized as E. sylvatica, whereas B. pinnatum and B. phoenicoides is infected by the wide-host-range species E. typhina [21,39,41]. Microsatellite data obtained in the current study suggest that Brachypodium infecting Epichloë endophytes are closely related but are differentiated into four distinct clusters. Isolates from B. sylvaticum, referred here to E. sylvatica, form three clusters that differ in their mode of reproduction (one asexual, two sexual), while isolates from B. pinnatum and B. phoenicoides form a fourth cluster most closely related to one of the sexual E. sylvatica clusters (Figure 2). As inferred from DNA sequence analysis, the European isolates from Calamagrostis grouped into distinct clades within a larger clade that encompassed five previously described Epichloë species. Based on these relationships, we propose the new species E. calamagrostidis associated with C. purpurea and C. villosa, and Epichloë ftanensis associated with C. arundinacea.
A remarkable finding of this study was that asexual isolates from B. sylvaticum showed very little variation across distant locations in Europe, which spanned over several thousand kilometers (ca. 2500 km) and represented most of the latitudinal distribution range of the host. This is in stark contrast to another woodland grass, H. europaeus, that was found to be infected by six different endophyte taxa along a longitudinal transect across Europe (Oberhofer and Leuchtmann 2012). This finding may be explained by the pattern of recolonization of land masses after the last glaciation [55] involving infected plants of B. sylvaticum that contained only one endophyte lineage. Rapid spread of B. sylvaticum plants with its seed transmitted endophyte is promoted by the awned seeds of the species which can easily attach to the fur of animals. Sexual isolates of E. sylvatica were genetically more variable and were often distinct from the asexual isolates, suggesting that for the sexual lineages either multiple colonization events have occurred or infections have been acquired later from long distance dispersal of ascospores. The genetic distinctness of sexual and asexual isolates of E. sylvatica has been previously reported in populations from Switzerland based on allozyme data [10]. The current study with a European wide sample making use of microsatellite data confirms this finding.
Sequence data of tubB and tefA genes were not concordant with microsatellite data and did not always resolve E. sylvatica isolates with different modes of reproduction in separate clades. Sexual and asexual isolates were included in the same clade or even had identical sequences, although some substructuring was evident. This may suggest that genetic differentiation of the asexual, clonally propagated isolates occurred relatively recently and that sexual and asexual lineages share a common origin.
Without exception all populations of B. sylvaticum that have been examined so far in Europe were endophyte infected, usually with an incidence of 100%. In light of the expectation that seed transmission is not perfect and at least a low percentage of uninfected seeds is produced [56]. This observation suggests that the symbiosis of this host with seed-transmitted E. sylvatica provides a selective advantage. However, it is still unclear what the benefits for the host of endophyte infection are, unlike in many other endophyte associations. For example, E. festucae and E. coenophiala (Morgan-Jones & W. Gams) C.W. Bacon & Schardl are known to produce a variety of alkaloids with distinct antiherbivore properties [57,58]. Previous analysis of E. sylvatica-infected B. sylvaticum plants indicated an absence of peramine (a pyrrolopyrazine), known lolines (aminopyrrolizidines) and ergopeptines (ergot alkaloids) [59]. Genome sequence analysis indicated no functional genes for ergot alkaloids, aminopyrrolizidines or indole-diterpenes, and a ppzA allele lacking the R domain for peramine (C.L. Schardl, unpublished data). However, ppzA genes likely determine biosynthesis of related pyrrolopyrazines, as recently described [60]. In an in vitro study using the model herbivore Spodoptera frugiperda, uninfected B. sylvaticum leaves were preferred over infected leaves suggesting that increased resistance to herbivores based on unknown factors could indeed play a role [61]. Such factors may include one or more pyrrolopyrazines that are products of the ppzA gene, compounds determined by other secondary metabolite gene clusters [62] or even an anti-insect protein such as is encoded by the mcf-like gene [63].
Additional endophyte benefits that could be involved and are documented for other endophyte-host systems include improved seedling establishment and growth promotion, which can improve competitive abilities of infected plants [64,65]. However, experiments testing competitive abilities of B. sylvaticum plants did not confirm this hypothesis, but rather endophyte infection had a negative effect on plant growth [66].
Another potential benefit of the endophyte is enhanced seed production or survival. It is worth noting that the Epichloë species (including E. sylvatica) apparently possess the fhb7 gene that was acquired by a wheat wild relative and encodes an enzyme for detoxification of trichothecenes which are virulence factors of the wheat head blight pathogen, Fusarium graminearum [67]. This raises the possibility that the endophyte may provide protection from such a pathogen, which prevents reduced seed yield.
Sequence data were used to infer the phylogenetic position of Brachypodium endophytes within the E. typhina complex. In the tubB phylogeny isolates from these hosts were distributed among two subclades, subclade sylvatica (I) and subclade pollinensis (IV) confirming previous findings [6]. The exception was placement of the isolate from B. phoenicoides in subclade poae (III), but without significant bootstrap support. Inspection of the alignment (Supplementary Materials File S4) suggested a possible intragenic recombination within tubB occurred between Brachypodium- and Poa-associated lineages. In the tefA tree, however, all isolates from Brachypodium hosts were in the same clade (I) and the subclade that included ssp. pollinensis was nested within that clade, suggesting that isolates from Brachypodium hosts share the same gene pool. The apparent paraphyly of Brachypodium-associated Epichloë in the tubB tree may suggest complicated origins of the Epichloë lineages occurring in symbiosis with Brachypodium species, or it may be a lineage sorting effect. Moreover, isolates from B. sylvaticum and B. pinnatum were not strictly grouped in separate subclades, indicating that endophytes of the two hosts may not be host specific, but rather can move between hosts. For example, sequences of an isolate from B. sylvaticum (8928/2) were identical with two isolates from B. pinnatum (9640/1, 9435/1) in the tefA phylogeny, but not in the tubB phylogeny.
Isolates found on Calamagrostis hosts formed two distinct clades in the Epichloë phylogeny, part of a larger clade that included E. amarillans, E. baconii, E. festucae and E. mollis, and one next to E. stromatolonga (Figure 3 and Figure 4). Epichloë stromatolonga is an asexual, stroma-forming species that has been described on C. epigejos from a single location in China [47]. Our isolates from C. arundinacea (Ca) were relatively close to this species based on tubB and tefA sequences and thus placed in the same subclade with 100% or 90% bootstrap support, respectively. However, given the distinct host relation and occurrence on different continents, we suggest that this endophyte represents an independent phylogenetic lineage that may have evolved in Europe. This endophyte should therefore be recognized as a distinct species, for which we propose the new name E. ftanensis. Mating tests have not been performed, but phylogenetic distance to other mating populations (MP) suggest that the new species (perhaps together with E. stromatolonga) represents a distinct MP. The new species has so far been found only at one site on C. arundinacea located in a remote valley of the Central Alps of Switzerland. More intensive sampling of C. arundinacea in a wider area would be necessary to assess its actual distribution.
The second clade with Calamagrostis-derived isolates contained isolates from C. villosa and C. purpurea. Isolates from C. villosa have been previously assigned to E. baconii based on their sexual compatibility with this species that otherwise infects Agrostis spp. [21]. Incidentally, isolates from C. villosa were genetically distinct both in allozyme patterns and sequences of three nuclear genes act1, tubB and tefA [46]. Our phylogenetic analyses of additional isolates from the same and from a second Calamagrostis host placed these isolates in a well resolved clade distinct from E. baconii (Figure 1 and Figure 2). The observed interfertility between E. baconii and Calamagrostis-derived isolates in mating tests poses a conflict between biological and phylogenetic species concepts. However, successful outcrossing observed experimentally does not necessarily mean that fertile mating between species occurs in nature. Agrostis and Calamagrostis hosts occupy different niches in grass- or woodlands and may not grow close enough for mutual gamete transfer by Botanophila flies. If rare mating still would occur, progeny may be lost due to lack of a compatible host because the two species are expected to be host adapted, as has been observed in other interfertile Epichloë species [68]. Strains from C. purpurea have not been tested and it is not known whether they are interfertile with E. baconii. The genetic distinctness of isolates from the Calamagrostis hosts as shown previously and in the current study suggests that there is no gene flow between E. baconii and E. calamagrostidis and that the Calamagrostis-associated strains represent a reproductively isolated phylogenetic species, despite not yet fully establishing sexual barriers.
5. Taxonomy
Epichloe calamagrostidis Leuchtm. & Schardl, sp. nov., Figure 5
MycoBank MB846049
Colonies on PDA white to tan, in the center cottony and raised, towards the margin flattened with sparse aerial mycelium, margin even or slightly wavy, reverse of colony distinctly brownish, moderate growing, attaining 23–26 mm diameter in 21 days at 24 °C. Sporulation in culture sparse to moderate. Conidiogenous cells phialidic, arising perpendicularly from hyphae, hyaline, 10–22 µm long, 1–2 µm wide at base and gradually at-tenuating to approximately 0.5 µm at the tip, with or without a basal septum. Conidia ellipsoidal to reniform, hyaline, aseptate, 3.3–4.9 (3.9) × 1.6–2.5 (2.0) µm. Stromata on flowering culms enclosing undeveloped inflorescence, leaves and sheath of flag leaf, cylindrical, 18–35 mm long and 2–3 mm wide, in early stage white and covered with a dense layer of conidiogenous cells producing conidia 4.0–5.6 × 2.0–3.0 µm in size. Upon fertilization stromata become orange to brownish with perithecia forming on top of the conidial stroma. Perithecia pyriform, approximately 140–160 µm wide, 240–280 µm high, light orange, 40–45 per mm2. Asci cylindrical with a short tapering stalk, bearing a distinct hemispherical apical cap with central pore, 200–280 × 4.8–7.2 µm, containing eight ascospores. Ascospores hyaline, filiform, 225–290 × 1.5–1.7 µm, non-septate within ascus, becoming multiseptated at full maturity, not disarticulating. Endophyte in infected flowering tillers not seed-transmitted.
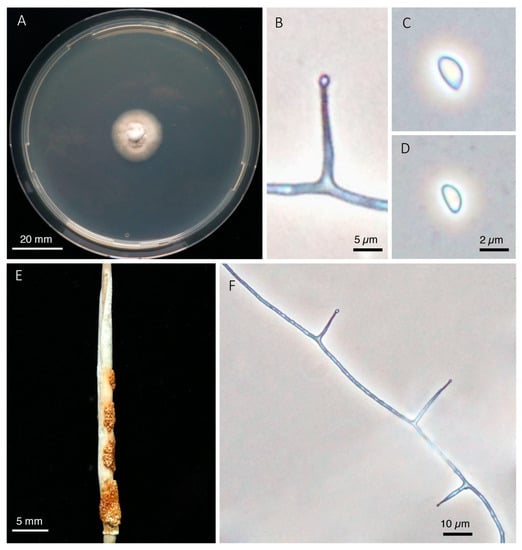
Figure 5.
Epichloë calamagrostidis. (A). Colony grown on PDA for 21 days at 24 °C. (B). Conidiogenous cell with emerging conidium. (C,D). Conidia. (E). Stroma with mature perithecia on C. varia (coll. H. Seitter). (F). Sporulating hypha in culture.
Etymology: Referring to the Host Genus Calamagrostis.
Holotype: SWITZERLAND, Ct. Grisons, Lavin, stromata on Calamagrostis villosa, 8 September 1990, leg. A. Leuchtmann (ZT Myc 99902); ex type cultures AL9039, ATCC 200745; GenBank accession nos. L78270 (tubB), AF231196 (tefA).
Additional specimens examined: SWITZERLAND, Ct. Grisons, Lavin, stromata on Calamagrostis villosa, 8 September 1990, leg. A. Leuchtmann, culture AL9040; Ct. Grisons, Bever, stromata on Calamagrostis villosa, 24 July 1996, 5 August 2022, leg. A. Leuchtmann, cultures AL9618, AL2016, CBS 147678; Ct. Grisons, Bregaglia, Roticcio, stromata on Calamagrostis villosa, 7 July 1997, leg. G. Meijer; Ct. Grisons, Bregaglia, Bondo, stromata on Calamagrostis villosa, 4 August 2022, leg. A. Leuchtmann; Ct. Obwalden, Lungern, Schönbüel, stromata on Calamagrostis varia, 2 August 1991, leg. A. Leuchtmann; Ct. Bern, Hasliberg, Wasserwendi, stromata on Calamagrostis varia, 6 August 1918, leg A. Thellung. LICHTENSTEIN: Triesen, Tuasswand, stromata on Calamagrostis varia, 6 June 1974, leg. H. Seitter. FINLAND, Oulu, Paltamo, stromata on Calamagrostis purpurea, July 2007, leg. P. Wäli, culture AL0908; Paltamo, Melalahti, on Calamagrostis purpurea, 30 August 1960, leg. T. Ulvinen.
Hosts: Calamagrostis purpurea, C. varia, C. villosa.
Known distribution: Switzerland, Finland.
Comments: Besides its genetic distinctness this species is characterized by relatively short stromata no longer than 35 mm, and short ascospores that do not disarticulate. Ascospores of the related E. baconii break into several part-spores while still within the ascus. We therefore consider Epichloë isolates from C. villosa and C. purpurea, and most likely also from C. varia that could not be sequenced, as distinct species and propose the name E. calamagrostidis, sp. nov. Epichloë calamagrostidis can form symptomless infections in Calamagrostis spp., because host grasses often do not flower (AL, personal observation). However, if infected plants occasionally form inflorescences instead of stromata the endophyte does not invade seeds, thus qualifying this association as type I [9].
An interesting observation was that stromata in populations of C. villosa and C. varia often remained unfertilized or, if fertilized, the perithecia were barren. Similar observations have been made for E. brachyelytri Schardl & Leuchtm. that rarely formed perithecia with ascospores [22], and in two other species, E. stromatolonga and E. scottii T. Thünen, Y. Becker, M.P. Cox & S. Ashrafi, fertilized stromata have never been observed [18,47]. A possible explanation for these observations is that endophytes at a particular site exist as clones that represent only one mating type, which prevents successful mating. Barren perithecia can be the result of interspecific matings with spermatia from other Epichloë species that may have been present in the area. Host grasses of C. villosa and C. varia do in fact propagate vegetatively through subterranean stolons resulting in large clonal patches. Alternatively, there may be no Botanophila flies present at these sites restricting efficient gamete transfer [69]. However, at least at the Bondo site where fertilized stromata on C. villosa occurred, signs of fly visitation (egg shells and feeding damage) on the stromata were observed suggesting that the first mentioned hypothesis is more likely.
Epichloe ftanensis Leuchtm. & A.D. Treindl, sp. nov., Figure 6
MycoBank MB846050
Colonies on PDA pure white, cottony, moderately raised, margin even or slightly frayed, reverse of colony light brown, moderate growing, attaining 26–32 mm diameter in 21 day at 24 °C. Sporulation in culture very sparse. Conidiogenous cells phialidic, arising perpendicularly from hyphae, hyaline, 16–42 µm long, 1.6–2 µm wide at base and gradually attenuating to approximately 0.5 µm at the tip, basal septum lacking. Conidia ellipsoidal to reniform, hyaline, aseptate, 3.3–4.8 (4.0) × 1.9–2.6 (2.3) µm. Stromata on flowering tillers enclosing undeveloped inflorescence, leaves and sheath of flag leaf, cylindrical, 19–45 mm long and 2.5–3 mm wide, in early stage white and covered with a dense layer of conidiogenous cells producing conidia 4.0–5.6 × 2.0–0–3.6 µm in size. Fertilized, mature stromata turn orange, forming perithecia on top of the conidial stroma, 12–16 per mm2. Individual perithecia pyriform, approximately 220 µm wide, 440 µm high, wall and neck light orange, embedded in a whitish tissue of globular cells. Asci cylindrical with a short tapering stalk, bearing a distinct hemispherical apical cap that may be flattened at maturity, 250–475 × 5.5–7 µm, containing eight ascospores. Ascospores hyaline, filiform 300–460 (380) × 1.5–1.6 (1.5) µm, with up to 20 septa at maturity, not disarticulating. Endophyte in infected flowering tillers not seed-transmitted.
Etymology: Referring to the Collection Site Near Village Ftan.
Holotype: SWITZERLAND, Ct. Grisons, Ftan, stromata on Calamagrostis arundinacea, 23 August 2020, leg. A. Leuchtmann & A.D. Treindl (ZT Myc 66903); ex type cultures AL2015, CBS 147676; GenBank accession nos. MW283354 (tubB), MW283391 (tefA).
Additional specimens examined: SWITZERLAND, Ct. Grisons, Ftan, stromata on Calamagrostis arundinacea, 5 September 2016, leg. A. Leuchtmann, cultures AL1614/1, AL1614/2.
Host: Calamagrostis arundinacea.
Known distribution: Switzerland.
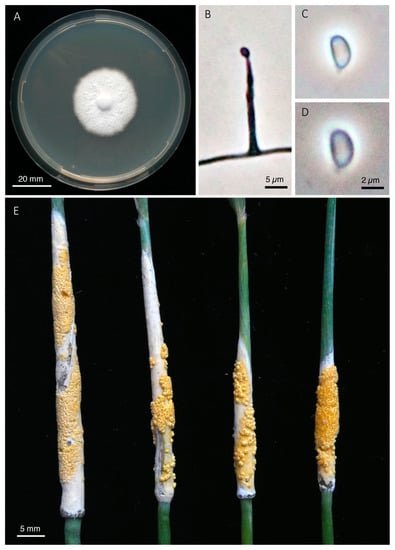
Figure 6.
Epichloë ftanensis. (A) Colony grown on PDA for 21 days at 24 °C. (B) Conidiogenous cell with emerging conidium. (C–D) Conidia. (E) Stromata with mature perithecia (holotype).
Comments: Distinctive morphological features of this taxon are the very long ascospores of up to 460 µm in length, and the size of the perithecia that are larger than in most other species. Only in E. elymi, a species native to North America, ascospores may occasionally be longer. Fertilization of stromata at the observed site was variable with perithecia covering 0% to 80% of the stroma surface. About half of the stromata showed signs of Botanophila fly visitations (egg shells or larvae) suggesting that flies take an active role in fertilization and outcrossing of this taxon [70]. Our isolates are genetically close to the earlier described species E. stromatolonga based on tubA and tefB sequences. However, collections of E. stromatolonga were reported to have stromata that always remain unfertilized and lack perithecia, thus the species is effectively asexual [47]. Furthermore, this taxon differs by its exceptionally long stromata (up 186 mm), by infecting a different host grass (C. epigejos) and by its geographic origin in China. Therefore, we treat our sexually reproducing specimens from C. arundinacea as distinct species and propose the name E. ftanensis, sp. nov.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/jof8101086/s1, Supplementary Materials File S1: Microsatellite alleles of Epichloë isolates at 16 loci; Supplementary Materials File S2: Principal Coordinates (PCoA) of asexual isolates from B. sylvaticum; Supplementary Materials File S3: Principal Coordinates (PCoA) of isolates from B. pinnatum and B. phoenicoides; Supplementary Materials File S4: Sequence alignement of tubB from B. phoenicoides (Bph) and selected ETC isolates.
Author Contributions
Conceptualization and formal analysis, A.L. and C.L.S.; writing—original draft preparation, A.L.; writing—review and editing, A.L. and C.L.S.; funding acquisition, A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Swiss National Science Foundation (SNF), Grant Number 31003A_138479 and 31003A_169269.
Data Availability Statement
DNA sequences are available at GenBank (www.ncbi.nlm.nih.gov).
Acknowledgments
Stuart Davies, Lena Jonsell, Dorothea Schmidt, Björn Ursing, Piipa Wääli and J.H.Willems provided plant or seed samples. We thank Artemis Treindl for valuable comments on the draft manuscript, Simon Crameri for statistical advise and Bea Arnold for laboratory assistance. We acknowledge the Genetic Diversity Centre of ETH Zurich (GDC), where microsatellite data and sequence data were generated.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Giraud, T.; Refrégier, G.; Le Gac, M.; de Vienne, D.M.; Hood, M.E. Speciation in fungi. Fungal Genet. Biol. 2008, 45, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Restrepo, S.; Tabima, J.F.; Mideros, M.F.; Grünwald, N.J.; Matute, D.R. Speciation in fungal and oomycete plant pathogens. Annu. Rev. Phytopathol. 2014, 52, 289–316. [Google Scholar] [CrossRef] [PubMed]
- Stukenbrock, E.H.; McDonald, B.A. The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 2008, 46, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Giraud, T.; Gladieux, P.; Gavrilets, S. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol. Evol. 2010, 25, 387–395. [Google Scholar] [CrossRef]
- Schardl, C.L. The epichloae, symbionts of the grass subfamily Poöideae. Ann. Missouri Bot. Gard. 2010, 97, 646–665. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Bacon, C.W.; Schardl, C.L.; White, J.F., Jr.; Tadych, M. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 2014, 106, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.G.; Nagabhyru, P.; Schardl, C.L. Epichloë festucae endophytic growth in florets, seeds, and seedlings of perennial ryegrass (Lolium perenne). Mycologia 2017, 109, 691–700. [Google Scholar] [CrossRef]
- Zhang, W.; Card, S.D.; Mace, W.J.; Christensen, M.J.; McGill, C.R.; Matthew, C. Defining the pathways of symbiotic Epichloë colonization in grass embryos with confocal microscopy. Mycologia 2017, 109, 153–161. [Google Scholar] [CrossRef]
- White, J.F., Jr. Endophyte-host associations in forage grasses. XI. A proposal concerning origin and evolution. Mycologia 1988, 80, 442–446. [Google Scholar] [CrossRef]
- Bucheli, E.; Leuchtmann, A. Evidence for genetic differentiation between choke-inducing and asymptomatic strains of the Epichloë grass endophyte from Brachypodium sylvaticum. Evolution 1996, 50, 1879–1887. [Google Scholar]
- Meijer, G.; Leuchtmann, A. The effects of genetic and environmental factors on disease expression (stroma formation) and plant growth in Brachypodium sylvaticum infected by Epichloë sylvatica. Oikos 2000, 91, 446–458. [Google Scholar] [CrossRef]
- Meijer, G.; Leuchtmann, A. Fungal genotype controls mutualism and sex in Brachypodium sylvaticum infected by Epichloë sylvatica. Acta Biol. Hung. 2001, 52, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Clarke, B.B.; Funk, C.R. Effect of fertilizer and fungicide applications on choke expression and endophyte transmission in chewings fescue. In Proceedings of the International Symposium on Acremonium/Grass Interactions, Baton Rouge, LA, USA, 3 November 1990; pp. 62–66. [Google Scholar]
- Funk, C.R.; Belanger, F.C.; Murphy, J.A. Role of endophytes in grasses used for turf and soil conservation. In Biotechnology of Endophytic Fungi of Grasses; Bacon, C.W., White, W.F., Jr., Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 201–209. [Google Scholar]
- Campbell, M.A.; Tapper, B.A.; Simpson, W.R.; Johnson, R.D.; Mace, W.; Ram, A.; Lukito, Y.; Dupont, P.Y.; Johnson, L.J.; Scott, D.B.; et al. Epichloë hybrida, sp nov., an emerging model system for investigating fungal allopolyploidy. Mycologia 2017, 109, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Shymanovich, T.; Charlton, N.D.; Musso, A.M.; Scheerer, J.; Cech, N.B.; Faeth, S.H.; Young, C.A. Interspecific and intraspecific hybrid Epichloë species symbiotic with the North American native grass Poa alsodes. Mycologia 2017, 109, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Leuchtmann, A.; Young, C.A.; Stewart, A.V.; Simpson, W.R.; Hume, D.E.; Scott, B. Epichloe novae-zelandiae, a new endophyte from the endemic New Zealand grass Poa matthewsii. N. Z. J. Bot. 2019, 57, 271–288. [Google Scholar] [CrossRef]
- Thünen, T.; Becker, Y.; Cox, M.P.; Ashrafi, S. Epichloë scottii sp. nov., a new endophyte isolated from Melica uniflora is the missing ancestor of Epichloë disjuncta. IMA Fungus 2022, 13, 2. [Google Scholar] [CrossRef]
- Tian, P.; Xu, W.B.; Li, C.J.; Song, H.; Wang, M.N.; Schardl, C.L.; Nan, Z.B. Phylogenetic relationship and taxonomy of a hybrid Epichloë species symbiotic with Festuca sinensis. Mycol. Prog. 2020, 19, 1069–1081. [Google Scholar] [CrossRef]
- Moon, C.D.; Craven, K.D.; Leuchtmann, A.; Clement, S.L.; Schardl, C.L. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Molec. Ecol. 2004, 13, 1455–1467. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Schardl, C.L. Mating compatibility and phylogenetic relationships among two new species of Epichloë and other congeneric European species. Mycol. Res. 1998, 102, 1169–1182. [Google Scholar] [CrossRef]
- Schardl, C.L.; Leuchtmann, A. Three new species of Epichloë symbiotic with North American grasses. Mycologia 1999, 91, 95–107. [Google Scholar] [CrossRef]
- White, J.F., Jr. Endophyte-host associations in grasses. XIX. A systematic study of some sympatric species of Epichloë in England. Mycologia 1993, 85, 444–455. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Schardl, C.L.; Siegel, M.R. Sexual compatibility and taxonomy of a new species of Epichloë symbiotic with fine fescue. Mycologia 1994, 86, 802–812. [Google Scholar] [CrossRef]
- Treindl, A.D.; Leuchtmann, A. Assortative mating in sympatric ascomycete fungi revealed by experimental fertilizations. Fungal Biol. 2019, 123, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Schirrmann, M.K.; Zoller, S.; Fior, S.; Leuchtmann, A. Genetic evidence for reproductive isolation among sympatric Epichloë endophytes as inferred from newly developed microsatellite markers. Microb. Ecol. 2015, 70, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.J.; Leuchtmann, A.; Rowan, D.D.; Tapper, B.A. Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (Festuca pratensis), and perennial rye-grass (Lolium perenne). Mycol. Res. 1993, 97, 1083–1092. [Google Scholar] [CrossRef]
- van Zijll de Jong, E.; Dobrowolski, M.P.; Bannan, N.R.; Stewart, A.V.; Smith, K.F.; Spangenberg, G.C.; Forster, J.W. Global genetic diversity of the perennial ryegrass fungal endophyte Neotyphodium lolii. Crop Sci. 2008, 48, 1487–1501. [Google Scholar] [CrossRef]
- Ekanayake, P.N.; Rabinovich, M.; Guthridge, K.M.; Spangenberg, G.C.; Forster, J.W.; Sawbridge, T.I. Phylogenomics of fescue grass-derived fungal endophytes based on selected nuclear genes and the mitochondrial gene complement. BMC Evol. Biol. 2013, 13, 270. [Google Scholar] [CrossRef]
- Clay, K.; Leuchtmann, A. Infection of woodland grasses by fungal endophytes. Mycologia 1989, 81, 805–811. [Google Scholar] [CrossRef]
- Oberhofer, M.; Leuchtmann, A. Genetic diversity in epichloid endophytes of Hordelymus europaeus suggests repeated host jumps and interspecific hybridizations. Molec. Ecol. 2012, 21, 2713–2726. [Google Scholar] [CrossRef]
- Afkhami, M.E. Fungal endophyte-grass symbioses are rare in the California floristic province and other regions with Mediterranean-influenced climates. Fungal Ecol. 2012, 5, 345–352. [Google Scholar] [CrossRef]
- Song, H.; Li, X.; Bao, G.; Song, Q.; Tian, P.; Nan, Z. Phylogeny of Neotyphodium endophyte from western Chinese Elymus species based on act sequences. Acta Microbiol. Sin. 2015, 55, 273–281. [Google Scholar]
- Davies, M.S.; Long, G.L. Performance of two contrasting morphs of Brachypodium sylvaticum transplanted into shaded and unshaded sites. J. Ecol. 1991, 79, 505–517. [Google Scholar] [CrossRef]
- Schippmann, U. Revision der europäischen Arten der Gattung Brachypodium Palisot de Beauvois (Poaceae); Conservatoire et jardin botaniques de Genève: Pregny-Chambesy, Switzerland, 1991; pp. 1–250. [Google Scholar]
- Eckblad, F.; Torkelsen, A. Epichloë typhina in Norway. Opera Bot. 1989, 100, 51–57. [Google Scholar]
- Raynal, G. Présence dans les graminées spontanées et prairiales de champignons endophytes du genre Acremonium. In Proceedings of the ANPP—Quatrième Conférence Internationale sur les Maladies des Plantes, Bordeaux, France, 6–8 December 1994; pp. 1125–1130. [Google Scholar]
- Enomoto, T.; Tsukiboshi, M.; Shimanuki, T. The gramineous plants in which Neotyphodium endophyes were found. J. Weed Sci. Technol. 1998, 43, 76–77. (In Japanese) [Google Scholar]
- Zabalgogeazcoa, I.; Vázquez de Aldana, B.R.; García Criado, B.; Gracía Ciudad, A. Fungal endophytes in natural grasslands of western Spain. In Book of Abstracts, Proceedings of the Grassland Conference, 4th International Neotyphodium/Grass Interactions Symposium, Soest, Germany, 27–29 September 2000; University of Paderborn: Soest, Germany, 2000; p. 126. [Google Scholar]
- Meijer, G.; Leuchtmann, A. Multistrain infections of the grass Brachypodium sylvaticum by its fungal endophyte Epichloë sylvatica. New Phytol. 1999, 141, 355–368. [Google Scholar] [CrossRef]
- Zabalgogeazcoa, I.; García Ciudad, G.; Leuchtmann, A.; Vázques de Aldana, B.R.; García Criado, B. Effects of choke disease in the grass Brachypodium phoenicoides. Plant Pathol. 2008, 57, 467–472. [Google Scholar] [CrossRef]
- Hess, H.E.; Landolt, E.; Hirzel, R. Flora der Schweiz und angrenzender Gebiete, 2nd ed.; Birkäuser: Basel, Switzerland, 1976; Volume 1, pp. 1–858. [Google Scholar]
- Iannone, L.J.; Novas, M.V.; Young, C.A.; De Battista, J.P.; Schardl, C.L. Endophytes of native grasses from South America: Biodiversity and ecology. Fungal Ecol. 2012, 5, 357–363. [Google Scholar] [CrossRef]
- White, J.F., Jr.; Halisky, P.M.; Sun, S.; Morgan-Jones, G.; Funk, C.R., Jr. Endophyte-host associations in grasses. XVI. Patterns of endophyte distribution in species of the tribe Agrostideae. Am. J. Bot. 1992, 79, 472–477. [Google Scholar] [CrossRef]
- White, J.F., Jr. Endophyte-host associations in grasses. XX. Structural and reproductive studies of Epichloë amarillans sp. nov. and comparisons to E. typhina. Mycologia 1994, 86, 571–580. [Google Scholar] [CrossRef]
- Craven, K.D.; Hsiau, P.T.W.; Leuchtmann, A.; Hollin, W.; Schardl, C.L. Multigene phylogeny of Epichloë species, fungal symbionts of grasses. Ann. Mo. Bot. Gard. 2001, 88, 14–34. [Google Scholar] [CrossRef]
- Ji, Y.L.; Zhan, L.H.; Kang, Y.; Sun, X.H.; Yu, H.S.; Wang, Z.W. A new stromata-producing Neotyphodium species symbiotic with clonal grass Calamagrostis epigeios (L.) Roth. grown in China. Mycologia 2009, 101, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Zuloaga, F.O.; Judziewicz, E.J.; Filgueiras, T.S.; Davis, J.I.; Morrone, O. A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 2015, 53, 117–137. [Google Scholar] [CrossRef]
- Roy, B.A.; Coulson, T.; Blaser, W.; Policha, T.; Stewart, J.L.; Blaisdell, G.K.; Güsewell, S. Population regulation by enemies of the grass Brachypodium sylvaticum: Demography in native and invaded ranges. Ecology 2011, 92, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Väre, H.; Itämies, J. Phorbia phrenione (Séguy) (Diptera: Anthomyiidae) in Finland. Sahlbergia 1995, 2, 119–124. [Google Scholar]
- Leuchtmann, A. Isozyme relationships of Acremonium endophytes from twelve Festuca species. Mycol. Res. 1994, 98, 25–33. [Google Scholar] [CrossRef]
- White, J.F., Jr.; Cole, G.T. Endophyte-host assiciations in forage grasses. II. Taxonomic observations on the endophyte of Fesutca arundinacea. Mycologia 1985, 77, 483–486. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony and Other Methods; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Afkhami, M.E.; Rudgers, J.A. Symbiosis lost: Imperfect vertical transmission of fungal endophytes in grasses. Am. Nat. 2008, 172, 405–416. [Google Scholar] [CrossRef]
- Siegel, M.R.; Bush, L.P. Defensive chemicals in grass-fungal endophyte associations. Recent Adv. Phytochem. 1996, 30, 81–119. [Google Scholar]
- Schardl, C.L.; Florea, S.; Pan, J.; Nagabhyru, P.; Bec, S.; Calie, P.J. The epichloae: Alkaloid diversity and roles in symbiosis with grasses. Curr. Opin. Plant Biol. 2013, 16, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Leuchtmann, A.; Schmidt, D.; Bush, L.P. Different levels of protective alkaloids in grasses with stroma-forming and seed-transmitted Epichloë/Neotyphodium endophytes. J. Chem. Ecol. 2000, 26, 1025–1036. [Google Scholar] [CrossRef]
- Berry, D.; Mace, W.; Grage, K.; Wesche, F.; Gore, S.; Schardl, C.L.; Young, C.A.; Dijkwel, P.P.; Leuchtmann, A.; Bode, H.B.; et al. Efficient nonenzymatic cyclization and domain shuffling drive pyrrolopyrazine diversity from truncated variants of a fungal NRPS. Proc. Natl. Acad. Sci. USA 2019, 116, 25614–25623. [Google Scholar] [CrossRef] [PubMed]
- Brem, D.; Leuchtmann, A. Epichloë grass endophytes increase herbivore resistance in the woodland grass Brachypodium sylvaticum. Oecologia 2001, 126, 522–530. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef]
- Ambrose, K.V.; Koppenhoefer, A.M.; Belanger, F.C. Horizontal gene transfer of a bacterial insect toxin gene into the Epichloë fungal symbionts of grasses. Sci. Rep. 2014, 4, 5562. [Google Scholar] [CrossRef]
- Clay, K. Effects of fungal endophytes on the seed and seedling biology of Lolium perenne and Festuca arundinacea. Oecologia 1987, 73, 358–362. [Google Scholar] [CrossRef]
- Clay, K. The impact of parasitic and mutualistic fungi on competitive interactions among plants. In Perspectives on Plant Competition; Grace, J.B., Tilman, D., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 391–412. [Google Scholar]
- Brem, D.; Leuchtmann, A. Intraspecific competition of endophyte infected vs uninfected plants of two woodland grass species. Oikos 2002, 96, 281–290. [Google Scholar] [CrossRef]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef]
- Schirrmann, M.K.; Leuchtmann, A. The role of host-specificity in the reproductive isolation of Epichloë endophytes revealed by reciprocal infections. Fungal Ecol. 2015, 15, 29–38. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Michelsen, V. Biology and evolution of the Epichloë-associated Botanophila species found in Europe (Diptera: Anthomyiidae). Insect Syst. Evol. 2016, 47, 1–14. [Google Scholar] [CrossRef]
- Bultman, T.L.; Leuchtmann, A. Biology of the Epichloë-Botanophila interaction: An intriguing association between fungi and insects. Fungal Biol. Rev. 2008, 22, 131–138. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).