Abstract
In the survey of mycobiota of mudflats in China, two new sexually reproducing Talaromyces sect. Talaromyces species were discovered and studied using a polyphasic approach. These species are named here Talaromyces haitouensis (ex-type AS3.160101T) and Talaromyces zhenhaiensis (ex-type AS3.16102T). Morphologically, T. haitouensis is distinguished by moderate growth, green-yellow gymnothecia, orange-brown mycelium, and echinulate ellipsoidal ascospores. T. zhenhaiensis is characterized by fast growth, absence of sporulation, cream yellow to naphthalene yellow gymnothecia and mycelium, and smooth-walled ellipsoidal ascospores with one equatorial ridge. The two novelties are further confirmed by phylogenetic analyses based on either individual sequences of BenA, CaM, Rpb2, and ITS1-5.8S-ITS2 or the concatenated BenA-CaM-Rpb2 sequences.
1. Introduction
The estimated number of global fungal species may be in a range of 2.2 to 3.8 million, whereas the number of described species only accounts for 3–8%, which is about 120,000 [1]. The less-explored environments, for instance the mudflat areas along coastlines, are thought to be the new habitats for discovery of novel fungal taxa [2,3]. With the application of molecular techniques in the studies on the mycobiota of coastal environments, the taxonomic panorama of marine-derived fungi (facultative marine fungi) is gradually emerging. Surprisingly, the predominant fungal genera from marine environments are those that have long been considered as ubiquitously terrestrial, such as Aspergillus, Penicillium, and Talaromyces [2,4], whose species are considered as extraordinary producers of bioactive secondary metabolites, such as alkaloids, lactones, polyketides, etc., some of which may have great pharmaceutical prospects and many of which are new compounds [5,6].
Compared with Aspergillus and Penicillium, the species of Talaromyces are relatively inadequately discovered in marine environments. For example, in a list of marine fungi, there are 47 species of Aspergillus, 39 species of Penicillium, and only 6 of Talaromyces [7]. However, Talaromyces is one of the most valuable reservoirs either for species diversity or for bioactive compounds (e.g., [5,6,8]). China has the vast tidal flat areas covering ca. 2 × 104 km2, which may possess various ecological habitats harboring new taxa of Talaromyces as well as the consequent bioactive compounds they produce [9].
Originally, the genus Talaromyces C. R. Benjam. was established to accommodate sexual species producing gymnothecial ascocarps [10], and these species had long been studied in the taxonomic schemes of Penicillium (e.g., [11,12]). However, the morphological, ecological, and molecular characteristics have implied the common intrinsic properties of the species in Talaromyces and those in subgen. Biverticillium Diercks of Pitt from those in subgen. Aspergilloides, Furcatum, and Penicillium, thus Talaromyces was regarded as the valid genus for these species irrespective of teleomorphs or anamorphs (e.g., [13,14,15,16,17,18,19]). In the study by Samson et al. [19], 71 species were listed in the genus Talaromyces. Houbraken et al. [20] introduced the genus Rasamsonia to accommodate thermotolerant and thermophilic Talaromyces species. Yilmaz et al. [21] recognized 88 species and divided Talaromyces into 7 sections, namely, sections Talaromyces, Helici, Purpurei, Trachyspermi, Bacillispori, Subinflati, and Islandici, and Sun et al. [22] subsequently described the new section Tenues in the genus. Up to 2021, about 175 species have been reported in Talaromyces, and as the largest section of the genus, sect. Talaromyces encompassed about 76 taxa [23,24].
In this paper, we propose two new sexual sect. Talaromyces species isolated from mudflats of China, namely, T. haitouensis sp. nov. and T. zhenhaiensis sp. nov., using the integration of morphological and molecular phylogenetic approaches.
2. Materials and Methods
2.1. Sampling and Fungal Isolation
Mudflat soil samples were collected from Haizhouwan Resort, Haitou Town, Lianyungang, Jiangsu Province (34°55′57″ N 119°11′56″ E, 3 m) and Zhenhai District, Ningbo, Zhejiang Province (29°58′19″ N, 121°48’19″ E, 8 m), China. Soil samples were kept in sterile centrifugal tubes (50 mL) and kept at 4 °C until used. The Talaromyces strains were isolated from these soil samples by using the improved dilution plating method of [25] with 0.1% agar water solution instead of water. Four distinctive strains were isolated in pure culture and have been deposited in China General Microbiological Culture Collection (CGMCC) as HR1-7 = AS3.16101; ZH3-18 = AS3.16102; WYZ25-10 = AS3.16103; NN72335 = AS3.15693.
2.2. Phenotypic Characterization of Strains
Colony characters were examined by inoculating the strains on the media proposed by Raper and Thom [11] (Czapek agar, Cz), Pitt [12] (Czapek yeast autolysate agar, CYA, yeast extract (Oxoid), Samson et al. [26] (5% malt extract agar, MEA, malt extract (Oxoid); Oatmeal agar, OA)), and Frisvad [27] (yeast extract sucrose agar, YES, yeast extract (Oxoid)), and incubating them at 25 °C. Colonies on CYA at 37 °C and 5 °C were also assessed. The names of colors were referenced to Ridgway [28]. To make wet mounts, a small amount of mycelium with reproductive structures was picked from the colonies on MEA and mounted in 85% lactic acid solution without staining. The microscopic characters were examined and photographed with an Axioplan2 imaging and Axiophot2 universal Microscope (Carl Zeiss Shanghai Co., Ltd., Shanghai, China).
2.3. DNA Extraction, Amplification, Sequencing, and Phylogenetic Analyses
Genomic DNA extraction was conducted according to Wang and Zhuang [29]. The partial β-tubulin gene (BenA) sequences were amplified with primers Bt2a or I2 and Bt2b [30,31], the partial calmodulin gene (CaM) sequences were obtained with the primers AD1, AD2 and Q1, Q2 [32]; the partial DNA-dependent RNA polymerase II second largest subunit gene (Rpb2) sequences were amplified with primers T1, T2 and E1, E2 [33], and the rDNA ITS1-5.8S-ITS2 (ITS) sequences were obtained with the primers ITS5 and ITS4 [34]. PCR was employed in 20 µL mixture containing 0.5 µL of each primer (10 pM/µL), 1.0 µL of genomic DNA (10 ng/µL), 10 µL of 2 × PCR MasterMix buffer (0.05 u/µL Taq polymerase, 4 mM MgCl2, 0.4 mM dNTPs), and 8 µL of double-distilled water (Tsingke Biotechnology Co., Ltd., Beijng, China). The amplification was conducted in an AB 2720 thermal cycler (Applied Biosystems, Drive Foster City, CA, USA) with the program consisting of 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s, and a final elongation at 72 °C for 5 min [32]. The amplicons of the anticipated length were purified and sequenced in double direction with an ABI 3700 DNA analyzer (Applied Biosystems, Waltham, MA, USA), the obtained raw sequences were proofread manually with Bioedit 5.0.9 [35], and the edited sequences were deposited in GenBank. Fifty-five strains of 49 taxa of sect. Talaromyces were included in the individual analysis of BenA, CaM, Rpb2, ITS sequences, and the concatenated BenA-CaM-Rpb2 sequences, with T. dendriticus CBS 660.80T of sect. Purpurei as the outgroup. The five sequence matrices were aligned and trimmed with “MUSCLE” implemented in MEGA version 6 to generate sequence matrices, then analyzed with the Maximum Likelihood (ML) method and subjected to 1000 bootstrap replications, with substitution model and rates among sites set as K2 + G for BenA, K2 + G + I for CaM, K2 + G + I for Rpb2, T92 + G + I for ITS and K2 + G + I for BenA-CaM-Rpb2 [36], and gaps were treated as partial deletion [37]. The sequence matrices were also subjected to Bayes Inference (BI) with MrBayes 3.2 to calculate the posterior probabilities (pp) of the clades [38]; the same substitution model and rates among sites were set as above. The resulting phylograms were viewed and adjusted with MEGA version 6 and then copied and pasted to Microsoft Office PowerPoint 2010 for further processing.
3. Results
PCR amplification generated amplicons of BenA about 420 bp or 660 bp, CaM about 650 bp, Rpb2 about 825 bp, and ITS about 560 bp. The trimmed alignments of BenA, CaM, Rpb2, ITS, and the combined BenA-CaM-Rpb2 sequences were 371, 521, 670, 456, and 1562 characters with gaps, respectively.
The phylogenetic trees generated by either the concatenated BenA-CaM-Rpb2 sequences or the four individual loci show three isolates (AS3.16101, AS3.16102, and AS3.15693) as two distinct species of sect. Talaromyces (Figure 1, Figure 2 and Figures S1–S3). T. haitouensis sp. nov. (ex-type AS3.16101T), together with T. aspriconidius and T. flavus forms a clade with 79%/1 and 72%/1 bootstrap/pp support in BenA-CaM-Rpb2 and BenA phylograms, respectively. In CaM phylogram, T. haitouensis and T. flavus are close related with 99%/1 bootstrap/pp support, but T. aspriconidius is not in the same clade with them. In Rpb2 phylogram, T. haitouensis and T. aspriconidius form a clade but without bootstrap support and T. flavus forms a separate clade from them. However, the ITS phylogram does not support the close relationship of these three species and T. haitouensis forms a solitary clade without any relatives. T. zhenhaiensis sp. nov. (ex-type AS3.16102T) and T. stipitatus are consistently clustered in one clade with strong bootstrap/pp support according to BenA-CaM-Rpb2, BenA, CaM, Rpb2, and ITS sequences.
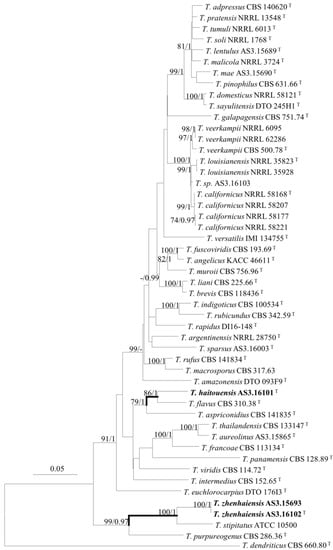
Figure 1.
ML phylogram inferred from the concatenated BenA-CaM-Rpb2 sequences. Bootstrap percentages over 70% derived from 1000 replicates and posterior probabilities over 0.95 of BI are indicated at the nodes. T indicates ex-type strains, strains belonging to new species are indicated in boldface. Scale Bar: number of substitutions per nucleotide position.
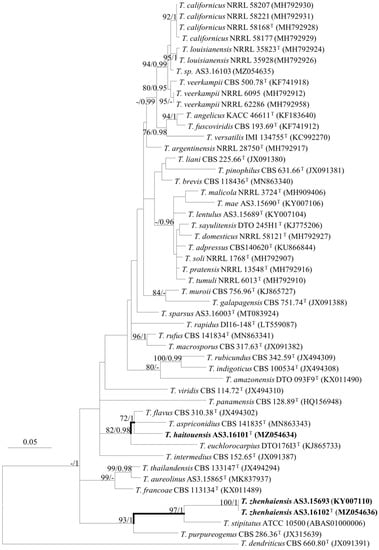
Figure 2.
ML phylogram inferred from partial BenA sequences. Bootstrap percentages over 70% derived from 1000 replicates and posterior probabilities over 0.95 of BI are indicated at the nodes. T indicates ex-type strains, strains belonging to new species are indicated in boldface. Scale Bars: number of substitutions per nucleotide position.
Description of New Taxa
Talaromyces haitouensis L. Wang, sp. nov. (Figure 3).
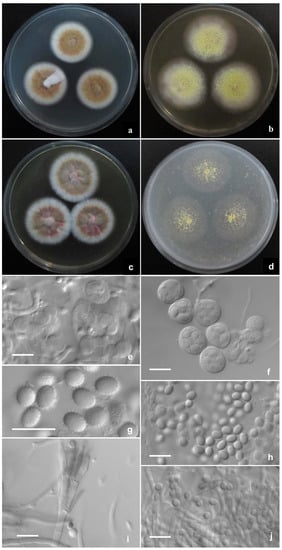
Figure 3.
Talaromyces haitouensis AS3.16101T incubated at 25 °C for 7 days. Colonies on (a) CYA. (b) MEA. (c) YES. (d) OA. (e) Initials. (f) Asci. (g,h) Ascospores. (i) Conidiophores. (j) Conidia. Scale Bars: 10 µm.
Fungal Names: FN570868.
Etymology: named after the location where the ex-type strain was isolated.
Holotype: HMAS 350335 (China, Jiangsu Province, Lianyungang, Haitou Town, Haizhouwan Resort, 34°55′57″ N 119°11′56″ E, 3 m, from the riverside soil, 5 May 2021, X-Y. Liu, ex-type strain: AS3.16101 = HR1-7; GenBank: ITS = MZ045695, BenA = MZ054634, CaM = MZ054637, Rpb2 = MZ054631).
CYA 25 °C 7 d: Colonies 22–25 mm diam, thin, radially plicate slightly, margins submerged, fimbriate; colony texture velutinous and granular due to limited gymnothecia, Pinard yellow (R. Pl. IV); sporulation absent or sparse; mycelium Mars yellow (R. Pl. III) mingled with Orange vinaceous (R. Pl. XXVII); exudate and soluble pigment absent; reverse Morocco red (R. Pl. I), Strawberry pink (R. Pl. I) at margins. MEA 25 °C 7 d: Colonies 48–51 mm diam, thin, plane, margins submerged, regular; colony texture granular due to abundant gymnothecia, Green-yellow (R. Pl. V); sporulation absent or limited, conidia en masse Olive-gray (R. Pl. LI); mycelium Green-yellow (R. Pl. V), white at margins; exudate and soluble pigment absent; reverse light Danube green (R. Pl. XXXII) centrally while Cinnamon buff (R. Pl. XXIX) at peripheries. YES 25 °C 7 d: Colonies 25–28 mm diam, thin, radially sulcate, margins on agar surface, fimbriate; colony texture velutinous and granular due to limited gymnothecia at centers, Pinard yellow (R. Pl. IV); sporulation absent or sparse, conidia en masse Light olive gray (R. Pl. LI); mycelium Pecan brown to Cacao brown (R. Pl. XXVIII), white at margins; exudate and soluble pigment absent; reverse Morocco red (R. Pl. I), Moral red (R. Pl. I) at margins. OA 25 °C 7 d: Colonies 33–35 mm diam, thin, plane, mostly submerged; colony texture slimy and sparsely granular due to limited gymnothecia in centers, Light greenish yellow (R. Pl. IV); sporulation absent; exudate and soluble pigment absent; reverse Sulphur yellow (R. Pl. V). Cz 25 °C 7 d: Colonies 14–17 mm diam, slightly thick, umbonate centrally, plane, margins submerged, fimbriate; colony texture velutinous and granular due to limited gymnothecia, Pinard yellow (R. Pl. IV); sporulation absent; mycelium Orange-vinaceous to Pale vinaceous (R. Pl. XXVII), white at margins; exudate limited, Strawberry pink (R. Pl. I); soluble pigment absent or limited, light red; reverse Nopal red (R. Pl. I). CYA 37 °C 7 d: Colonies 18–20 mm diam, slightly thick, irregularly plicate slightly, margins on agar surface, regular; sporulation absent; mycelium Shell pink (R. Pl. XXVIII); exudates and soluble pigment absent; reverse Ochraceous-salmon (R. Pl. XV). CYA 5 °C 7 d: Growth absent.
Ascomata after 14 d, globose, (400–) 450–480 μm; Asci globose, 12.5–13 μm to elliposoidal, 15 × 12–13 μm; Ascospores echinulate, broadly ellipsoidal, 5–6 × 4 μm; Conidiophores from aerial hyphae, biverticillate, stipes smooth-walled, 20–60 × 2–2.5 μm; Metulae 2–4 per stipe, 10–13 × 2–2.5 μm; Phialides 2–4 per metula, 9–12 × 2–2.5 μm; Conidia smooth-walled, pyriform to ellipsoidal, 2.5–3 × 2–2.5 μm.
Notes: T. haitouensis is characterized by normal growth at 25 °C and 37 °C, green-yellow gymnothecia, orange-brown mycelium, and echinulate broadly ellipsoidal ascospores.
Talaromyces zhenhaiensis L. Wang, sp. nov. (Figure 4).
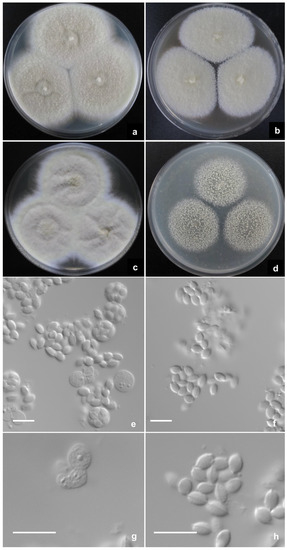
Figure 4.
Talaromyces zhenhaiensis AS3.16102T incubated at 25 °C for 7 days. Colonies on (a) CYA. (b) MEA. (c) YES. (d) OA. (e,g) Asci and ascospores. (f,h) Ascospores. Scale Bars: 10 µm.
Fungal Names: FN570869.
Etymology: named after the location where the ex-type strain was isolated.
Holotype: HMAS 350336 (China, Zhejiang Province, Ningbo, Zhenhai District, 29°58′19″ N 121°48′19″ E, 8 m, from mudflat soil, 10 September 2019, F-H Song, ex-type strain: AS3.16102 = ZH3-18; GenBank: ITS = MZ045697, BenA = MZ054636, CaM = MZ054639, Rpb2 = MZ054633).
CYA 25 °C 7 d: Colonies 65–67 mm diam, plane, thin, margins submerged, fimbriate, umbonate centrally; colony texture velutinous, overlaid with sparse floccose mycelium; sporulation absent; mycelium Sulfur yellow (R. Pl. V) to Cartridge buff (R. Pl. XXX), white at margins; exudate and soluble pigment absent; reverse Honey yellow (R. Pl. XXX). MEA 25 °C 7 d: Colonies 43–45 mm diam, thin, plane, margins on agar surface, fimbriate; colony texture velutinous; sporulation absent, mycelium pale yellow, near Massicot yellow (R. Pl. IV), white at margins; immature gymnothecia abundant, white to Massicot yellow (R. Pl. IV); exudate and soluble pigment absent; reverse pale Orange yellow (R. Pl. XXX). YES 25 °C 7 d: Colonies 61–62 mm diam, thin, irregularly sulcate, margins on agar surface, fimbriate; colony texture velutinous with sparse floccose mycelium overlaid; sporulation absent; mycelium Cartridge buff (R. Pl. XXX) to Naphthalene yellow (R. Pl. XVI), white at margins; exudate and soluble pigment absent; reverse Raw sienna (R. Pl. III). OA 25 °C 7 d: Colonies 37–40 mm diam, thin, plane, margins on agar surface, regular; colony texture granular due to abundant gymnothecia, cream yellow to Naphthalene yellow (R. Pl. XVI); sporulation absent; mycelium white; exudate and soluble pigment absent; reverse Sulphur yellow (R. Pl. V). Cz 25 °C 7 d: Colonies 38–40 mm diam, plane, thin, margins submerged, fimbriate; colony texture velutinous; sporulation absent; mycelium pale yellow, near Martius yellow (R. Pl. IV) to Marguerite yellow (R. Pl. XXX), white at margins; immature gymnothecia abundant, white to Marguerite yellow (R. Pl. XXX); exudate and soluble pigment absent; reverse Aniline yellow (R. Pl. IV). CYA 37 °C 7 d: Colonies 56–60 mm diam, thin, plane, margins on agar surface, regular; colony texture velutinous; sporulation absent; mycelium white; exudate and soluble pigment absent; reverse Augus brown (R. Pl. III). CYA 5 °C 7 d: Growth absent.
Ascomata after 14 d, globose, 300–500 μm; Asci globose, 8–10 μm, borne in short chains; Ascospores flattened ellipsoidal, smooth-walled, with one equatorial ridge, 4–5 × 3 μm.
Additional strains examined: Hainan Province, Ledong, Mount Jianfengling, 18°42′36″ N 108°49′48″ E, 800 m, from soil, 8 November 2015, X-Z Jiang; culture AS3.15693 = NN072335; GenBank: ITS = KY007094, BenA = KY007110, CaM = KY007102, Rpb2 = KY112592.
Notes: T. zhenhaiensis is characterized by fast growth at 25 °C and 37 °C, naphthalene-yellow gymnothecia and mycelium, and smooth-walled ellipsoidal ascospores with one equatorial ridge.
4. Discussion
Sexual sect. Talaromyces species such as T. flavus, T. liani, and T. aureolinus usually produce broadly ellipsoidal ascospores with echinulate walls, but some produce ellipsoidal ascospores with one or several ridges, such as T. stipitatus and T. viridis [21,24]. In the two novel species proposed here, ascospores of T. haitouensis are with echinulate walls and those of T. zhenhaiensis have ridged walls.
The phenotypic features, such as moderate growth, aureous ascomata and mycelium, and echinulate, broadly ellipsoidal ascospores of T. haitouensis are also shared by T. flavus and T. aureolinus, but T. haitouensis generally grows faster than T. flavus at 25 °C (CYA: 22–25 mm vs. 9–10 mm, MEA: 48–51mm vs. 31–32 mm, YES: 25–28 mm vs. 24–26 mm, OA: 33–35 mm vs. 30–32 mm), and in contrast to T. aureolinus, the new species grows well at 37 °C and shows slimy colony texture with sparse ascomata and mycelium on OA, but T. aureolinus does not grow at 37 °C and forms normal colonies about 33–35 mm with abundant ascomata on OA. Moreover, T. haitouensis produces orange-brown mycelium on CYA and YES, while T. aureolinus commonly produces lemon-yellow mycelium on all culture media. In micro-morphology, the initials of T. haitouensis consist of irregularly swollen cells, while those of T. flavus comprise the ascogonia encircled with antheridia, and T. haitouensis produces much larger gymnothecia, asci and ascospores than those of T. flavus (400–480 μm vs. 150–400 μm, 12.5–13 μm or 15 × 12–13 μm vs. 9.5–13.5 × 8–11.5 μm, 5–6 × 4 μm vs. 4–5.5 × 3–3.5 μm, respectively). T. haitouensis produces short stipes (20–60 μm), but T. aureolinus much longer ones (300–450 μm) from surface and substratum hyphae [11,21,24].
T. zhenhaiensis forms a clade with T. stipitatus in all phylograms inferred from individual genes and the concatenated BenA-CaM-Rpb2 sequences with strong support (Figure 1, Figure 2 and Figures S1–S3). The morphological differences between them are subtle, but still tangible, for instance, T. zhenhaiensis grows much faster than T. stipitatus at 25 °C (CYA: 65–67 mm vs. 32–38 mm, YES: 61–62 mm vs. 40–45 mm, OA: 37–40 mm vs. 30–35 mm) and at 37 °C (56–60 mm vs. 28–32 mm), and the color of the mycelium and gymnothecia of the new species is near cream yellow to Naphthalene yellow, which is lighter than that of T. stipitatus which is bright yellow with a slight greenish tint. Microscopically, though the shapes and dimensions of gymnothecia, asci, and ascospores of the two species are similar, the asci of T. zhenhaiensis are larger than those of T. stipitatus (asci: 8–10 μm vs. 6–8 μm) [11,21].
The intertidal mudflats along the coastlines consist of transitional ecological habitats from terrestrial to marine ones, which are complicated with various and variable environmental factors either chemical or physical, such as salinity, carbon, nitrogen, and phosphorus nutrients due to domestic, agricultural, and industrial sewage and wastes, anaerobic soil, and the mixture of fresh and sea water, as well as the ebb and flow of the tide. Some categories of ubiquitous fungi, for example, certain species of Aspergillus, Penicillium, and Talaromyces possess the robust metabolic plasticity to interact with the hash environmental factors either abiotic or biotic (e.g., [39]). Though the marine-derived strains are not distinct from their terrestrial counterparts taxonomically, the marine habitats provide the selective sampling locales to isolate certain species that are rare or difficult to be discovered from terrestrial sites, moreover, their metabolites are so diverse that they are considered as the treasure trove for discovery of new compounds of pharmaceutical interest (e.g., [40]).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof8010036/s1. Figure S1: ML phylogram inferred from partial CaM sequences. Bootstrap percentages over 70% derived from 1000 replicates and posterior probabilities of BI over 0.95 are indicated at the nodes. T indicates ex-type strains, strains belonging to new species are indicated in boldface. Scale bars: number of substitutions per nucleotide position. Figure S2. ML phylogram inferred from partial Rpb2 sequences. Bootstrap percentages over 70% derived from 1000 replicates and posterior probabilities of BI over 0.95 are indicated at the nodes. T indicates ex-type strains, strains belonging to new species are indicated in boldface. Scale bars: number of substitutions per nucleotide position. Figure S3. ML phylogram inferred from partial ITS sequences. Bootstrap percentages over 70% derived from 1000 replicates and posterior probabilities of BI over 0.95 are indicated at the nodes. T indicates ex-type strains, strains belonging to new species are indicated in boldface. Scale bars: number of substitutions per nucleotide position.
Author Contributions
Conceptualization, L.W.; methodology, L.W.; formal analysis, P.-J.H., J.-Q.S.; writing—original draft preparation, P.-J.H. and J.-Q.S.; writing—review and editing, L.W.; supervision, L.W.; funding acquisition, P.-J.H. and L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work is funded by the National Natural Science Foundation of China, grant numbers U20A20101, 31750001, 32170011, the National Project on Scientific Groundwork, Ministry of Science and Technology of China, grant number 2019FY100700, and the Key Research Program of Frontier Science, Chinese Academy of Sciences, grant number QYZDY-SSW-SMC029.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences newly generated in this study can be found in the NCBI database.
Acknowledgments
Fu-Hang Song, Xiao-Yong Liu, and Xian-Zhi Jiang collected soil samples.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, FUNK-0052-2016. [Google Scholar] [CrossRef]
- Heo, Y.M.; Lee, H.; Kim, K.; Sun, L.K.; Kim, J.J. Fungal diversity in intertidal mudflats and abandoned solar salterns as a source for biological resources. Mar. Drugs 2019, 17, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.S.; Oh, S.Y.; Fong, J.J.; Houbraken, J.; Lim, Y.W. The diversity and ecological roles of Penicillium in intertidal zones. Sci. Rep. 2019, 9, 13540. [Google Scholar] [CrossRef] [Green Version]
- Nicoletti, R.; Vinale, F. Bioactive compounds from marine-derived Aspergillus, Penicillium, Talaromyces and Trichoderma species. Mar. Drugs 2018, 16, 408. [Google Scholar] [CrossRef] [Green Version]
- Zhai, M.M.; Li, J.; Jiang, C.X.; Shi, Y.P.; Di, D.L.; Crews, P.; Wu, Q.X. The bioactive secondary metabolites from Talaromyces species. Nat. Prod. Bioprospect. 2016, 6, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Nicoletti, R.; Trincone, A. Bioactive compounds produced by strains of Penicillium and Talaromyces of marine origin. Mar. Drugs 2016, 14, 37. [Google Scholar] [CrossRef] [Green Version]
- Jones, E.G.; Suetrong, S.; Sakayaroj, J.; Bahkali, A.H.; Abdel-Wahab, M.A.; Boekhout, T.; Pang, K.L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015, 73, 1–72. [Google Scholar] [CrossRef]
- Nicoletti, R.; Salvatore, M.M.; Andolfi, A. Secondary metabolites of mangrove-associated strains of Talaromyces. Mar. Drugs 2018, 16, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, X.H.; Liu, L.P.; Shao, T.Y.; Shao, H.B.; Liu, Z.P. Developing and sustainably utilize the coastal mudflat areas in China. Sci. Total Environ. 2016, 569–570, 1077–1086. [Google Scholar] [CrossRef]
- Benjiamin, C.R. Ascocarps of Aspergillus and Penicillium. Mycologia 1955, 47, 669–687. [Google Scholar] [CrossRef]
- Raper, K.B.; Thom, C. A Manual of the Penicillia; Williams and Wilkins: Baltimore, MD, USA, 1949; pp. 1–875. [Google Scholar]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic states Eupenicillium and Talaromyces; Academic Press: London, UK, 1980; pp. 1–634. [Google Scholar]
- Malloch, D. The Trichocomaceae: Relationships with other ascomycetes. In Advances in Penicillium and Aspergillus Systematics; Samson, R.A., Pitt, J.I., Eds.; Plenum Press: New York, NY, USA, 1985; pp. 365–382. [Google Scholar]
- LoBuglio, K.F.; Pitt, J.I.; Taylor, J.W. Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologia 1993, 85, 592–604. [Google Scholar] [CrossRef]
- Berbee, M.L.; Yoshimura, A.; Sugiyama, J.; Taylor, J.W. Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia 1995, 87, 210–222. [Google Scholar] [CrossRef]
- Wang, L.; Zhuang, W.-Y. Phylogenetic analyses of penicillia based on partial calmodulin gene sequences. BioSystems 2007, 88, 113–126. [Google Scholar] [CrossRef]
- Houbraken, J.; Samson, R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 2011, 70, 1–51. [Google Scholar] [CrossRef]
- Samson, R.A.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.A.; Peterson, S.W.; Varga, J.; Frsivad, J.C. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef] [Green Version]
- McNeill, J.; Barrie, F.R.; Buck, W.R.; Demoulin, V.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Marhold, K.; Prado, J.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code). Regnum Vegetabile 154; Koeltz Scientific Books: Koenigstein, Germany, 2012; pp. 1–257. [Google Scholar]
- Houbraken, J.; Spierenburg, J.H.; Frisvad, J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Anton. Leeuw. 2012, 101, 403–421. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.D.; Chen, A.J.; Houbraken, J.; Frisvad, J.C.; Wu, W.P.; Wei, H.L.; Zhou, Y.G.; Jiang, X.Z.; Samson, R.A. New section and species in Talaromyces. MycoKeys 2020, 68, 75–113. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsube, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Samson, R.A.; Frisvad, J.C. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Xu, X.; Wang, L. Four new species of Talaromyces sect. Talaromyces discovered in China. Mycologia 2021, 113, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Malloch, D. Moulds Their Isolation, Cultivation and Identification; University of Toronto Press: Toronto, ON, Canada, 1981; pp. 1–97. [Google Scholar]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2010; pp. 1–390. [Google Scholar]
- Frisvad, J.C. Physiological criteria and mycotoxin production as aids in identification of common asymmetric penicillia. Appl. Environ. Microbiol. 1981, 41, 568–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridgway, R. Color Standards and Color Nomenclature; Published by the Author: Washington, DC, USA, 1912; pp. 1–53. [Google Scholar]
- Wang, L.; Zhuang, W.Y. Designing primer sets for amplification of partial calmodulin genes from penicillia. Mycosystema 2004, 23, 466–473. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wang, L. Penicillium kongii, a new terverticillate species isolated from plant leaves in China. Mycologia 2013, 105, 1547–1554. [Google Scholar] [CrossRef]
- Wang, L. Four new records of Aspergillus section Usti from Shandong Province, China. Mycotaxon 2012, 120, 373–384. [Google Scholar] [CrossRef]
- Jiang, X.Z.; Yu, Z.D.; Ruan, Y.M.; Wang, L. Three new species of Talaromyces sect. Talaromyces discovered from soil in China. Sci. Rep. 2018, 8, 4932. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols—A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nuc. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Primavera, J.H.; Dahdouh-Guebas, F.; Mckee, K.; Bosire, J.O.; Cannicci, S.; Diele, K.; Fromard, F.; Koedam, N.; Marchand, C.; et al. Ecological role and services of tropical mangrove ecosystems: A reassessment. Global Ecol. Biogeogr. 2014, 23, 726–743. [Google Scholar] [CrossRef]
- Overy, D.P.; Bayman, P.; Kerr, R.G.; Bills, G.F. An assessment of natural product discovery from marine (sensu strictu) and marine-derived fungi. Mycology 2014, 5, 145–167. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).