Abstract
Agaricus bisporus is a globally cultivated mushroom with high economic value. Despite its widespread cultivation, commercial button mushroom strains have little genetic diversity and discrimination of strains for identification and breeding purposes is challenging. Molecular markers suitable for diversity analyses of germplasms with similar genotypes and discrimination between accessions are needed to support the development of new varieties. To develop cleaved amplified polymorphic sequences (CAPs) markers, single nucleotide polymorphism (SNP) mining was performed based on the A. bisporus genome and resequencing data. A total of 70 sets of CAPs markers were developed and applied to 41 A. bisporus accessions for diversity, multivariate, and population structure analyses. Of the 70 SNPs, 62.85% (44/70) were transitions (G/A or C/T) and 37.15% (26/70) were transversions (A/C, A/T, C/G, or G/T). The number of alleles per locus was 1 or 2 (average = 1.9), and expected heterozygosity and gene diversity were 0.0–0.499 (mean = 0.265) and 0.0–0.9367 (mean = 0.3599), respectively. Multivariate and cluster analyses of accessions produced similar groups, with F-statistic values of 0.134 and 0.153 for distance-based and model-based groups, respectively. A minimum set of 10 markers optimized for accession identification were selected based on high index of genetic diversity (GD, range 0.299–0.499) and major allele frequency (MAF, range 0.524–0.817). The CAPS markers can be used to evaluate genetic diversity and population structure and will facilitate the management of emerging genetic resources.
1. Introduction
Button mushroom (Agaricus bisporus) is a popular edible mushroom that is consumed worldwide. A. bisporus extracts have high antioxidant activity and are known to improve cardiovascular health [1,2]. Global mushroom production increased more than 30-fold during 1978–2013, and total production value increased to $63 billion [3]. A. bisporus production is approximately 4.4 million tons per year, constituting 15% of global mushroom production [3]. Despite the high economic value of button mushrooms, genetic diversity is low. Although new strains can be produced through phenotypic selection and limited parent strain crossing, high similarity remains among varieties [4]. Evaluation of the genetic characteristics of mushrooms with similar phenotypes is needed to facilitate the introduction of novel traits into commercial varieties. Molecular resources are also needed to support the efficient selection of accessions, collection and preservation of strains, diversity assessment, and population structure analysis [5]. The rapid development of next-generation sequencing technologies has enabled large-scale sequencing projects in a variety of organisms. Molecular markers that are stable, highly polymorphic, and provide valuable diversity information are preferred over phenotype-based markers that are subject to environmental effects [6]. Polymerase chain reaction (PCR)-based molecular markers, such as simple sequence repeats (SSR) and single nucleotide polymorphisms (SNPs), can be used for molecular genetic studies of culture collections. Genetic diversity in A. bisporus has been examined using several similar approaches, including analysis with restriction fragment length polymorphisms (RFLP) [7], discrimination analysis with random amplified polymorphic DNA (RAPD) [8], genetic diversity analysis using SSR markers [9,10,11], phylogenetic analysis using SNP markers [12], SNP genotyping [13,14], and quantitative trait locus (QTL) mapping using SNP markers [15,16]. However, molecular markers that can be used for the analysis of population structure and diversity of accession collections are biased toward SSR markers, and new SNP genotyping markers are needed.
SNPs, which are differences at single nucleotide positions between or within species, are the most widely distributed genetic variants in the genome. SNPs found in coding and non-coding regions are classified as transitions (Ts) (C/T or G/A) or transversions (Tv) (C/G, A/T, C/A, or T/G), depending on the type of nucleotide substitution. If present in coding regions, SNPs can change the structure or function of proteins, causing phenotypic differences [17]. Thus, compared with methods using other molecular markers, SNP genotyping-based diversity assessments can more accurately and specifically explain phenotypic differences. SNP markers are highly reproducible co-dominant markers that can be used to distinguish between homozygosity and heterozygosity and to discriminate accessions, and they can also be used for association mapping and analysis of genetic diversity and group structure [18,19,20]. Two main types of SNP-based markers are cleaved amplified polymorphic sequences (CAPS) and derived cleaved amplified polymorphic sequences (dCAPS), the latter of which offers increased utility for genotyping [21,22]. CAPS marker, also known as the PCR–RFLP marker, combined with primers that can amplify specific regions is popularly used for molecular genetic studies in fungus, owing to its advantage in detecting secondary polymorphisms that cannot be directly detected by PCR amplification [23,24]. Moreover, since the CAPS marker system consists of PCR and restriction enzyme treatment, it is much easier and less time-consuming than conventional RFLP based on Southern hybridization.
In this study, CAPS markers were developed for the analysis of genetic diversity and population structure in A. bisporus accessions. The newly developed CAPS markers will support the efficient evaluation and management of new and existing accessions, thereby facilitating further studies on genetic diversity and population structure in button mushrooms.
2. Materials and Methods
2.1. A. bisporus Genetic Resources and DNA Extraction
For the genetic diversity and population structure analysis, we used 41 A. bisporus strains provided by the Korean Mushroom Culture Collection (KMCC) at the National Institute of Horticultural and Herbal Science (NIHHS) of Rural Development Administration (RDA) in Eumseong, Republic of Korea. Geographic origins and KMCC numbers of 41 A. bisporus strains are given in Table 1.

Table 1.
A. bisporus accessions used for validation of CAPS markers.
A. bisporus strains were plated on compost dextrose agar (CDA) medium (Kisanbio, Seoul, Korea), incubated at 25 °C in the dark for 60 days, and lyophilized. DNA extraction was performed using a Plant SV mini kit (GeneAll, Seoul, Korea) according to the manufacturer protocol. Extracted DNA concentrations were quantified using an Epoch microplate spectrophotometer (BioTek, Winooski, VT, USA).
2.2. Primer Construction and PCR
To develop CAP markers, SNP mining was performed based on the A. bisporus genome and resequencing data derived from our previous study [10]. Software (dCAPS Finder 2.0) [25] was used to identify restriction enzyme recognition sites based on the resulting SNPs, and a set of 70 CAPS markers was produced (Table 2). PCR reactions (20 µL) contained 10 µL of Excel TB 2× Taq premix (Inclone Biotech, Yongin, Korea), 2 µL of 10 pmol primer (forward/reverse), 5 µL of distilled water, and 3 µL of DNA. DNA amplification was performed as follows: initial denaturation at 95 °C for 2 min; followed by 30 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 40 s, and extension at 72 °C for 45 s; and a final extension at 72 °C for 10 min. Amplified PCR products were digested using 38 restriction enzymes. Restriction reactions (10 µL) contained 1 unit of restriction enzyme, 1 µL 10 × NEBuffer™, and 5 µL PCR product. Reactions were incubated at an appropriate temperature (Table 2) for 60 min. Digested PCR products were analyzed using 2.5% agarose gel electrophoresis.

Table 2.
CAPS markers developed using SNPs mined from A. bisporus resequencing data.
2.3. Data Analysis
Using the Power Marker version 3.25 software package [26], the diversity of each CAPS marker was analyzed on the basis of five statistical parameters including major allele frequency (MAF), number of genotypes (NG), number of alleles (NA), genetic diversity (GD), and heterozygosity (He). Genetic distance was calculated using ‘’Nei’s standard’’ [27] followed by phylogeny reconstruction using rooted unweighted pair group method with arithmetic mean (UPGMA) as implemented in MEGA version 7 [28].
To visualize the relationship between the sample genotypes among the 41 A. bisporus accessions, principle coordinate analysis (PCoA) was conducted using GenALEx 6.5 software [29]. It was chosen to complement the UPGMA cluster analysis.
A non-hierarchical analysis of molecular variance (AMOVA) (1000 permutations) based on the degree of genetic divergence among populations was performed using GenALEx 6.5 software [29]. Population structure was analyzed based on Bayesian clustering using STRUCTURE 2.3.1 [30]. The populations number (K) was set from 1 to 10, and the populations set as location priors (LOCPRIOR) [31] under the admixture model were used to run the Markov chain Monte Carlo (MCMC) simulation algorithm. The length of the burn-in period was set to 10,000 iterations. The Delta K value was obtained by the method of Evanno [30]. The 10 runs for the optimal delta K were averaged by using the programs STRUCTURE HARVESTER [32]. Next, a hierarchical AMOVA, which was calculated considering the main groups obtained from the STRUCTURE analysis, was implemented by the software GenALEx 6.5 [29]. The statistical significance was also tested using a nonparametric approach described in Excoffier et al. (1992) with 1000 permutations [33].
The minimum number of marker sets (n) needed to distinguish each accession was determined using the accumulation curve approach of the “poppr package” in R [34]. For minimum marker set combinations, initial markers were selected with high GD, NA, and NG values. From the second to the last marker, markers were sequentially selected based on their ability to subdivide the highest number of accessions according to the genotyping data. Grouping based on genotyping data was performed in Microsoft Excel. After selection of minimum marker set combinations, phylogenetic analysis was used to confirm whether the accession could be fully distinguished using the minimum marker (Figure 1).
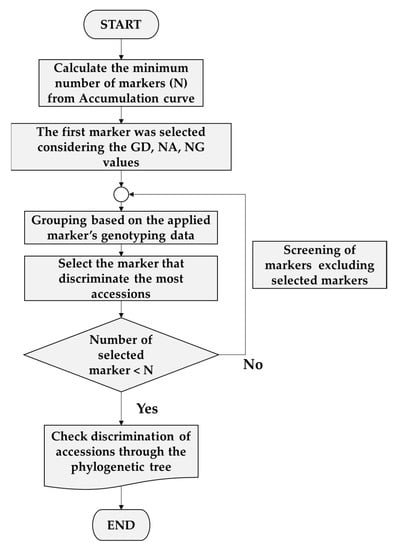
Figure 1.
Pipeline used for minimum marker set selection.
3. Results
3.1. Genotyping and Marker Diversity
Seventy CAPS markers were developed and used to assess 41 A. bisporus accessions. Ts polymorphisms were more common than Tv polymorphisms. The most common Ts difference was G→A, which occurred 14 times, followed by 11 C→T, 10 A→G, and 9 T→C. The most common Tv differences were C→A and C→G (four instances), followed by three A→C, A→T, G→C, G→T, T→A, and T→G changes (Table 2). The diversity index of each marker is show in Table 3. Of the 70 CAPS markers, 64 were polymorphic and 6 were monomorphic (MAF = 1). Excluding the six monomorphic markers, MAF ranged from 0.5 (AB-gCAPS-036) to 0.984 (AB-gCAPS-093), with an average of 0.698. Two NAs and two (n = 28) and three (n = 36) NGs were identified with the polymorphic markers. Similarly, excluding the six markers with one allele, He ranged from 0 (12 accessions) to 0.826 (AB-gCAPS-003), with an average of 0.290. GD ranged from 0.031 (AB-gCAPS-093) to 0.05 (AB-gCAPS-036), with an average of 0.394.

Table 3.
Genetic diversity index of 41 Agaricus bisporus accessions assessed with 70 CAPS markers.
To assess whether the CAPS markers developed in this study were suitable for evaluating diversity and population structures, diversity was determined using the SSR diversity index, a widely used metric in population genetic studies [9,10,11,35]. GD, a representative diversity index, is influenced by the allele frequency. However, as SSR and SNP markers have multiple and single target locus characteristics, respectively, allele frequencies tend to differ and theoretically calculated GD index values also differ. As it is difficult to compare diversity between SNP and SSR markers using the GD index, an appropriate alternative comparison based on scaling to the maximum GD index value of each marker was used, with the following equation: . When the SNP marker had a maximum of three alleles, the maximum GD value was 0.66, and the SSR marker reached a maximum GD value of 0.99 as the number of alleles increased. The average GD value of the CAPS markers in this study was 0.3599, and the minimum and maximum GD values of polymorphic markers were 0.031 and 0.5, respectively. When compared with SSR values, the average corrected SSR value, based on the maximum, was 0.540, and the minimum and maximum GD values were 0.046 and 0.750, respectively. Polymorphism frequencies were lower than average GD values of 0.548, 0.619, and 0.6807 from previously developed SSR markers. However, polymorphism levels were higher than the average GD value of 0.395 from monospores of limited accessions [9,10,11,35].
3.2. Grouping Based on Data Analysis and AMOVA
Multivariate and population structure analyses were performed to understand accession and population characteristics. Multivariate analysis included phylogenetic cluster analysis and principal coordinates analysis (PCoA), and population structure analysis was performed using a model-based structure. Phylogenetic tree analysis produced three groups: Group 1 (CHN 2, DEU 1, JPN 1, KOR 7, NLD 3, and USA 2), Group 2 (DEU 2, JPN 1, KOR 3, NLD 2, and USA 3), and Group 3 (CHN 3, DEU 2, JPN 4, KOR 3, NLD 1, and USA 1) (Figure 2). PCoA analysis also revealed three groups: P1 (CHN 2, DEU 1, JPN 1, KOR 7, NLD 3, and USA 2), P2 (DEU 2, JPN 1, KOR 3, NLD 1, and USA 2), and P3 (CHN 3, DEU 2, JPN 4, KOR 3, NLD 1, and USA 1) (Figure 3).
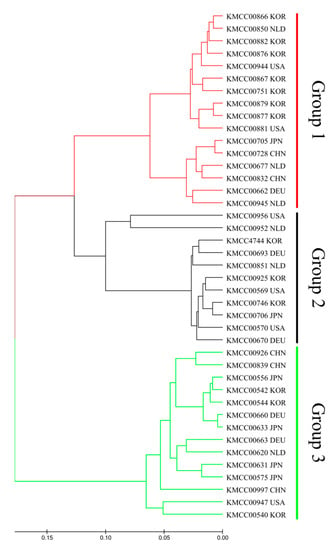
Figure 2.
Phylogenetic tree constructed using the unweighted pair pop method with arithmetic mean based on Nei’s genetic distance.
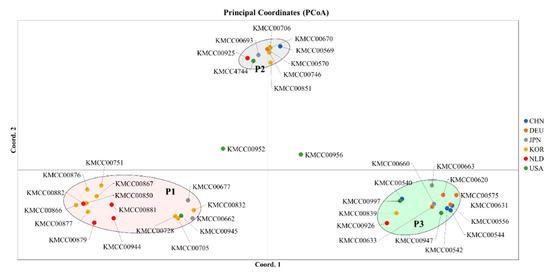
Figure 3.
Principal coordinates analysis to identify the characteristics and members of each cluster. Accessions were divided into three main groups, with two central accessions not classified.
Model-based structure analysis produced two groups: POP 1 (CHN 2, DEU 1, JPN 1, KOR 7, NLD 3, and USA 2) and POP 2 (CHN 3, DEU 2, JPN 4, KOR 3, NLD 1, and USA 1), and the remaining accessions were classified as an Admix group. Population structure was revealed by classification of accessions into each group using an unrooted tree (Figure 4). Groupings were largely consistent across the three methods: Group 1, P3, and POP 1; and Group 3, P1, and POP 2 had the same accessions. Finally, Group 2 and Admix had the same accessions, with P2 having all except two of the same accessions.
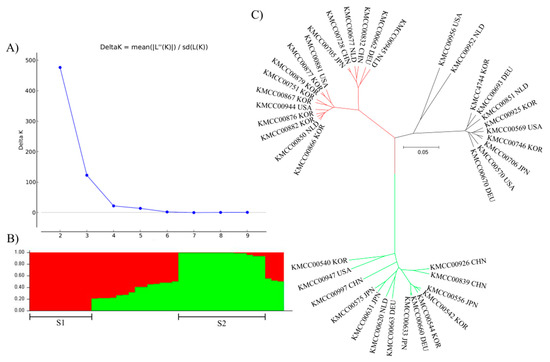
Figure 4.
Group structure analysis of Agaricus bisporus accessions. The most appropriate number of groups was determined by repeating five iterations using Structure Burn-in and Markov Chain Monte Carlo (MCMC) with 100,000 iterations. (A) Delta K values were calculated using Structure Harvester. (B) Probability of each accession belonging to a group. (C) Unrooted tree confirmation of population structure.
To determine the degree of genetic variation and differentiation among groups, AMOVA was performed with two group types as follows: distance-based groups (Groups 1, 2, and 3) and model-based groups (POP 1, POP 2, and Admix). The variation in the population level of the two groups was 13% in the distance-based groups and 15% in the model-based groups, and the variation on an individual level was 31% and 30% among individuals in distance-based and model groups, respectively, and 55% within individuals in both groups. The F-statistic (FST) value was 0.134 in the distance-based group and 0.153 in the model-based group (Table 4).

Table 4.
AMOVA analysis of distance- and model-based clustering.
3.3. Selection of Minimum Markers for Discrimination
A minimal marker set for accession discrimination was developed using an accumulation curve (Figure 5A) according to the pipeline shown in Figure 1. AB-gCAPS-059 was selected as the first marker. From the second marker onwards, the phylogenetic tree was used to select markers that provided discrimination of the largest numbers of accessions. The 10 markers that were selected (AB-gCAPS-017, AB-gCAPS-022, AB-gCAPS-026, AB-gCAPS-033, AB-gCAPS-038, AB-gCAPS-039, AB-gCAPS-042, AB-gCAPS-059, AB-gCAPS-061, and AB-gCAPS-066) were able to distinguish among the 41 A. bisporus accessions, as confirmed using a phylogenetic tree (Figure 5B).

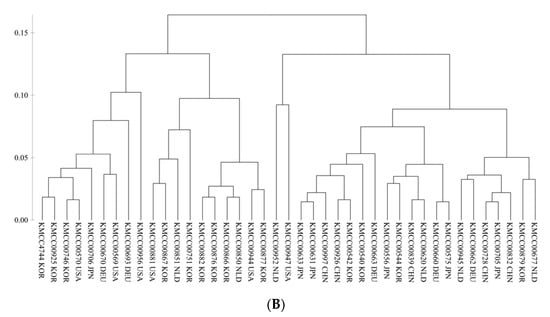
Figure 5.
(A) The number of markers sufficient to serve as a minimum marker was determined using an accumulation curve calculated using the poppr package in R. (B) Phylogenetic tree illustrating discrimination of 41 Agaricus bisporus accessions using 10 selected markers.
4. Discussion
4.1. Polymorphism Did Not Differ According to SNP Mutation Type
SNPs occur throughout the genome, with differing effects depending on the polymorphism type and location. SNPs can affect protein amino acid sequences if they occur within a coding region and introduce a codon change (non-synonymous change). Two types of SNP are found: Ts and Tv. SNP differences within the purines (A, G) or pyrimidine (C, T) nucleotides are Ts SNPs, and those that change from purine to pyrimidine or vice versa are Tv SNPs. [17]. Although there are four possible Ts changes and eight possible Tv changes, Ts SNPs occur at higher frequency than Tv SNPs [36]. Research suggests this is due to the higher number of possibly damaging non-synonymous changes resulting from Tv mutations compared with Ts mutations. Thus, Tv changes have a greater physicochemical impact on amino acid sequences and are not favored during natural selection [37]. The tv is considered to be a more drastic change than a ts, because substitution of one-ring to two-ring chemical structure or vice versa (Tv) requires more energy than substitution without change in the ring structure (Ts) [38]. The Ts/Tv ratio has been used as an important parameter in bacteria studies such as phylogenetic tree reconstruction and estimation of divergence [39].
Numbers of Ts and Tv polymorphisms were compared in the A. bisporus CAPS markers developed in this study. Of the 70 markers, 44 markers were Ts SNPs, and 26 markers were SNPs, a ratio of 1.7:1 for Ts:Tv. Average GD values were 0.363 (Ts) and 0.354 (Tv), showing no substantial differences. Thus, it can be inferred that the SNP type (Ts or Tv) in the A. bisporus marker set was not biased.
4.2. Diversity of Developed SNP Markers
Assessments based on molecular markers can be divided into population genetic assessments and trait assessments. SSR markers, with relatively more alleles, show a high level of diversity and are frequently used for population genetics, whereas SNP markers are frequently used to identify specific traits or determine lineage and population [40,41].
SSR and SNP markers exhibit different polymorphism traits, such as the number of repeats in the sequence, mutations of a single nucleotide, and genome-wide mutations; and the ability to obtain different types of information in the same study may facilitate the combined use of the two marker types [41,42]. SSRs were found to be better suited for detecting structure in populations at a small spatial scale with a systematic and continuous sampling design. SNP markers rather reflect ancient divergence of distant and naturally separated populations, being less sensitive to sampling design [43]. Despite these differences, both marker types were suitable for detecting the genetic structure of the fungal populations considered.
Previous studies of mushroom diversity and population structures using SSR and SNP markers focused on differences in the diversity index for the same marker [9,10,11,12,13,14,44]. However, direct comparison of diversity was difficult because of differences in the target loci of the two marker types. To solve these problems, the diversity index of the two markers was calculated and compared using an equation based on the maximum diversity index of each marker. This comparison method can be applied to a range of markers to facilitate comparative analysis of mushroom diversity and population structure.
4.3. Population Structure Analysis Using SNP Markers
In previous studies, SNP markers showed a similar grouping pattern and more accurate lineage classification of the neighbor joining tree in population structure analysis compared to SSR markers [41,42]. In particular, the explanatory value for the first main coordinate in PCoA analysis was higher for SNP markers, and this was a common phenomenon regardless of accession [41,42]. No common characteristics were observed in the clustered accessions of all groups with the three clustering methods used in this study, and collection location did not correlate with accessions in the clusters. This may be because the SNP markers classified lineage, and the current commercial accessions were divided into relatively few lineages. In addition, accessions in geographically separated regions had similar sequences, consistent with the genetically similar nature of accessions cultivated worldwide [4].
Population structure analysis is used to identify characteristics by composing clusters according to accession similarities. This led us to speculate that similarities among sequences would have a strong impact on accession clustering, and we therefore used AMOVA to confirm the extent to which genetic variation between populations and accessions were affected by one another. AMOVA of a model-based population revealed slight difference between groups and accessions by collection area (13% and 86%, respectively) and genotype (15% and 85%, respectively). Population variation according to genotype and collection area did not differ substantially, consistent with most commercially cultivated button mushrooms being derived from similar strains.
4.4. Selection of Minimum Markers for Accession Identification
Development of a minimum set of molecular markers to distinguish accessions provides the basis for future evaluation of existing new A. bisporus resources. An accumulation curve, in which SNP loci from the 70 CAPS markers were randomly identified and calculated [34], was used to determine the minimum number of loci required to distinguish between accessions. This approach can be extended to determine minimum marker numbers for future applications. In this study, 10 SNP markers were sufficient to distinguish 41 A. bisporus accessions. Previous research also developed minimum marker sets for accession differentiation: SSR markers were used to distinguish all accessions by using 4 of 26 accessions [10] and 6 of 171 accessions [11]. Discrimination using SNP markers tends to require more markers, and distinguish fewer accessions, than SSR markers. SNP markers may therefore be less efficient when examining large numbers of accessions; however, linkages determined using SNP markers may be more stable than linkages established using SSR markers.
Currently, studies using SNP markers in button mushrooms focus on the evaluation of accessions by direct comparison of SNPs by sequencing or through QTL mapping related to characteristics such as mushroom color and robustness [13,14,15,16]. Advances in genomic analysis have facilitated the efficient use of SSR and SNP markers for population genetic studies of accession collections. However, studies of genetic diversity and population structure using CAPS markers in mushrooms are limited. The SNP markers developed in this study will facilitate the comparison and evaluation of existing A. bisporus accessions and will provide the basis of future analysis and management of new accessions.
5. Conclusions
Consumer demand for new button mushroom varieties has increased alongside the recent growth of the mushroom industry. Optimal selection of breeding materials through the evaluation and management of accession collections is important for the development of new varieties. Several molecular markers have been used to evaluate crops with restrictive genotypes, such as button mushrooms. Of these, SNP markers are widely used for association mapping, accession discrimination, and analysis of genetic diversity and population structure. However, only limited numbers of molecular markers are available to support A. bisporus breeding strategies. In this study, a set of 70 CAPS markers was developed to analyze the diversity and population structure of 41 A. bisporus accessions. Of the 70 markers, a set of 10 minimum markers was identified that was able to identify all 41 A. bisporus accessions. The developed CAPS markers will be useful for analysis of button mushroom diversity and population structures, and will also be useful for variety identification.
Author Contributions
Conception and design: I.-H.J., and J.-W.C.; performed experiment: H.A.; data analysis: H.-Y.L., D.S., S.H.C., H.C., and T.K.H.; data interpretation: all authors; writing: H.A.; review: I.-H.J., and J.-W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Golden Seed Project (Grant no. 213007–05-5-SBH20) from the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data for this manuscript are available at NCBI GenBank numbers CP039873–CP039885.
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Yun, M.-J.; Oh, S.-I.; Lee, M.-S. Antioxidative and antimutagenic effects of Agaricus bisporus ethanol extracts. J. Korean Soc. Food Sci. Nutr. 2009, 38, 19–24. [Google Scholar] [CrossRef][Green Version]
- Jeong, S.C.; Jeong, Y.T.; Yang, B.K.; Islam, R.; Koyyalamudi, S.R.; Pang, G.; Cho, K.Y.; Song, C.H. White button mushroom (Agaricus bisporus) lowers blood glucose and cholesterol levels in diabetic and hypercholesterolemic rats. Nutr. Res. 2010, 30, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Royse, D.J.; Baars, J.; Tan, Q. Current overview of mushroom production in the world. In Edible and Medicinal Mushrooms: Technology and Applications; Zied, D.C., Pardo-Giménez, A., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2017; Chapter 2; pp. 5–13. [Google Scholar]
- Sonnenberg, A.; Baars, J.; Hendrickx, P.; Lavrijssen, B.; Gao, W.; Weijn, A.; Mes, J. Breeding and strain protection in the button mushroom Agaricus bisporus. In Proceedings of the 7th International Conference of the World Society for Mushroom Biology and Mushroom Products, Arcachon, France, 4–7 October 2011; pp. 7–15. [Google Scholar]
- Bretting, P.; Widrlechner, M.P. Genetic markers and plant genetic accession management. In Plant Breeding Reviews; NCRPIS Publications and Papers: Ames, IA, USA, 1995; Volume 13, pp. 11–86. [Google Scholar]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Loftus, M.; Moore, D.; Elliott, T. DNA polymorphisms in commercial and wild strains of the cultivated mushroom, Agaricus bisporus. Theor. Appl. Genet. 1988, 76, 712–718. [Google Scholar] [CrossRef]
- Moore, A.; Challen, M.; Warner, P.; Elliott, T. RAPD discrimination of Agaricus bisporus mushroom cultivars. Appl. Microbiol. Biotechnol. 2001, 55, 742–749. [Google Scholar] [CrossRef]
- Foulongne-Oriol, M.; Spataro, C.; Savoie, J.-M. Novel microsatellite markers suitable for genetic studies in the white button mushroom Agaricus bisporus. Appl. Microbiol. Biotechnol. 2009, 84, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Raveendar, S.; An, H.; Oh, Y.-L.; Jang, K.-Y.; Kong, W.-S.; Ryu, H.; So, Y.-S.; Chung, J.-W. Development of polymorphic simple sequence repeat markers using high-throughput sequencing in button mushroom (Agaricus bisporus). Mycobiology 2018, 46, 421–428. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Jo, I.-H.; Oh, Y.-L.; Jang, K.-Y.; Kong, W.-S.; Sung, J.-K.; So, Y.-S.; Chung, J.-W. Molecular characterization of 170 new gDNA-SSR markers for genetic diversity in button mushroom (Agaricus bisporus). Mycobiology 2019, 47, 527–532. [Google Scholar] [CrossRef]
- Oh, Y.-L.; Choi, I.-G.; Kong, W.-S.; Jang, K.-Y.; Oh, M.J.; Im, J.-H. Evaluating genetic diversity of Agaricus bisporus accessions through phylogenetic analysis using single-nucleotide polymorphism (SNP) markers. Mycobiology 2021, 49, 61–68. [Google Scholar] [CrossRef]
- Sonnenberg, A.S.; Gao, W.; Lavrijssen, B.; Hendrickx, P.; Sedaghat-Tellgerd, N.; Foulongne-Oriol, M.; Kong, W.-S.; Schijlen, E.G.; Baars, J.J.; Visser, R.G. A detailed analysis of the recombination landscape of the button mushroom Agaricus bisporus var. bisporus. Fungal Genet. Biol. 2016, 93, 35–45. [Google Scholar] [CrossRef]
- Sonnenberg, A.S.; Baars, J.J.; Gao, W.; Visser, R.G. Developments in breeding of Agaricus bisporus var. bisporus: Progress made and technical and legal hurdles to take. Appl. Microbiol. Biotechnol. 2017, 101, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Weijn, A.; Baars, J.J.; Mes, J.J.; Visser, R.G.; Sonnenberg, A.S. Quantitative trait locus mapping for bruising sensitivity and cap color of Agaricus bisporus (button mushrooms). Fungal Genet. Biol. 2015, 77, 69–81. [Google Scholar] [CrossRef]
- Gao, W.; Baars, J.J.; Maliepaard, C.; Visser, R.G.; Zhang, J.; Sonnenberg, A.S. Multi-trait QTL analysis for agronomic and quality characters of Agaricus bisporus (button mushrooms). AMB Express 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Edwards, D.; Forster, J.W.; Chagné, D.; Batley, J. What Are SNPs? In Association Mapping in Plants; Oraguzie, N.C., Rikkerink, E.H.A., Gardiner, S.E., De Silva, H.N., Eds.; Springer: New York, NY, USA, 2007; pp. 41–52. [Google Scholar]
- Moon, S.; Lee, H.-Y.; Ka, K.-H.; Koo, C.-D.; Ryu, H. Development of a CAPS marker for the identification of the Lentinula edodes cultivar, ‘Sanmaru 2ho’. J. Mushroom 2018, 16, 51–56. [Google Scholar]
- Konieczny, A.; Ausubel, F.M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993, 4, 403–410. [Google Scholar] [CrossRef]
- Blair, M.W.; Cortés, A.J.; Penmetsa, R.V.; Farmer, A.; Carrasquilla-Garcia, N.; Cook, D.R. A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2013, 126, 535–548. [Google Scholar] [CrossRef]
- Parsons, B.L.; Heflich, R.H. Genotypic selection methods for the direct analysis of point mutations. Mutat. Res. Rev. Mutat. Res. 1997, 387, 97–121. [Google Scholar] [CrossRef]
- Neff, M.M.; Neff, J.D.; Chory, J.; Pepper, A.E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998, 14, 387–392. [Google Scholar] [CrossRef]
- Arlotta, C.; Ciacciulli, A.; Strano, M.C.; Cafaro, V.; Salonia, F.; Caruso, P.; Licciardello, C.; Russo, G.; Smith, M.W.; Cuenca, J.; et al. Disease resistant citrus breeding using newly developed high resolution melting and CAPS protocols for Alternaria brown spot marker assisted selection. Agronomy 2020, 10, 1368. [Google Scholar] [CrossRef]
- Choi, Y.R.; Lee, J.Y.; Hwang, S.; Kim, H.U. PCR-based InDel marker associated with powdery mildew-resistant MR-1. Agronomy 2020, 10, 1274. [Google Scholar] [CrossRef]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. TRENDS Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Pamilo, P.; Nei, M. Relationships between gene trees and species trees. Mol. Biol. Evol. 1988, 5, 568–583. [Google Scholar]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes-application to human mitochondrial-DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Singh, N.; Choudhury, D.R.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.; Singh, N.; Singh, R. Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS ONE 2013, 8, e84136. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Ingvarsson, P.K. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol. Biol. Evol. 2009, 27, 650–660. [Google Scholar] [CrossRef]
- Wang, J.; Raskin, L.; Samuels, D.C.; Shyr, Y.; Guo, Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics 2015, 31, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.L.; Lee, H.; Popodi, E.; Townes, J.P.; Tang, H. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2015, 112, E5990–E5999. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Van Inghelandt, D.; Melchinger, A.E.; Lebreton, C.; Stich, B. Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor. Appl. Genet. 2010, 120, 1289–1299. [Google Scholar] [CrossRef]
- Yang, X.; Xu, Y.; Shah, T.; Li, H.; Han, Z.; Li, J.; Yan, J. Comparison of SSRs and SNPs in assessment of genetic relatedness in maize. Genetica 2011, 139, 1045. [Google Scholar] [CrossRef]
- Tsykun, T.; Rellstab, C.; Dutech, C.; Sipos, G.; Prospero, S. Comparative assessment of SSR and SNP markers for inferring the population genetic structure of the common fungus Armillaria cepistipes. Heredity 2017, 119, 371–380. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, X.; Li, D.; Liu, Y.; Song, B.; Zhang, C.; Wang, Q.; Chen, M.; Zhang, Z.; Li, Y. Identification of resistance to wet bubble disease and genetic diversity in wild and cultivated strains of Agaricus bisporus. Int. J. Mol. Sci. 2016, 17, 1568. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).