Abstract
Antifungal susceptibility testing (AST) has come to establish itself as a mandatory routine in clinical practice. At the same time, the mycological diagnosis seems to have headed in the direction of non-culture-based methodologies. The downside of these developments is that the strains that cause these infections are not able to be studied for their sensitivity to antifungals. Therefore, at present, the mycological diagnosis is correctly based on laboratory evidence, but the antifungal treatment is undergoing a growing tendency to revert back to being empirical, as it was in the last century. One of the explored options to circumvent these problems is to couple non-cultured based diagnostics with molecular-based detection of intrinsically resistant organisms and the identification of molecular mechanisms of resistance (secondary resistance). The aim of this work is to review the available molecular tools for antifungal resistance detection, their limitations, and their advantages. A comprehensive description of commercially available and in-house methods is included. In addition, gaps in the development of these molecular technologies are discussed.
1. Introduction
Antifungal susceptibility testing is an essential tool in different clinical scenarios. Standardized protocols (from the Clinical and Laboratory Standards Institute (CLSI)and from the European Committee on Antimicrobial Susceptibility Testing (EUCAST)) and commercially available methods (some of them automated) are able to detect resistant fungal strains, to guide antifungal therapies, and to offer reliable epidemiological data on antifungal resistance [1]. When these methodologies seemed to have come to establish themselves as a mandatory routine in any clinical microbiology laboratory, the mycological diagnosis seems to have headed in the direction of non-culture-based methodologies. These techniques, which include serological and molecular-based tools, improve mycological diagnosis in speed, sensitivity, and specificity. A proven invasive mycosis can be diagnosed by amplifying fungal DNA from a paraffin-embedded tissue [2] or by detecting a fungal antigen (e.g., Cryptococcus spp.) with a speed previously dreamed of [3]. Moreover, there are commercially available molecular-based methods able to detect fungal DNA with good sensitivity and speed [4]. The downside of these developments is that the strains that cause these infections are not available to study their sensitivity to antifungals. Therefore, at present, the mycological diagnosis is correctly based on laboratory evidence, but the antifungal treatment is undergoing a growing tendency to revert back to being empirical, as it was in the last century. Additionally, reports of clinical resistance to antifungals are steadily increasing, leaving mycologists with the dilemma of having higher rates of clinical resistance and fewer isolates to study. One of the explored options to circumvent these problems is to couple non-cultured based diagnostics with molecular-based detection of intrinsically resistant organisms and the identification of molecular mechanisms of resistance (secondary resistance). This idea would also overcome one of the main drawbacks of conventional AST techniques, the time needed to get confident results (>24 h) [5,6,7,8].
The aim of this work is to review the available molecular tools for antifungal resistance detection, their limitations, advantages, and development gaps.
2. Intrinsic Resistance Detection
As with other microorganisms, antifungal resistance is a broad concept that can be divided into clinical and microbiological resistance. The former was defined as the lack of inhibition of a microorganism in the infection site and it is related to different factors dependent on the drug, the patient, or both, rather than with the microorganism that causes the infection [9,10]. On the other hand, microbiological resistance depends on the particular characteristics of the microorganism and it can be subdivided into primary or intrinsic and secondary or acquired resistance. The results obtained in the AST give an idea of both microbiological resistance types.
Intrinsic microbiological resistance is the innate ability of a fungal species to resist the activity of a particular antifungal drug due to its inborn functional or structural features (e.g., absence of the drug target, inaccessibility of the drug into the cell). This resistance is exhibited by all strains of the same species of a fungus and is not related to exposure to the antifungal. On the other hand, secondary microbiological resistance is developed after antifungal treatment (in vivo or after environmental exposure) and is observed in particular strains of a normally susceptible species. These resistance phenotypes are due to genetic alterations that are manifested in a stable or in a transitory way [11,12]. Intrinsic and secondary resistance usually share the same molecular mechanisms. As an example, we can state the intrinsic fluconazole (FLC) resistance in Aspergillus fumigatus due to the naturally occurring T301I substitution at its Cyp51Ap [13] and the secondary mechanism of FLC resistance in Candida albicans attributable to an equivalent substitution (T315A) at its Erg11p [14].
Intrinsic resistance was defined by CLSI as “inherent or innate (not acquired) antimicrobial resistance which is reflected in wild-type antimicrobial patterns of all or almost all representatives of a species. Intrinsic resistance is so common that susceptibility testing is unnecessary” [15]. EUCAST lists a species as intrinsically resistant to an agent when “all or a vast majority of their strains exhibit minimal inhibitory concentration (MIC) values that are so high that the agent should not be considered for either therapy or clinical susceptibility testing” [16]. These definitions coincide in that AST is not necessary to be performed since all the strains of the species are resistant to that particular drug. Thus, in some microorganisms/drug combinations, taxonomy would act as an AST subrogate marker. On this topic, the CLSI already included in M60 document the sentence “Isolates of Candida krusei are assumed to be intrinsically resistant to FLC, so their MICs should not be interpreted using this scale.” (referring with scale to susceptible, susceptible dose-dependent, and resistant) [17]. Moreover, other species as Aspergillus fumigatus or other groups of fungi (Basidiomycetes and Mucorales) are being evaluated to be considered intrinsically resistant to FLC and to echinocandins, respectively.
There are powerful tools able to accurately identify intrinsically resistant fungal species from culture as Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) [18,19,20,21]. However, the identification of filamentous fungi is often compromised by difficulties in the extraction steps and by the fact that some cryptic resistant species are not yet included in the databases. The first problem could be resolved by performing extended extraction procedures [22] or using special culture media [23]. Despite the usefulness of MALDI-TOF as a technique, molecular-based identification (DNA sequencing) is still the gold-standard for fungal taxonomy [24].
Since the beginning, DNA-based identification methods in medical mycology faced a huge problem: choosing a gene (or a portion of a gene or genome region) useful in a clinical laboratory. To fulfill this objective, this hypothetical gen would have the following characteristics: it has to (i) have high inter-species and low intra-species variability, (ii) have a short sequence with high discriminatory power, (iii) be unique for all fungal species, (iv) make use of universal primers (same pair of primers for all species), and (v) be easily amplified [24]. These five points were condensed on the DNA barcoding concept that was aimed to allow accurate and fast species identification [25,26,27,28,29,30]. However, the truthfulness of these molecular-based identification procedures depends on the correct previous taxonomic classification of the used control strain [31]. This fact leads us to a paradox where molecular taxonomy’s objective is to improve classical taxonomy, but the former needs the last to achieve this goal.
There are several reports published regarding which gene to choose. However, any of the described molecular markers fulfill all the described criteria. The one that came closest to be the standard for fungi is the gene region known as ribosomal DNA internal transcribed spacers regions (ITS) [26,32,33]. This region is being successfully used for Candida spp. identification. Several reports used ITS-based PCRs to rapidly identify Candida spp. at the species level, including intrinsically resistant (e.g., C. krusei/FLC) [34] and less susceptible cryptic species [35,36,37,38]. Similarly, for Mucorales, ITS alone is a correct DNA marker with enough discriminative power to identify the currently accepted morphospecies of Mucor, Lichtheimia, and Rhizopus [39,40]. For other genera, ITS sequencing or ITS-based PCR identification is not enough or is not the correct method. For Trichosporon spp., intergenic spacer regions (IGS1) (and not ITS) sequencing unambiguously identify all Trichosporon species [41], while for most pathogenic filamentous fungi, a multilocus DNA-barcoding approach is needed [42,43]. Aspergillus spp. identification to sections/complexes level is based on sequencing of ITS regions [44,45] but a secondary marker is needed to allow the identification at the species level. For this genus, calmodulin (CaM), Beta-tubulin (BenA), and the second-largest subunit of the RNA polymerase II (RPB2) were proposed as taxonomy secondary markers. The last is quite difficult to amplify in certain species. For BenA, a different number of introns and paralogous genes were described making the amplification with one set of primers difficult. On the other hand, CaM can be amplified easily with the same primer set in most of the species making this gene the proposed secondary marker for Aspergillus spp. identification [46,47,48]. For Fusarium species, its identification is based on the sequencing of the ITS regions plus RBP2 and a portion of the translation elongation factor 1 alpha (TEF1) [49].
The described knowledge allowed the development of several techniques able to identify intrinsically resistant and cryptic (less susceptible) species. Most of these techniques and procedures designed to identify fungal pathogens were developed in research laboratories (in-house PCRs) and were barely used in clinical settings. On the other hand, there are few commercially available molecular-based methods in clinical use.
2.1. Intrinsic Resistance Detection by Commercially Available Molecular Taxonomy-Based Method
There are few FDA-cleared molecular-based methods capable of identifying fungal pathogens [50]. Some of them are able to identify them directly from positive blood culture bottles (e.g., FilmArray, Biofire–Biomerieux; Candida PNA FISH assay, OpGen; T2Candida-Biosystems and SeptiFast-Roche) [51] or from other clinical samples [52]. The major common limitation of these methods is the narrow coverage for fungal pathogens and the low impact in the selection of specific antifungal treatment [4,51]. The newest version of Biofire Filmarray is able to detect C. albicans, C. parapsilosis sensu stricto, C. glabrata sensu stricto, C. tropicalis, C. krusei (intrinsic FLC-resistant), Cryptococcus neofromans/gattii complex (intrinsic echinocandin resistant), and C. auris (multidrug-resistant). This last species showed a high prevalence of FLC resistance (>90% of the strains are considered FLC resistant). Despite that almost all C. auris strains show this phenotype, it was demonstrated that this resistance is acquired [53]. T2Candida panel (Magnetic resonance-based from T2 Biosystems) and LightCycler® SeptiFast MGRADE system (Real-Time PCR from Roche Diagnostics) are able to rapidly (<5 h) diagnose candidemia utilizing whole blood directly from patients with a great level of detection (1CFU/mL for T2Candida) [54,55], a good specificity (>95% and >72%, respectively) and sensitivity (>60% and >90%, respectively) in real-life clinical settings [56,57]. However, both methods are able to identify only the most commonly isolated Candida spp. (C. albicans, C. tropicalis, C. parapsilosis, C. krusei, and C. glabrata) and Aspergillus fumigatus sensu stricto (Septifast only) [52,57]. The described diagnostic capability allows the clinicians to rapidly start an antifungal therapy helping with antifungal stewardship but it gives false-negative results when none of the named Candida spp. (or A. fumigatus) are the etiological agent of the infection (a huge problem especially given the shift in Candida spp. epidemiology) [52]. Moreover, considering that the current first-line treatment for candidemia are echinocandins, the identification to species level of these five common Candida spp. has little impact on the choice of antifungal treatment since none are intrinsically resistant to this class of antifungals (with the exception of Biofire that can detect Cryptococcus spp.). However, the Infectious Diseases Society of America (IDSA)guidelines propose echinocandins as the first treatment option for deep Candida infections (including candidemias) caused by any species of this genus. [58]. This guideline only suggests to study the echinocandin susceptibility of the strains isolated from patients who had a prior echinocandin treatment and in those patients with C. glabrata or C. parapsilosis infections. In this case, none of the described methods (filmarrays, T2, Septicheck, etc) is helpful since no strain is available to perform the required AST (as they are non-culture based methods). On the other hand, both methods are able to identify the intrinsically FLC-resistant C. krusei (Biofire can identify C. auris also) and C. glabrata (higher FLC doses as treatment).
Another important development in fungal diagnostics is the real-time PCR kit, commercialized by Pathonostics, designed to diagnose aspergillosis and to detect markers of secondary azole resistance named AsperGenius®. The tests are divided into two panels, one able to identify the etiological agent and the other to detect azole resistance. The first panel identifies aspergillosis caused by A. fumigatus, Aspergillus terreus, and Aspergillus spp. [59,60]. This test was evaluated clinically with good results in serum (sensitivity > 78% and specificity of 100% for species with a limit of detection >10 genomic units of A. fumigatus) [60], plasma (80% sensitivity and >77% specificity) [61], and in broncoalveolar lavage (BAL) (>80% sensitivity and >90% specificity for hematological and ICU patients) [62,63]. The biggest advantage of this diagnostic kit is its ability to detect the intrinsic amphotericin B resistant A. terreus and the fact that it has no false-negative results for aspergillosis caused by non-A. fumigatus and non-A. terreus (other available kits are able to detect only A. fumigatus) [64]. Moreover, and as an unexpected result since the kit was not designed to do so, AsperGenius® can detect intrinsically azole and amphotericin B resistant cryptic species of the Aspergillus section Fumigati as A. lentulus and A. felis. These results were obtained by using the AsperGenius® resistance panel where the TR34 target (see details in the next section) was negative for these two cryptic species and the melting curves for the CYP51A mutations were also different [65].
After this short summary of the main commercially available molecular-based method able to detect intrinsically resistant fungi, it is clear that a bigger effort is needed to include more species to the existing panels or to design more comprehensive ones. Efforts should be focused on the differentiation of the fungal groups of species with known intrinsic resistance to certain antifungals directly from clinical samples. The main groups of fungi to be differentiated in order to select a correct antifungal treatment and to have a real impact on mortality [66] are: (i) ascomycetous from basidiomycetous yeasts, (ii) agents of hialohifomycoses from Mucorales, and (iii) Candida spp. from filamentous fungi since the last of each of these pairs of pathogens are intrinsically resistant to echinocandins, voriconazole, and FLC, respectively (Figure 1). One promising tool able to fulfill at least in part these requirement seems to be the PCR-electrospray ionization mass spectrometry that is being tested for mycoses diagnostics [67,68,69,70,71].
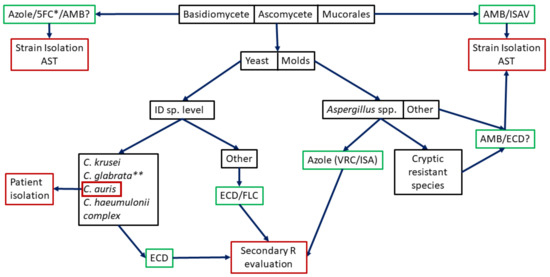
Figure 1.
Potential algorithm for intrinsic resistance studies. Black boxes indicate where potential molecular methods would be used (most not designed yet). Green boxes indicate used empirical treatments. Red boxes indicate needed actions (patient isolation if C. auris is identified, the need of secondary resistance evaluation or the strain isolation and AST). 5FC: 5-fluorocytosine, AMB: amphotericin B, ISAV: isavuconazole, ID: Identification, ECD: echinocandins, FLC: fluconazole, VRC: voriconazole. * Combined with AMB for Cryptococcus spp. ** FLC Susceptible Dose-Dependent/Resistant.
2.2. Intrinsic Resistance Detection by in-House Molecular-Based Method
Several reports of in-house PCRs demonstrated the feasibility and the practical potential of the different methods to uncover intrinsic resistant or less susceptible fungal species, both starting from colonies and from clinical samples. Within the in-house methods developed, the ones that stand out are able to detect cryptic Candida spp. that showed reduced susceptibilities to different antifungal agents as the C. glabrata, C. parapsilosis, and C. albicans species complexes. Different formats were used including a multiplex PCR able to detect all nine species (C. glabrata sensu stricto, C. nivariensis, C. bracarensis, C. parapsilosis sensu stricto, C. orthopsilosis, C. metapsilosis, C. albicans, C. dubliniensis, and C. africana) [72], several multiplex PCRs for the detection of each species of one of the complexes [38,73,74], PCRs coupled to restriction enzyme digestions [75,76], high resolution melting curves [77], a multiplex real-time PCR using molecular beacons [75], etc. Some of these methods were successfully used to evaluate the prevalence of these cryptic species in strain collections [36,78,79,80]. In response to the need for molecular tools to identify C. auris due to its high FLC resistance rate (>90%), a classical and a real-time PCRs based on ITS amplification were published. The first included a one single tube PCR able to uncover C. auris and C. haeumulonii with an internal reaction control [81] and a specific PCR for C. auris DNA amplification [82]. The published real-time PCRs are capable to detect C. auris alone or C. auris, C. haeumulonii, C. duobushaemulonii, and C. lusitaniae by analyzing melting curves [82].
Very recently, qPCRs (Sybrgreen and with TaqMan probes) targeting the C. auris ITS2 region were designed in order to detect C. auris DNA from skin and surveillance samples (limits of detection of 4 and 1 C. auris CFU/PCR, respectively) [83,84,85]. Moreover, these qPCRs can be coupled with a second panel of primers to uncover markers of secondary resistance as ERG11 and FKS1 mutations responsible for FLC and echinocandin resistance, respectively [86]. These C. auris detection tools are examples of the ideality of molecular diagnostics of antifungal resistance using clinically important samples. They showed the potential, sensitivity, and usefulness of these tools. However, as in many other molecular tools, a suspicion of the presence in a sample of a particular species is needed in order to select the molecular method to detect them. Thus, these tools are useful in confirming an infection or colonization caused by the suspected agent and have a great negative predictive value.
For Mucorales, most of the in-house methods are designed to be used directly from clinical samples since it is difficult to isolate these fungi from the culture [87,88]. The usefulness of these molecular methods is that a positive result is the needed evidence to start an antifungal treatment with amphotericin B. These methods are mainly based on the amplification and subsequent sequencing of rDNA using total DNA isolated from formalin-fixed and fresh tissues [89,90,91,92,93]. Moreover, there were reports of using different molecular targets as a semi-nested PCR able to identify several Mucorales species [94], an RFLP-PCR used in biopsies [22,95], a pan-Mucorales PCR based on the amplification of the spore coating protein (CotH) [96], a qPCR able to detect Rhizomucor spp., Mucor spp. Rhizopus spp. and Lichtheimia spp. in broncoalveolar lavage samples [97], a qPCR based on ITS amplification [98], etc.
2.3. Is Species Identification Enough as Surrogate Marker of Intrinsic Resistance or Should We Go Further?
There are some examples of fungal species that were divided into clades, varieties, or types that showed differences in their antifungal susceptibility. One of the first noted examples was the differences in 5-fluorocytosine (5FC) susceptibilities of different C. albicans clades (resistance to 5FC seems restricted to clade I) [99]. Other clinically relevant examples to be cited are Cryptococcus neoformans/gattii complex species, C. auris, and Trichophyton mentagrophytes. The named basidiomyceteous yeasts are divided into genetic varieties that have different epidemiological cut-off values for multiple antifungal agents [17,100,101]. C. auris was firstly considered intrinsically resistant to FLC (MIC > 64 µg/mL) but after studying a more geographically diverse collection of strains it was established that this phenotype was shown by strains of the clade I, III, and IV, while most of the strains of the clade II showed lower FLC MICs [102,103]. Similarly, T. mentagrophytes was divided into different genetic types. Type VIII isolates (from India) showed a high level of terbinafine resistance [104].
Looking at this data, we can assume that the identification of clades, types, and varieties of particular species showing reduced antifungal susceptibility would be a useful surrogate marker of resistance. However, to do so, continuous antifungal surveillance and molecular epidemiology studies are needed to increase the number of species to be included in this list.
3. Secondary Resistance Detection
The number of described secondary antifungal resistance mechanisms differs between drugs. For amphotericin B, there are few reports on secondary antifungal resistance. It is so rare that it has raised questions about the ability of AST methods to detect it [105,106]. These secondary resistance mechanisms were described 40 years ago or were barely studied [107,108]. For azole drugs, there is a wide range of different mechanisms [10] while for echinocandins, secondary clinical resistance seems mainly linked to amino acid substitutions at its target (Fksp) [10,11]. It should be also stated that clinically important secondary resistance is related to genetically stable mutants selected during treatment in a multistep process. This process involves a cell stress step produced by the drug, followed by an adaptation step that ends up in a stable escape mutant. These adaptation steps include the overexpression of genes that compensate for the drug-produced alterations, overexpression of stress response pathways, chromosome rearrangements, etc. These processes are usually reversible if the drug pressure is reduced or is eliminated. On the other hand, stable escape mutants retain the phenotypic traits that classified the strain as resistant (MICs surpassing the clinical breakpoint) whether the drug pressure is maintained or not [109]. These last kinds of mutants are the ones that shall be detected by molecular tools to confirm a resistant phenotype and in some cases, its detection is considered an independent risk factor for treatment failure [110,111].
The ways in which fungi acquire the ability to escape the action of antifungals can be divided into three groups of mechanisms: (i) alteration of drug-target interaction (target modification and target hyper-production), (ii) reduction of the cytoplasmic concentration of the drug (overexpression of efflux pumps and reduction of drug penetration), and (iii) metabolic by-pass (Figure 2). The prevalence of each of these mechanisms depends on the drug/fungi combination. Briefly, for azole agents, overexpression of efflux pumps followed by mutations at the azole drug target (ERG11) and the overexpression of ERG11 are the main resistance mechanisms in Candida spp. On the other hand, CYP51A substitutions followed by CYP51A overexpression coupled with CYP51A substitutions are the main mechanisms of azole resistance in Aspergillus spp. Conversely, Cryptococcus spp. azole resistance mechanisms seem to be related mainly with ERG11 mutations and chromosome rearrangements that lead to the overexpression of ERG11 and transcription factors genes that increase the expression of efflux pumps. Turning to echinocandins, target modification (FKS mutations) is the most important mechanism of echinocandin resistance despite the studied fungal species. For a more detailed description of molecular mechanisms of antifungal resistance, interested readers are referred to the following references [11,72,112,113,114,115,116,117,118].
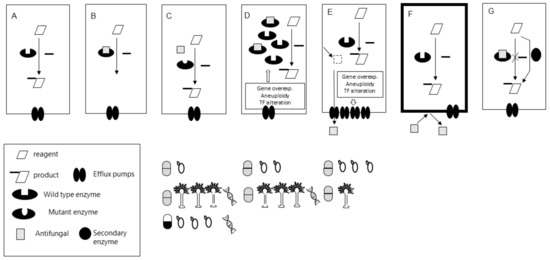
Figure 2.
(A) Essential enzymatic reaction for a fungal cell. (B) Inhibition of the reaction by the antifungal drug. (C,D) Alterations of the interaction drug-enzyme. (C) Mutation on the drug target (less efficient or non-interaction). (D) Overexpression of the drug-target. In the box are the most commonly described mechanisms leading to overexpression. (E,F) Reduction of the cytoplasmic drug concentration. (E) Overexpression of efflux pumps (e.g., CDRs) by different underlying mechanisms (described in the box). (F) Impermeability. The drug is not trespassing the membrane (e.g., no transporter available as happens with 5-fluorocytosine and Histoplasma capsulatum. (G) Metabolic bypass. Grey and black and White pill symbols represent azole and echinocandin antifungals resistance mechanisms, respectively. Yeast and Aspergillus graphics under each of the mechanisms represent the description of each particular mechanism in Candida spp. and Aspergillus spp. respectively. The number of Yeasts and Aspergillus graphs show the relative prevalence of each of the mechanisms in Candida and Aspergillus spp. clinical strains, respectively. DNA graphs represent molecular tools’ availability to detect that particular mechanism in a drug/fungi combination.
3.1. The Bottlenecks of Secondary Resistance Molecular Detection
For intrinsic resistance detection, a wide range of taxonomy markers are available. As mentioned, one of the most used is the ITS region which is a multicopy universal region of the fungal genome. These characteristics imply that one pair of primers can be used to detect multiple fungal species with high sensitivity (each fungal cell can carry between >10 to >1000 copies of this DNA region) [119]. Oppositely, for the case of resistance markers, there are several bottlenecks in terms of sensitivity and methodological complexity. The first is obviously the low sensitivity since the genes to be evaluated are a single copy (two in diploid organisms). The second is that we should uncover mutations in a gen rather than detecting the presence of a gen. The third is that different mechanisms involving different genes and/or mutations are responsible for similar or equal phenotypes. The fourth is that in some cases we must evaluate overexpression and not just the presence and/or existence of mutations. Fifth, in the case of diploid organisms, there are mechanisms that are dominant (a mutant allele gives the phenotype) limiting the use of classic PCR. Sixth, there are no universal primers (each species uses different oligonucleotides), so we must identify the etiological agent before detecting resistance mechanisms. The seventh is that some phenotypes are the result of a combination of mechanisms. The eighth is that we can only detect known mechanisms, as opposed to phenotypic or whole-cell methods such as MIC assessment that detect all mechanisms.
3.2. Available Molecular Tools. Which Secondary Mechanisms Are We Able to Detect?
Having in mind the described bottlenecks, in-house and commercially available tools were designed to detect the mechanisms regarded as the unique responsibilities of particular resistance phenotypes. Thus, most of the published tools are able to detect some of the CYP51A and FKS mutations linked with triazole and echinocandin resistance in Aspergillus fumigatus and Candida spp., respectively.
There are no standard methods to detect alterations on gene coding antifungal targets. However, several methods were published to be used mostly from isolates. The first reports simply used PCR amplification followed by sequencing [120,121,122,123,124,125]. Later specific PCR-based methods designed for the detection of particular mutations were published using different methods and formats (Table 1 and Table 2).

Table 1.
Molecular tools for the detection of mechanisms of triazole resistance in Aspergillus spp.

Table 2.
Molecular tools for the detection of mechanisms of azole and echinocandin resistance in Candida spp.
3.2.1. Triazole Secondary Resistance in Aspergillus spp.
The development of molecular tools for the detection of triazole resistance mechanisms in A. fumigatus was less complicated than for other fungal species since fewer mechanisms were described. Resistance linked to mutations at CYP51A has been detected by in-house classical PCRs (followed by sequencing, digestion, minisequencing-SnaPshot) [131,132,133,134,135], astringent classical PCRs [139], real-time PCRs (coupled with taqman probes, molecular beacons probes, locked primers, sybrgreen followed by melting curves analysis and FRET probes) [126,127,128,129,130,136], loop-mediated isothermal amplification (LAMP) [138], whole-genome sequencing (WGS) [140], etc. The majority of these methods are able to detect the most common mechanisms as the promoter alterations (TR34-L98H and TR46-Y121F) (Table 1). The resistance mechanisms that include promoter alterations are also detected by the two commercially available methods (not FDA approved yet) named AsperGenius [61,62,63,141] and MycoGENIE [142]. Both diagnostic kits share the format (multiplex real-time PCR) and the capacity of detection of TR34-L98H mutations. As described earlier, AsperGenius is able to detect DNA of intrinsically resistant species while MycoGENIE is able to detect only wild-type and resistant A. fumigatus sensu stricto isolates. AsperGenius also covers the second most common mechanism of triazole resistance (TR46-Y121F-T289A). One of the few molecular-based methods capable to detect other mutations (G54, M220, and G138C) is an in-house real-time PCR coupled with molecular beacons designed in a two-panel format. The first panel detects itraconazole or triazole cross-resistance while the second panel can differentiate G54W (ITC-PSC cross-resistance) from other substitutions at the residue 54 conferring ITC resistance. Moreover, in the same panel, molecular beacons that confirm resistance mechanisms were included [126]. For less prevalent mechanisms, quantitative real time PCRs and PCR followed by minisequencing were reported as feasible tools to detect the overexpression of efflux pumps and the uncommon mutations at CYP51B, respectively [129,144] (Figure 3).
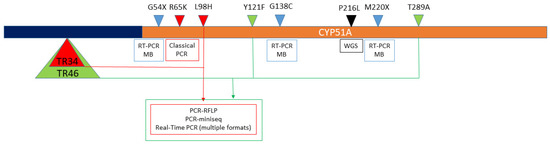
Figure 3.
Mutations in A. fumigatus cyp51A (brown bar) and its promoter (5´UTR) region (dark blue bar) detected by the available molecular methods. Boxes indicate the molecular methods used to detect the mechanisms that are over them. Arrow heads represent the mutations linked with triazole resistance. Green and red arrow heads show the combined mechanisms that include CYP51A mutations together with promoter alterations described as tandem repetitions (TR). Light blue arrow and black heads represent single-nucleotide mutations detected by RT-PCR coupled with molecular beacons (RT-PCR MB) and whole-genome sequencing (WGS).
Another point to consider is that five out of 15 (33%) described methods were tested using clinical samples (sputum, BAL, Formalin-fixed, and paraffin-embedded tissues). Two of them are commercially available methods (see Table 1).
3.2.2. Azole Resistance in Candida spp.
Turning to Candida spp., the panorama is different. Molecular mechanisms of azole resistance in Candida spp. are relatively well studied. However, its detection is complicated since each species can have different mechanisms and because an individual mechanism is not always strictly correlated with strain´s MIC. Firstly, some Erg11p amino acid substitutions in Candida spp. were described but not validated as exclusive causatives of azole resistance [125,153]. Secondly, in the majority of the cases, the resistance phenotype is due to a combination of mechanisms. They can include two or more of the following: (i) mutations at different genes of the ergosterol biosynthesis pathway (especially in ERG11 but also in ERG3) [125,153,154], (ii) overexpression of ERG11 due to the gain of function mutations in transcription factors or due to aneuploidies (complete or partial chromosome duplications) [155,156], overexpression of efflux pumps [157], etc. (Figure 1). Additionally, it was demonstrated that aneuploidies can lead to tolerance or resistance to multiple unrelated drugs [158]. This multiplicity of mechanisms makes the use of a multiplex format mandatory for the molecular detection of resistance to azoles in Candida spp., meaning, the sequencing of several genes coupled with the expression evaluation of others.
Some of the latest proposed approaches are the use of next-generation sequencing (NSG) and whole-genome sequencing (WGS) to detect several mutations in different genes of the ergosterol biosynthesis pathway (ERG11 and ERG3) and transcription factors that regulate efflux pump expression (e.g., TAC1 and PDR1) together with FKS mutations [102,149]. These techniques are able to detect novel DNA alterations but they are limited to reference labs due to their high cost (equipment, reagents, and the complexity of data analysis) [149]. In the few published reports that used NGS, strain collection was studied, but standardized AST was firstly used as a screening tool. Thus, NGS was used to uncover the mechanism responsible for the already known resistance phenotype and not as a diagnostic tool of antifungal resistance [102,150].
The most important exception to what was stated is the detection of the FLC resistance mechanism in C. auris. Unlike other Candida spp., FLC resistance in C. auris was linked only to a limited number of Erg11p substitutions (mostly Y132F, K143R, F126T, I466M, and Y501H). The prevalence of these mutations is different in each of the described 4 geographical clades of C. auris and some substitutions are almost exclusively found in one clade (e.g., I446M in clade IV–South American) [102,151]. This fact makes feasible the detection of C. auris resistant strains by molecular tools. Xin Hou et al. reported an asymmetric real-time PCR coupled with molecular beacons able to identify Y132F and K143R Erg11p substitutions together with an FKS mutation (S639F) responsible for echinocandin resistance [152].
3.2.3. Echinocandin Resistance in Candida spp.
As for C. auris, the molecular detection of echinocandin resistance in Candida spp. can be performed using relatively simple methods. This detection is aided by the fact that resistance to echinocandins has two fundamental characteristics: it is almost exclusively linked to a limited number of mutations [11,159] and its presence is an independent risk factor for echinocandin therapy failure [111]. Although echinocandin resistance was described in several Candida spp., its prevalence is higher in C. glabrata and in C. albicans. This fact is reflected in the published technological developments. The reported molecular tools for the detection of C. glabrata and C. albicans FKS mutants include classical PCRs [36,144,147], Sanger [150], and next-generation sequencing for both species [149]. There are methods designed exclusively for the detection of C. albicans FKS mutants as a real-time PCR coupled with molecular beacons [148]. On the other hand, asymmetric real-time PCR coupled with molecular beacons with melting curve analysis [143], Luminex Mag-Pix assay [146], and whole-genome sequencing were used to detect C. glabrata FKS mutants [145]. More details on the methodologies including their limitations are described in Table 2.
Most of the described tools detect the most prevalent mutations that are responsible for the most pronounced phenotype (e.g., at the residues S629 and S663 in C. glabrata Fks1p and Fks2p). On the other hand, the two reported classical PCRs are also able to detect less common mutations that showed lower MIC values as the substitutions at the residues D648, P649, and R1361 at C. albicans Fks1p and D632 at C. glabrata Fks1p. However, these methods showed intrinsic limitations of classical PCRs as their inability to detect heterozygous mutants [36,144,147]. All PCR-based methods (classical and real-time) have common problems with the design of primers and allele-specific probes. The first is the presence of synonymous or silent polymorphisms at C. albicans and C. glabrata FKS hot spots that were partially fixed by the introduction of a wobble base in the probe (or degenerated probe) [148] while others designed multiple primer combinations trying to avoid the residues where polymorphisms were reported [144,147].
4. Conclusions
The clinical predictive value (or lack thereof) of the uncovering of a molecular resistance mechanism may be relative. The “90–60 rule” applies to both molecular and whole-cell antifungal susceptibility evaluation. This “rule” roughly states that ~90% of infections due to susceptible isolates respond to the correct antifungal treatment, whereas ~60% of the infections caused by resistant isolates (or infections treated with an incorrect drug) respond to therapy [160].
Molecular methods can be used to detect intrinsic and secondary resistance. There are in-house and commercially available methods. The major common limitation of these methods is the narrow coverage for fungal pathogens and the low impact in the selection of specific antifungal treatments.
The commercially available diagnostic tools have a low impact on the selection of specific antifungal treatments. A bigger effort is needed to include more species in the existing panels. Efforts should focus on the differentiation of the fungal groups of species with known intrinsic resistance to certain antifungals directly from clinical samples.
The rDNA (ITS) is a good marker for intrinsic resistance detection for Mucorales and for Candida spp. On the other hand, for most pathogenic filamentous fungi, a multilocus DNA-barcoding approach is needed.
As in any other molecular tools, a suspicion of the presence of a particular species in a sample is needed in order to select a particular molecular method. Thus, these tools are useful in confirming an infection or colonization and have a great negative predictive value.
The identification of clades, types, and varieties of particular species showing reduced antifungal susceptibility would be a useful surrogate marker of resistance.
There are more bottlenecks in terms of sensitivity and methodological complexity for the detection of secondary than for intrinsic resistance by molecular methods. These limitations include single copy vs. multiple copy genes, mutation detection and or expression evaluation vs. presence of a gene, several mechanisms with the same phenotype vs. one gene same diagnosis, no universal primers vs. universal primers, etc.
Molecular methods can only detect known mechanisms, as opposed to phenotypic or whole-cell methods such as MIC assessment that detect all mechanisms.
There are no molecular methods able to detect amphotericin B (AMB) resistance.
It is difficult to detect azole resistance in Candida spp. due to the multiplicity of mechanisms involved. NSG and WGS were used in reference labs to confirm and study already known resistance phenotypes.
Most of the published secondary resistance detection tools are able to detect some of the CYP51A and FKS mutations linked with triazole and echinocandin resistance in Aspergillus fumigatus and Candida spp. (specially C. albicans, C. glabrata, and C. auris), respectively
A. fumigatus triazole resistance molecular diagnosis is mostly limited to the detection of CYP51A promoter alterations (TR34 and TR46).
There are multiple technological options to detect echinocandin resistance mechanisms in C. albicans and C. glabrata.
There are clinical settings where resistance mechanism detection would be valuable as places where triazole resistance in A. fumigatus prevalence surpass 10%, high use of empirical treatment, etc.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflict of interest.
References
- Berkow, E.L.; Lockhart, S.R.; Ostrosky-Zeichner, L. Antifungal susceptibility testing: Current approaches. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef]
- Ganesan, A.; Wells, J.; Shaikh, F.; Peterson, P.; Bradley, W.; Carson, M.L.; Petfield, J.L.; Klassen-Fischer, M.; Akers, K.S.; Downing, K.; et al. Molecular Detection of Filamentous Fungi in Formalin-Fixed Paraffin-Embedded Specimens in Invasive Fungal Wound Infections Is Feasible with High Specificity. J. Clin. Microbiol. 2019, 58. [Google Scholar] [CrossRef]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 2020, 71, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Wickes, B.L.; Wiederhold, N.P. Molecular diagnostics in medical mycology. Nat. Commun. 2018, 9, 5135. [Google Scholar] [CrossRef]
- Clinical and Laboratory Strandards Institute CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 3rd ed.; CLSI Standard M38; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard-Third Edition M27-A3; CLSI (Clinical and Laboratory Standard Institute): Wayne, PA, USA, 2008. [Google Scholar]
- EUCAST. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds; EUCAST Definitive Document E.DEF 9.3.2.; EUCAST. 2020. Available online: https://www.aspergillus.org.uk/wp-content/uploads/2016/03/EUCAST_E_Def_9_3_Mould_testing_definitive_0.pdf (accessed on 9 March 2021).
- EUCAST. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts; EUCAST Document E.DEF 7.3.2. Clin. Microbiol. Infect. 2008, 14, 398–405. [Google Scholar] [CrossRef] [PubMed]
- White, T.C.; Marr, K.A.; Bowden, R.A. Clinical, Cellular, and Molecular Factors That Contribute to Antifungal Drug Resistance. Clin. Microbiol. Rev. 1998, 11, 382–402. [Google Scholar] [CrossRef]
- Arastehfar, A.; Lass-Flörl, C.; Garcia-Rubio, R.; Daneshnia, F.; Ilkit, M.; Boekhout, T.; Gabaldon, T.; Perlin, D.S. The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J. Fungi 2020, 6, 138. [Google Scholar] [CrossRef]
- Perlin, D.S. Echinocandin Resistance in Candida. Clin. Infect. Dis. 2015, 61, S612–S617. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Coste, A.; Ferrari, S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 2009, 9, 1029–1050. [Google Scholar] [CrossRef] [PubMed]
- Leonardelli, F.; Macedo, D.; Dudiuk, C.; Cabeza, M.S.; Gamarra, S.; Garcia-Effron, G. Aspergillus fumigatus Intrinsic Fluconazole Resistance Is Due to the Naturally Occurring T301I Substitution in Cyp51Ap. Antimicrob. Agents Chemother. 2016, 60, 5420–5426. [Google Scholar] [CrossRef]
- Lamb, D.C.; Kelly, D.E.; Schunck, W.-H.; Shyadehi, A.Z.; Akhtar, M.; Lowe, D.J.; Baldwin, B.C.; Kelly, S.L. The Mutation T315A in Candida albicans Sterol 14α-Demethylase Causes Reduced Enzyme Activity and Fluconazole Resistance through Reduced Affinity. J. Biol. Chem. 1997, 272, 5682–5688. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; M100Ed30; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- EUCAST. EUCAST Expert Rules Version 2 2020. Available online: https://www.eucast.org/expert_rules_and_intrinsic_resistance/ (accessed on 1 March 2020).
- Clinical and Laboratory Strandards Institute CLSI. Performance Standards for Antifungal Susceptibility Testing of Yeasts, 2nd ed.; M60; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- De Carolis, E.; Posteraro, B.; Lass-Flörl, C.; Vella, A.; Florio, A.R.; Torelli, R.; Girmenia, C.; Colozza, C.; Tortorano, A.M.; Sanguinetti, M.; et al. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 2012, 18, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Iriart, X.; Lavergne, R.-A.; Fillaux, J.; Valentin, A.; Magnaval, J.-F.; Berry, A.; Cassaing, S. Routine Identification of Medical Fungi by the New Vitek MS Matrix-Assisted Laser Desorption Ionization–Time of Flight System with a New Time-Effective Strategy. J. Clin. Microbiol. 2012, 50, 2107–2110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bille, E.; Dauphin, B.; Leto, J.; Bougnoux, M.-E.; Beretti, J.-L.; Lotz, A.; Suarez, S.; Meyer, J.; Join-Lambert, O.; Descamps, P.; et al. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin. Microbiol. Infect. 2012, 18, 1117–1125. [Google Scholar] [CrossRef]
- Wilkendorf, L.S.; Bowles, E.; Buil, J.B.; Van Der Lee, H.A.L.; Posteraro, B.; Sanguinetti, M.; Verweij, P.E. Update on Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Identification of Filamentous Fungi. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Gamarra, S.; Chaves, M.; Cabeza, M.; Macedo, D.; Leonardelli, F.; Franco, D.; Boleas, M.; Garcia-Effron, G. Mucormycosis outbreak due to Rhizopus microsporus after arthroscopic anterior cruciate ligament reconstruction surgery evaluated by RAPD and MALDI-TOF Mass spectrometry. J. Med Mycol. 2018, 28, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Robert, M.G.; Romero, C.; Dard, C.; Garnaud, C.; Cognet, O.; Girard, T.; Rasamoelina, T.; Cornet, M.; Maubon, D. Evaluation of ID Fungi Plates Medium for Identification of Molds by MALDI Biotyper. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Guarro, J.; Gené, J.; Stchigel, A.M. Developments in Fungal Taxonomy. Clin. Microbiol. Rev. 1999, 12, 454–500. [Google Scholar] [CrossRef]
- Gregory, T.R. DNA barcodes an adjunct to linnaean taxonomy. Nature 2005, 434, 1067. [Google Scholar] [CrossRef]
- Irinyi, L.; Lackner, M.; De Hoog, G.S.; Meyer, W. DNA barcoding of fungi causing infections in humans and animals. Fungal Biol. 2016, 120, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Schindel, D.E.; Miller, S.E. Benefits of DNA barcoding. Nature 2005, 435, 17. [Google Scholar] [CrossRef] [PubMed]
- Will, K.W.; Rubinoff, D. Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics 2004, 20, 47–55. [Google Scholar] [CrossRef]
- Marshall, E. Will DNA bar codes breathe life into classification? Science 2005, 307, 1037. [Google Scholar] [CrossRef] [PubMed]
- Ebach, M.C.; Holdrege, C. DNA barcoding is no substitute for taxonomy. Nat. Cell Biol. 2005, 434, 697. [Google Scholar] [CrossRef]
- Meyer, C.P.; Paulay, G. DNA Barcoding: Error Rates Based on Comprehensive Sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in Silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Li, Y.L.; Leaw, S.N.; Chen, J.-H.; Chang, H.C.; Chang, T.C. Rapid Identification of Yeasts Commonly Found in Positive Blood Cultures by Amplification of the Internal Transcribed Spacer Regions 1 and 2. Eur. J. Clin. Microbiol. Infect. Dis. 2003, 22, 693–696. [Google Scholar] [CrossRef]
- Borman, A.M.; Petch, R.; Linton, C.J.; Palmer, M.D.; Bridge, P.D.; Johnson, E.M. Candida nivariensis, an Emerging Pathogenic Fungus with Multidrug Resistance to Antifungal Agents. J. Clin. Microbiol. 2008, 46, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Morales-López, S.; Dudiuk, C.; Vivot, W.; Szusz, W.; Córdoba, S.B.; Garcia-Effron, G. Phenotypic and Molecular Evaluation of Echinocandin Susceptibility of Candida glabrata, Candida bracarensis, and Candida nivariensis Strains Isolated during 30 Years in Argentina. Antimicrob. Agents Chemother. 2017, 61, e00170-17. [Google Scholar] [CrossRef]
- Cartier, N.; Chesnay, A.; N’Diaye, D.; Thorey, C.; Ferreira, M.; Haillot, O.; Bailly, É.; Desoubeaux, G. Candida nivariensis: Identification strategy in mycological laboratories. J. Med. Mycol. 2020, 30, 101042. [Google Scholar] [CrossRef]
- Dudiuk, C.; Morales-López, S.E.; Podesta, V.; Macedo, D.; Leonardelli, F.; Vitale, R.G.; Tosello, M.E.; Cabeza, M.S.; Biasoli, M.; Gamarra, S.; et al. Multiplex PCR designed to differentiate species within the Candida glabrata complex. Rev. Iberoam. Micol. 2017, 34, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Walther, G.; Pawłowska, J.; Alastruey-Izquierdo, A.; Wrzosek, M.; Rodriguez-Tudela, J.; Dolatabadi, S.; Chakrabarti, A.; De Hoog, G. DNA barcoding in Mucorales: An inventory of biodiversity. Pers. Mol. Phylogeny Evol. Fungi 2013, 30, 11–47. [Google Scholar] [CrossRef]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic Species Recognition and Species Concepts in Fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Tudela, J.L.; Diaz-Guerra, T.M.; Mellado, E.; Cano, V.; Tapia, C.; Perkins, A.; Gomez-Lopez, A.; Rodero, L.; Cuenca-Estrella, M. Susceptibility Patterns and Molecular Identification of Trichosporon Species. Antimicrob. Agents Chemother. 2005, 49, 4026–4034. [Google Scholar] [CrossRef]
- Roe, A.D.; Rice, A.V.; Bromilow, S.E.; Cooke, J.E.K.; Sperling, F.A.H. Multilocus species identification and fungal DNA barcoding: Insights from blue stain fungal symbionts of the mountain pine beetle. Mol. Ecol. Resour. 2010, 10, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Chaverri, P.; Branco-Rocha, F.; Jaklitsch, W.; Gazis, R.; Degenkolb, T.; Samuels, G.J. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 2015, 107, 558–590. [Google Scholar] [CrossRef]
- Balajee, S.A.; Gribskov, J.L.; Hanley, E.; Nickle, D.; Marr, K.A. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 2005, 4, 625–632. [Google Scholar] [CrossRef]
- Gautier, M.; Normand, A.-C.; Ranque, S. Previously unknown species of Aspergillus. Clin. Microbiol. Infect. 2016, 22, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Hubka, V.; Kolarik, M. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: Primer specificity testing and taxonomic consequences. Pers. Mol. Phylogeny Evol. Fungi 2012, 29, 1–10. [Google Scholar] [CrossRef]
- Peterson, S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 2008, 100, 205–226. [Google Scholar] [CrossRef]
- Samson, R.; Visagie, C.; Houbraken, J.; Hong, S.-B.; Hubka, V.; Klaassen, C.; Perrone, G.; Seifert, K.; Susca, A.; Tanney, J.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef]
- Van Diepeningen, A.D.; Feng, P.; Ahmed, S.; Sudhadham, M.; Bunyaratavej, S.; De Hoog, G.S. Spectrum ofFusariuminfections in tropical dermatology evidenced by multilocus sequencing typing diagnostics. Mycoses 2014, 58, 48–57. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Nucleic Acid Based Tests. FDA. Available online: https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests (accessed on 28 December 2020).
- Simor, A.E.; Porter, V.; Mubareka, S.; Chouinard, M.; Katz, K.; Vermeiren, C.; Fattouh, R.; Matukas, L.M.; Tadros, M.; Mazzulli, T.; et al. Rapid Identification of Candida Species from Positive Blood Cultures by Use of the Film Array Blood Culture Identification Panel. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Zacharioudakis, I.M.; Zervou, F.N.; Mylonakis, E. T2 Magnetic Resonance Assay: Overview of Available Data and Clinical Implications. J. Fungi 2018, 4, 45. [Google Scholar] [CrossRef]
- Biomerieux Enhanced BIOFIRE® Blood Culture Identification 2 (BCID2). Available online: https://www.rapidmicrobiology.com/news/enhanced-biofire-bcid2-panel-submitted-for-fda-clearance (accessed on 26 December 2020).
- Clancy, C.J.; Nguyen, M.H. Finding the “Missing 50%” of Invasive Candidiasis: How Nonculture Diagnostics Will Improve Understanding of Disease Spectrum and Transform Patient Care. Clin. Infect. Dis. 2013, 56, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Kasper, D.C.; Altiok, I.; Mechtler, T.P.; Böhm, J.; Straub, J.; Langgartner, M.; Pollak, A.; Herkner, K.R.; Berger, A. Molecular Detection of Late-Onset Neonatal Sepsis in Premature Infants Using Small Blood Volumes: Proof-of-Concept. Neonatology 2013, 103, 268–273. [Google Scholar] [CrossRef]
- Bomkamp, J.P.; Sulaiman, R.; Hartwell, J.L.; Desai, A.; Winn, V.C.; Wrin, J.; Kussin, M.L.; Hiles, J.J. Evaluation of a Rapid Fungal Detection Panel for Identification of Candidemia at an Academic Medical Center. J. Clin. Microbiol. 2019, 58. [Google Scholar] [CrossRef]
- Straub, J.; Paula, H.; Mayr, M.; Kasper, D.; Assadian, O.; Berger, A.; Rittenschober-Böhm, J. Diagnostic accuracy of the ROCHE Septifast PCR system for the rapid detection of blood pathogens in neonatal sepsis—A prospective clinical trial. PLoS ONE 2017, 12, e0187688. [Google Scholar] [CrossRef]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2015, 62, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- PathoNostics AsperGenius®. PathoNostics. Available online: https://www.pathonostics.com/product/aspergenius (accessed on 26 December 2020).
- Pelzer, B.W.; Seufert, R.; Koldehoff, M.; Liebregts, T.; Schmidt, D.; Buer, J.; Rath, P.-M.; Steinmann, J. Performance of the AsperGenius® PCR assay for detecting azole resistant Aspergillus fumigatus in BAL fluids from allogeneic HSCT recipients: A prospective cohort study from Essen, West Germany. Med. Mycol. 2019, 58, 268–271. [Google Scholar] [CrossRef]
- White, P.L.; Posso, R.B.; Barnes, R.A. Analytical and Clinical Evaluation of the PathoNostics AsperGenius Assay for Detection of Invasive Aspergillosis and Resistance to Azole Antifungal Drugs during Testing of Serum Samples. J. Clin. Microbiol. 2015, 53, 2115–2121. [Google Scholar] [CrossRef]
- Chong, G.M.; Van Der Beek, M.T.; Borne, P.A.V.D.; Boelens, J.; Steel, E.; Kampinga, G.A.; Span, L.F.R.; Lagrou, K.; Maertens, J.A.; Dingemans, G.J.H.; et al. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: A multicentre validation of the AsperGenius assay ® in 201 patients with haematological disease suspected for invasive aspergillosis. J. Antimicrob. Chemother. 2016, 71, 3528–3535. [Google Scholar] [CrossRef]
- Chong, G.-L.M.; Van De Sande, W.W.J.; Dingemans, G.J.H.; Gaajetaan, G.R.; Vonk, A.G.; Hayette, M.-P.; Van Tegelen, D.W.E.; Simons, G.F.M.; Rijnders, B.J.A. Validation of a New Aspergillus Real-Time PCR Assay for Direct Detection of Aspergillus and Azole Resistance of Aspergillus fumigatus on Bronchoalveolar Lavage Fluid. J. Clin. Microbiol. 2015, 53, 868–874. [Google Scholar] [CrossRef]
- Ademtech MycoGENIE®. Real-Time PCR Kits—Ademtech. Available online: https://www.ademtech.com/molecular-diagnostic/mycology/real-time-pcr-kit/ (accessed on 26 December 2020).
- Chong, G.; Vonk, A.; Meis, J.; Dingemans, G.; Houbraken, J.; Hagen, F.; Gaajetaan, G.; Van Tegelen, D.; Simons, G.; Rijnders, B. Interspecies discrimination of A. fumigatus and siblings A. lentulus and A. felis of the Aspergillus section Fumigati using the AsperGenius® assay. Diagn. Microbiol. Infect. Dis. 2017, 87, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Morrell, M.; Fraser, V.J.; Kollef, M.H. Delaying the Empiric Treatment of Candida Bloodstream Infection until Positive Blood Culture Results Are Obtained: A Potential Risk Factor for Hospital Mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–3645. [Google Scholar] [CrossRef]
- Ecker, D.J.; Sampath, R.; Massire, C.; Blyn, L.B.; Hall, T.A.; Eshoo, M.W.; Hofstadler, S.A. Ibis T5000: A universal biosensor approach for microbiology. Nat. Rev. Genet. 2008, 6, 553–558. [Google Scholar] [CrossRef]
- Wolk, D.M.; Kaleta, E.J.; Wysocki, V.H. PCR-electrospray ionization mass spectrometry: The potential to change infectious disease diagnostics in clinical and public health laboratories. J. Mol. Diagn. 2012, 14, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Uhl, J.R.; Hall, L.; Weber, M.M.; Walchak, R.C.; Buckwalter, S.; Wengenack, N.L. Broad-Range Direct Detection and Identification of Fungi by Use of the PLEX-ID PCR-Electrospray Ionization Mass Spectrometry (ESI-MS) System. J. Clin. Microbiol. 2013, 51, 1699–1706. [Google Scholar] [CrossRef][Green Version]
- Shin, J.H.; Ranken, R.; Sefers, S.E.; Lovari, R.; Quinn, C.D.; Meng, S.; Carolan, H.E.; Toleno, D.; Li, H.; Lee, J.N.; et al. Detection, Identification, and Distribution of Fungi in Bronchoalveolar Lavage Specimens by Use of Multilocus PCR Coupled with Electrospray Ionization/Mass Spectrometry. J. Clin. Microbiol. 2012, 51, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Garcia-Hermoso, D.; Mercier-Delarue, S.; Lanternier, F.; Gits-Muselli, M.; Menotti, J.; Denis, B.; Bergeron, A.; Legrand, M.; Lortholary, O.; et al. Molecular identification of Mucorales in human tissues: Contribution of PCR electrospray-ionization mass spectrometry. Clin. Microbiol. Infect. 2015, 21, 594.e1–594.e5. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Fang, W.; Pan, W.; Liao, W.; Yan, L.; Boekhout, T. Identification of nine cryptic species of Candida albicans, C. glabrata, and C. parapsilosis complexes using one-step multiplex PCR. BMC Infect. Dis. 2018, 18, 480. [Google Scholar] [CrossRef]
- Reyes-Montes, M.D.R.; Acosta-Altamirano, G.; Duarte-Escalante, E.; Salazar, E.G.; Martínez-Herrera, E.; Arenas, R.; González, G.; Frías-De-León, M.G. Usefulness of a multiplex PCR for the rapid identification of Candida glabrata species complex in Mexican clinical isolates. Rev. Inst. Med. Trop. São Paulo 2019, 61, e37. [Google Scholar] [CrossRef]
- Romeo, O.; Criseo, G. First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene. Diagn. Microbiol. Infect. Dis. 2008, 62, 230–233. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Canton, E.; Peman, J.; Dilger, A.; Romá, E.; Perlin, D.S. Assessment of Two New Molecular Methods for Identification of Candida parapsilosis Sensu Lato Species. J. Clin. Microbiol. 2011, 49, 3257–3261. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, A.; Davidson, A.D.; Gow, N.A.R.; Maiden, M.C.J.; Odds, F.C. Candida orthopsilosis and Candida metapsilosis spp. nov. to Replace Candida parapsilosis Groups II and III. J. Clin. Microbiol. 2005, 43, 284–292. [Google Scholar] [CrossRef]
- Cai, S.; Xu, J.; Shao, Y.; Gong, J.; Zhao, F.; He, L.; Shan, X. Rapid identification of the Candida glabrata species complex by high-resolution melting curve analysis. J. Clin. Lab. Anal. 2020, 34, e23226. [Google Scholar] [CrossRef] [PubMed]
- Theill, L.; Dudiuk, C.; Morano, S.; Gamarra, S.; Nardin, M.E.; Méndez, E.; Garcia-Effron, G. Prevalence and antifungal susceptibility of Candida albicans and its related species Candida dubliniensis and Candida africana isolated from vulvovaginal samples in a hospital of Argentina. Rev. Argent. Microbiol. 2016, 48, 43–49. [Google Scholar] [CrossRef]
- Arastehfar, A.; Daneshnia, F.; Salehi, M.-R.; Zarrinfar, H.; Khodavaisy, S.; Haas, P.-J.; Roudbary, M.; Najafzadeh, M.-J.; Zomorodian, K.; Charsizadeh, A.; et al. Molecular characterization and antifungal susceptibility testing of Candida nivariensis from blood samples—An Iranian multicentre study and a review of the literature. J. Med Microbiol. 2019, 68, 770–777. [Google Scholar] [CrossRef]
- Morales-López, S.E.; Taverna, C.G.; Bosco-Borgeat, M.E.; Maldonado, I.; Vivot, W.; Szusz, W.; Garcia-Effron, G.; Córdoba, S.B. Candida glabrata species complex prevalence and antifungal susceptibility testing in a culture collection: First description of Candida nivariensis in Argentina. Mycopathologia 2016, 181, 871–878. [Google Scholar] [CrossRef]
- Theill, L.; Dudiuk, C.; Morales-Lopez, S.; Berrio, I.; Rodríguez, J.Y.; Marin, A.; Gamarra, S.; Garcia-Effron, G. Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev. Iberoam. Micol. 2018, 35, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and Accurate Molecular Identification of the Emerging Multidrug-Resistant Pathogen Candida auris. J. Clin. Microbiol. 2017, 55, 2445–2452. [Google Scholar] [CrossRef] [PubMed]
- Georgacopoulos, O.; Nunnally, N.S.; Le, N.; Lysen, C.; Welsh, R.M.; Kordalewska, M.; Perlin, D.S.; Berkow, E.L.; Sexton, D.J. Performance Evaluation of Culture-Independent SYBR Green Candida auris Quantitative PCR Diagnostics on Anterior Nares Surveillance Swabs. J. Clin. Microbiol. 2020, 58, 58. [Google Scholar] [CrossRef]
- Leach, L.; Zhu, Y.; Chaturvedi, S. Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J. Clin. Microbiol. 2017, 56. [Google Scholar] [CrossRef]
- Sexton, D.J.; Kordalewska, M.; Bentz, M.L.; Welsh, R.M.; Perlin, D.S.; Litvintseva, A.P. Direct Detection of Emergent Fungal Pathogen Candida auris in Clinical Skin Swabs by SYBR Green-Based Quantitative PCR Assay. J. Clin. Microbiol. 2018, 56, e01337-18. [Google Scholar] [CrossRef]
- Kordalewska, M.; Lee, A.; Zhao, Y.; Perlin, D.S. Detection of Candida auris Antifungal Drug Resistance Markers Directly from Clinical Skin Swabs. Antimicrob. Agents Chemother. 2019, 63, 63. [Google Scholar] [CrossRef] [PubMed]
- Millon, L.; Scherer, E.; Rocchi, S.; Bellanger, A.-P. Molecular Strategies to Diagnose Mucormycosis. J. Fungi 2019, 5, 24. [Google Scholar] [CrossRef]
- Dannaoui, E. Molecular tools for identification of Zygomycetes and the diagnosis of zygomycosis. Clin. Microbiol. Infect. 2009, 15, 66–70. [Google Scholar] [CrossRef]
- Hammond, S.P.; Bialek, R.; Milner, D.A.; Petschnigg, E.M.; Baden, L.R.; Marty, F.M. Molecular Methods to Improve Diagnosis and Identification of Mucormycosis: Fig. 1. J. Clin. Microbiol. 2011, 49, 2151–2153. [Google Scholar] [CrossRef]
- Schwarz, P.; Bretagne, S.; Gantier, J.-C.; Garcia-Hermoso, D.; Lortholary, O.; Dromer, F.; Dannaoui, E. Molecular Identification of Zygomycetes from Culture and Experimentally Infected Tissues. J. Clin. Microbiol. 2006, 44, 340–349. [Google Scholar] [CrossRef]
- Dannaoui, E.; Schwarz, P.; Slany, M.; Loeffler, J.; Jorde, A.T.; Cuenca-Estrella, M.; Hauser, P.M.; Shrief, R.; Huerre, M.; Freiberger, T.; et al. Molecular Detection and Identification of Zygomycetes Species from Paraffin-Embedded Tissues in a Murine Model of Disseminated Zygomycosis: A Collaborative European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Fungal Infection Study Group (EFISG) Evaluation. J. Clin. Microbiol. 2010, 48, 2043–2046. [Google Scholar] [CrossRef] [PubMed]
- Gudiol, C.; Bodro, M.; Simonetti, A.; Tubau, F.; González-Barca, E.; Cisnal, M.; Domingo-Domenech, E.; Jiménez, L.; Carratalà, J. Changing aetiology, clinical features, antimicrobial resistance, and outcomes of bloodstream infection in neutropenic cancer patients. Clin. Microbiol. Infect. 2013, 19, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Chen, S.; Sorrell, T.; Carter, D.; Malik, R.; Martin, P.; Halliday, C. Development and Clinical Application of a Panfungal PCR Assay to Detect and Identify Fungal DNA in Tissue Specimens. J. Clin. Microbiol. 2006, 45, 380–385. [Google Scholar] [CrossRef]
- Bialek, R.; Konrad, F.; Kern, J.; Aepinus, C.; Cecenas, L.; Gonzalez, G.M.; Just-Nübling, G.; Willinger, B.; Presterl, E.; Lass-Flörl, C.; et al. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J. Clin. Pathol. 2005, 58, 1180–1184. [Google Scholar] [CrossRef] [PubMed]
- Machouart, M.-C.; Larche, J.; Burton, K.; Collomb, J.; Maurer, P.; Cintrat, A.; Biava, M.F.; Greciano, S.; Kuijpers, A.F.A.; Contet-Audonneau, N.; et al. Genetic Identification of the Main Opportunistic Mucorales by PCR-Restriction Fragment Length Polymorphism. J. Clin. Microbiol. 2006, 44, 805–810. [Google Scholar] [CrossRef]
- Baldin, C.; Soliman, S.S.M.; Jeon, H.H.; Alkhazraji, S.; Gebremariam, T.; Gu, Y.; Bruno, V.M.; Cornely, O.A.; Leather, H.L.; Sugrue, M.W.; et al. PCR-Based Approach Targeting Mucorales-Specific Gene Family for Diagnosis of Mucormycosis. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Scherer, E.; Iriart, X.; Bellanger, A.P.; Dupont, D.; Guitard, J.; Gabriel, F.; Cassaing, S.; Charpentier, E.; Guenounou, S.; Cornet, M.; et al. Quantitative PCR (qPCR) Detection of Mucorales DNA in Bronchoalveolar Lavage Fluid to Diagnose Pulmonary Mucormycosis. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Bernal-Martínez, L.; Buitrago, M.J.; Castelli, M.V.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Development of a single tube multiplex real-time PCR to detect the most clinically relevant Mucormycetes species. Clin. Microbiol. Infect. 2013, 19, E1–E7. [Google Scholar] [CrossRef]
- Pujol, C.; Pfaller, M.A.; Soll, D.R. Flucytosine Resistance Is Restricted to a Single Genetic Clade of Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 262–266. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Chowdhary, A.; Gonzalez, G.M.; Guinea, J.; Hagen, F.; Meis, J.F.; Thompson, G.R.; Turnidge, J. Multicenter Study of Isavuconazole MIC Distributions and Epidemiological Cutoff Values for the Cryptococcus neoformans-Cryptococcus gattii Species Complex Using the CLSI M27-A3 Broth Microdilution Method. Antimicrob. Agents Chemother. 2014, 59, 666–668. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Aller, A.I.; Canton, E.; Castañón-Olivares, L.R.; Chowdhary, A.; Cordoba, S.; Cuenca-Estrella, M.; Fothergill, A.; Fuller, J.; Govender, N.; et al. Cryptococcus neoformans-Cryptococcus gattii Species Complex: An International Study of Wild-Type Susceptibility Endpoint Distributions and Epidemiological Cutoff Values for Fluconazole, Itraconazole, Posaconazole, and Voriconazole. Antimicrob. Agents Chemother. 2012, 56, 5898–5906. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Nenoff, P.; Verma, S.B.; Ebert, A.; Süß, A.; Fischer, E.; Auerswald, E.; Dessoi, S.; Hofmann, W.; Schmidt, S.; Neubert, K.; et al. Spread of Terbinafine-Resistant Trichophyton mentagrophytes Type VIII (India) in Germany–“The Tip of the Iceberg?”. J. Fungi 2020, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Kanafani, Z.A.; Perfect, J.R. Resistance to Antifungal Agents: Mechanisms and Clinical Impact. Clin. Infect. Dis. 2008, 46, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Arendrup, M.; Cantón, E.; Cordoba, S.; Dannaoui, E.; García-Rodríguez, J.; Gonzalez, G.M.; Govender, N.P.; Martin-Mazuelos, E.; Lackner, M.; et al. Multicenter Study of Method-Dependent Epidemiological Cutoff Values for Detection of Resistance in Candida spp. and Aspergillus spp. to Amphotericin B and Echinocandins for the Etest Agar Diffusion Method. Antimicrob. Agents Chemother. 2016, 61. [Google Scholar] [CrossRef]
- Dick, J.D.; Merz, W.G.; Saral, R. Incidence of polyene-resistant yeasts recovered from clinical specimens. Antimicrob. Agents Chemother. 1980, 18, 158–163. [Google Scholar] [CrossRef]
- Sokol-Anderson, M.L.; Brajtburg, J.; Medoff, G. Amphotericin B-Induced Oxidative Damage and Killing of Candida albicans. J. Infect. Dis. 1986, 154, 76–83. [Google Scholar] [CrossRef]
- Healey, K.R.; Perlin, D.S. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. J. Fungi 2018, 4, 105. [Google Scholar]
- Bienvenu, A.L.; Leboucher, G.; Picot, S. Comparison of fks gene mutations and minimum inhibitory concentrations for the detection of Candida glabrata resistance to micafungin: A systematic review and meta-analysis. Mycoses 2019, 62, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Kwa, A.L.; Cheng, S.; Du, C.; Clancy, C.J. The Presence of anFKSMutation Rather than MIC Is an Independent Risk Factor for Failure of Echinocandin Therapy among Patients with Invasive Candidiasis Due to Candida glabrata. Antimicrob. Agents Chemother. 2012, 56, 4862–4869. [Google Scholar] [CrossRef]
- Rivero-Menendez, O.; Alastruey-Izquierdo, A.; Mellado, E.; Cuenca-Estrella, M. Triazole Resistance in Aspergillus spp.: A Worldwide Problem? J. Fungi 2016, 2, 21. [Google Scholar] [CrossRef]
- Zhang, J.; Zoll, J.; Engel, T.; Heuvel, J.V.D.; Verweij, P.E.; Debets, A.J.M. The Medical Triazole Voriconazole Can Select for Tandem Repeat Variations in Azole-Resistant Aspergillus Fumigatus Harboring TR34/L98H via Asexual Reproduction. J. Fungi 2020, 6, 277. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.G.; Meis, J.F. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; Cuenca-Estrella, M.; Mellado, E. Triazole Resistance in Aspergillus Species: An Emerging Problem. Drugs 2017, 77, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Sionov, E.; Chang, Y.C.; Garraffo, H.M.; Dolan, M.A.; Ghannoum, M.A.; Kwon-Chung, K.J. Identification of a Cryptococcus neoformans Cytochrome P450 Lanosterol 14α-Demethylase (Erg11) Residue Critical for Differential Susceptibility between Fluconazole/Voriconazole and Itraconazole/Posaconazole. Antimicrob. Agents Chemother. 2011, 56, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Sionov, E.; Chang, Y.C.; Kwon-Chung, K.J. Azole Heteroresistance in Cryptococcus neoformans: Emergence of Resistant Clones with Chromosomal Disomy in the Mouse Brain during Fluconazole Treatment. Antimicrob. Agents Chemother. 2013, 57, 5127–5130. [Google Scholar] [CrossRef]
- Chang, M.; Sionov, E.; Lamichhane, A.K.; Kwon-Chung, K.J.; Chang, Y.C. Roles of Three Cryptococcus neoformans and Cryptococcus gattii Efflux Pump-Coding Genes in Response to Drug Treatment. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Lofgren, L.A.; Uehling, J.K.; Branco, S.; Bruns, T.D.; Martin, F.; Kennedy, P.G. Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 2019, 28, 721–730. [Google Scholar] [CrossRef]
- Mellado, E.; Garcia-Effron, G.; Alcázar-Fuoli, L.; Melchers, W.J.G.; Verweij, P.E.; Cuenca-Estrella, M.; Rodríguez-Tudela, J.L. A New Aspergillus fumigatus Resistance Mechanism Conferring In Vitro Cross-Resistance to Azole Antifungals Involves a Combination of cyp51A Alterations. Antimicrob. Agents Chemother. 2007, 51, 1897–1904. [Google Scholar] [CrossRef]
- Mellado, E.; Garcia-Effron, G.; Alcazar-Fuoli, L.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Substitutions at Methionine 220 in the 14α-Sterol Demethylase (Cyp51A) of Aspergillus fumigatus Are Responsible for Resistance In Vitro to Azole Antifungal Drugs. Antimicrob. Agents Chemother. 2004, 48, 2747–2750. [Google Scholar] [CrossRef]
- Diaz-Guerra, T.M.; Mellado, E.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. A Point Mutation in the 14α-Sterol Demethylase Gene cyp51A Contributes to Itraconazole Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2003, 47, 1120–1124. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Effron, G.; Lee, S.; Park, S.; Cleary, J.D.; Perlin, D.S. Effect of Candida glabrata FKS1 and FKS2 Mutations on Echinocandin Sensitivity and Kinetics of 1,3-β-d-Glucan Synthase: Implication for the Existing Susceptibility Breakpoint. Antimicrob. Agents Chemother. 2009, 53, 3690–3699. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Park, S.; Perlin, D.S. Correlating Echinocandin MIC and Kinetic Inhibition of fks1 Mutant Glucan Synthases for Candida albicans: Implications for Interpretive Breakpoints. Antimicrob. Agents Chemother. 2008, 53, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Marichal, P.; Koymans, L.; Willemsens, S.; Bellens, D.; Verhasselt, P.; Luyten, W.; Borgers, M.; Ramaekers, F.C.S.; Odds, F.C.; Bossche, H.V. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 1999, 145, 2701–2713. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Effron, G.; Dilger, A.; Alcazar-Fuoli, L.; Park, S.; Mellado, E.; Perlin, D.S. Rapid Detection of Triazole Antifungal Resistance in Aspergillus fumigatus. J. Clin. Microbiol. 2008, 46, 1200–1206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Der Linden, J.W.M.; Snelders, E.; Arends, J.P.; Daenen, S.M.; Melchers, W.J.G.; Verweij, P.E. Rapid Diagnosis of Azole-Resistant Aspergillosis by Direct PCR Using Tissue Specimens. J. Clin. Microbiol. 2010, 48, 1478–1480. [Google Scholar] [CrossRef]
- Denning, D.W.; Park, S.; Lass-Florl, C.; Fraczek, M.G.; Kirwan, M.; Gore, R.; Smith, J.; Bueid, A.; Moore, C.B.; Bowyer, P.; et al. High-frequency Triazole Resistance Found in Nonculturable Aspergillus fumigatus from Lungs of Patients with Chronic Fungal Disease. Clin. Infect. Dis. 2011, 52, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Klaassen, C.H.W.; De Valk, H.A.; Curfs-Breuker, I.M.; Meis, J.F. Novel mixed-format real-time PCR assay to detect mutations conferring resistance to triazoles in Aspergillus fumigatus and prevalence of multi-triazole resistance among clinical isolates in the Netherlands. J. Antimicrob. Chemother. 2010, 65, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Kathuria, S.; Randhawa, H.S.; Gaur, S.N.; Klaassen, C.H.; Meis, J.F. Isolation of multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR/L98H mutations in the cyp51A gene in India. J. Antimicrob. Chemother. 2011, 67, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Spiess, B.; Seifarth, W.; Merker, N.; Howard, S.J.; Reinwald, M.; Dietz, A.; Hofmann, W.-K.; Buchheidt, D. Development of Novel PCR Assays to Detect Azole Resistance-Mediating Mutations of theAspergillus fumigatus cyp51AGene in Primary Clinical Samples from Neutropenic Patients. Antimicrob. Agents Chemother. 2012, 56, 3905–3910. [Google Scholar] [CrossRef] [PubMed]
- Spiess, B.; Postina, P.; Reinwald, M.; Cornely, O.A.; Hamprecht, A.; Hoenigl, M.; Lass-Flörl, C.; Rath, P.-M.; Steinmann, J.; Miethke, T.; et al. Incidence of Cyp51 A Key Mutations in Aspergillus fumigatus—A Study on Primary Clinical Samples of Immunocompromised Patients in the Period of 1995–2013. PLoS ONE 2014, 9, e103113. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Khan, Z.; Hagen, F.; Meis, J.F. Simple, Low-Cost Molecular Assays for TR34/L98H Mutations in the cyp51A Gene for Rapid Detection of Triazole-Resistant Aspergillus fumigatus Isolates. J. Clin. Microbiol. 2014, 52, 2223–2227. [Google Scholar] [CrossRef] [PubMed]
- Dudakova, A.; Spiess, B.; Tangwattanachuleeporn, M.; Sasse, C.; Buchheidt, D.; Weig, M.; Groß, U.; Bader, O. Molecular Tools for the Detection and Deduction of Azole Antifungal Drug Resistance Phenotypes in Aspergillus Species. Clin. Microbiol. Rev. 2017, 30, 1065–1091. [Google Scholar] [CrossRef]
- Araújo, R.; Gungor, O.; Amorim, A. Single-tube PCR coupled with mini-sequencing assay for the detection of cyp51A and cyp51B polymorphisms in Aspergillus fumigatus. Futur. Microbiol. 2015, 10, 1797–1804. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, F.; Hashemi, S.J.; Zoll, J.; Melchers, W.J.G.; Rafati, H.; Dehghan, P.; Rezaie, S.; Tolooe, A.; Tamadon, Y.; Van Der Lee, H.A.; et al. Quantitative Analysis of Single-Nucleotide Polymorphism for Rapid Detection of TR34/L98H and TR46/Y121F/T289A-Positive Aspergillus fumigatus Isolates Obtained from Patients in Iran from 2010 to 2014. Antimicrob. Agents Chemother. 2015, 60, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Fraczek, M.G.; Bromley, M.; Buied, A.; Moore, C.B.; Rajendran, R.; Rautemaa, R.; Ramage, G.; Denning, D.W.; Bowyer, P. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013, 68, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-S.; Rodriguez-Manzano, J.; Malpartida-Cardenas, K.; Sewell, T.; Bader, O.; Armstrong-James, D.; Fisher, M.C.; Georgiou, P. Rapid and Sensitive Detection of Azole-Resistant Aspergillus fumigatus by Tandem Repeat Loop-Mediated Isothermal Amplification. J. Mol. Diagn. 2019, 21, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Macedo, D.; Devoto, T.B.; Pola, S.; Finquelievich, J.L.; Cuestas, M.L.; García-Effron, G. A Novel Combination of CYP51A Mutations Confers Pan-Azole Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2020, 64, 64. [Google Scholar] [CrossRef]
- Hagiwara, D.; Takahashi, H.; Watanabe, A.; Takahashi-Nakaguchi, A.; Kawamoto, S.; Kamei, K.; Gonoi, T. Whole-Genome Comparison of Aspergillus fumigatus Strains Serially Isolated from Patients with Aspergillosis. J. Clin. Microbiol. 2014, 52, 4202–4209. [Google Scholar] [CrossRef]
- Montesinos, I.; Argudín, M.A.; Hites, M.; Ahajjam, F.; Dodémont, M.; Dagyaran, C.; Bakkali, M.; Etienne, I.; Jacobs, F.; Knoop, C.; et al. Culture-Based Methods and Molecular Tools for Azole-Resistant Aspergillus fumigatus Detection in a Belgian University Hospital. J. Clin. Microbiol. 2017, 55, 2391–2399. [Google Scholar] [CrossRef]
- Dannaoui, E.; Gabriel, F.; Gaboyard, M.; Lagardere, G.; Audebert, L.; Quesne, G.; Godichaud, S.; Verweij, P.E.; Accoceberry, I.; Bougnoux, M.-E. Molecular Diagnosis of Invasive Aspergillosis and Detection of Azole Resistance by a Newly Commercialized PCR Kit. J. Clin. Microbiol. 2017, 55, 3210–3218. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Nagasaki, Y.; Kordalewska, M.; Press, E.G.; Shields, R.K.; Nguyen, M.H.; Clancy, C.J.; Perlin, D.S. Rapid Detection ofFKS-Associated Echinocandin Resistance in Candida glabrata. Antimicrob. Agents Chemother. 2016, 60, 6573–6577. [Google Scholar] [CrossRef]
- Dudiuk, C.; Gamarra, S.; Leonardeli, F.; Jimenez-Ortigosa, C.; Vitale, R.G.; Afeltra, J.; Perlin, D.S.; Garcia-Effron, G. Set of Classical PCRs for Detection of Mutations in Candida glabrata FKS Genes Linked with Echinocandin Resistance. J. Clin. Microbiol. 2014, 52, 2609–2614. [Google Scholar] [CrossRef]
- Biswas, C.; Chen, S.C.-A.; Halliday, C.; Martínez, E.; Rockett, R.J.; Wang, Q.; Timms, V.J.; Dhakal, R.; Sadsad, R.; Kennedy, K.J.; et al. Whole Genome Sequencing of Candida glabrata for Detection of Markers of Antifungal Drug Resistance. J. Vis. Exp. 2017, 56714, e56714. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.D.; Bolden, C.B.; Kuykendall, R.J.; Lockhart, S.R. Development of a Luminex-Based Multiplex Assay for Detection of Mutations Conferring Resistance to Echinocandins in Candida glabrata. J. Clin. Microbiol. 2013, 52, 790–795. [Google Scholar] [CrossRef][Green Version]
- Dudiuk, C.; Gamarra, S.; Jiménez-Ortigosa, C.; Leonardelli, F.; Macedo, D.; Perlin, D.S.; Garcia-Effron, G. Quick Detection of FKS1Mutations Responsible for Clinical Echinocandin Resistance in Candida albicans. J. Clin. Microbiol. 2015, 53, 2037–2041. [Google Scholar] [CrossRef]
- Balashov, S.V.; Park, S.; Perlin, D.S. Assessing Resistance to the Echinocandin Antifungal Drug Caspofungin in Candida albicans by Profiling Mutations in FKS1. Antimicrob. Agents Chemother. 2006, 50, 2058–2063. [Google Scholar] [CrossRef] [PubMed]
- Garnaud, C.; Botterel, F.; Sertour, N.; Bougnoux, M.-E.; Dannaoui, E.; Larrat, S.; Hennequin, C.; Guinea, J.; Cornet, M.; Maubon, D. Next-generation sequencing offers new insights into the resistance of Candida spp. to echinocandins and azoles. J. Antimicrob. Chemother. 2015, 70, 2556–2565. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Deshpande, L.M.; Davis, A.P.; Rhomberg, P.R.; Pfaller, M.A. Monitoring Antifungal Resistance in a Global Collection of Invasive Yeasts and Molds: Application of CLSI Epidemiological Cutoff Values and Whole-Genome Sequencing Analysis for Detection of Azole Resistance in Candida albicans. Antimicrob. Agents Chemother. 2017, 61, e00906-17. [Google Scholar] [CrossRef]
- Healey, K.R.; Kordalewska, M.; Ortigosa, C.J.; Singh, A.; Berrío, I.; Chowdhary, A.; Perlin, D.S. Limited ERG11 Mutations Identified in Isolates of Candida auris Directly Contribute to Reduced Azole Susceptibility. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Lee, A.; Jiménez-Ortigosa, C.; Kordalewska, M.; Perlin, D.S.; Zhao, Y. Rapid Detection of ERG11-Associated Azole Resistance and FKS-Associated Echinocandin Resistance in Candida auris. Antimicrob. Agents Chemother. 2018, 63, e01811-18. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Loge, C.; Besse, B.; Hennequin, C.; Le Pape, P. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: New substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 2010, 66, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Rolley, N.; Kelly, D.E.; Kelly, S.L. Identification and Characterization of Four Azole-Resistant erg3 Mutants of Candida albicans. Antimicrob. Agents Chemother. 2010, 54, 4527–4533. [Google Scholar] [CrossRef]
- Selmecki, A.; Forche, A.; Berman, J. Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science 2006, 313, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.; Bhattacharya, S.; Wakabayashi, H.; Bellaousov, S.; Kravets, A.; Welle, S.L.; Myers, J.; Hayes, J.J.; Bulger, M.; Rustchenko, E. Transcriptional Regulation on Aneuploid Chromosomes in Divers Candida albicans Mutants. Sci. Rep. 2018, 8, 1630. [Google Scholar] [CrossRef] [PubMed]
- Coste, A.; Turner, V.; Ischer, F.; Morschhäuser, J.; Forche, A.; Selmecki, A.; Berman, J.; Bille, J.; Sanglard, D. A Mutation in Tac1p, a Transcription Factor Regulating CDR1 and CDR2, Is Coupled with Loss of Heterozygosity at Chromosome 5 to Mediate Antifungal Resistance in Candida albicans. Genetics 2006, 172, 2139–2156. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Kravets, A.; Bethlendy, G.; Welle, S.; Rustchenko, E. Chromosome 5 Monosomy of Candida albicans Controls Susceptibility to Various Toxic Agents, Including Major Antifungals. Antimicrob. Agents Chemother. 2013, 57, 5026–5036. [Google Scholar] [CrossRef]
- Perlin, D.S. Antifungal drug resistance: Do molecular methods provide a way forward? Curr. Opin. Infect. Dis. 2009, 22, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.H.; Pfaller, M.A. Has antifungal susceptibility testing come of age? Clin. Infect. Dis. 2002, 35, 982–989. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).