Abstract
Penicillium brevicompactum is a filamentous ascomycete used in the pharmaceutical industry to produce mycophenolic acid, an immunosuppressant agent. To extend options for genetic engineering of this fungus, we have tested two resistance markers that have not previously been applied to P. brevicompactum. Although a generally available phleomycin resistance marker (ble) was successfully used in DNA-mediated transformation experiments, we were not able to use a commonly applicable nourseothricin resistance cassette (nat1). To circumvent this failure, we constructed a new nat gene, considering the codon bias for P. brevicompactum. We then used this modified nat gene in subsequent transformation experiments for the targeted disruption of two nuclear genes, MAT1-2-1 and flbA. For MAT1-2-1, we obtained deletion strains with a frequency of about 10%. In the case of flbA, the frequency was about 4%, and this disruption strain also showed reduced conidiospore formation. To confirm the deletion, we used ble to reintroduce the wild-type genes. This step restored the wild-type phenotype in the flbA deletion strain, which had a sporulation defect. The successful transformation system described here substantially extends options for genetically manipulating the biotechnologically relevant fungus P. brevicompactum.
1. Introduction
Before the advent of DNA-mediated transformation systems in filamentous fungi, random mutagenesis and strain screening programs were the only genetic approaches to strain improvement in biotechnologically relevant fungi. However, these mutagenesis methods often resulted in strains that had deleterious effects on fungal growth, sporulation, and genome stability. In the past 40 years or so, different directed transformation systems have been established to overcome these problems [1]. Such systems deciphered diverse gene functions, based on targeted gene disruption or overexpression experiments [2]. Previously, advanced transformation strategies were developed for filamentous fungi, such as CaCl2/polyethylene glycol, electroporation, particle bombardment, and Agrobacterium tumefaciens–mediated transformation [3,4,5]. Moreover, RNA interference and CRISPR/Cas9 systems have been described as suitable approaches to genetic manipulation of fungi [6,7,8].
An efficient transformation strategy requires a suitable screening system and effective selectable markers. Antibiotic resistance genes of bacterial origin are the most frequently used markers in filamentous fungi because no mutant recipient strains are needed to select transformants. In contrast, auxotrophic marker systems require construction of appropriate recipients. The hygromycin B resistance (hph) gene is an applicable selection marker for most fungi. However, because some Aspergillus species are relatively resistant to this antibiotic, the phleomycin resistance gene (ble) has been described as an alternative dominant selectable marker for these species [2,9]. Application of antibiotics neomycin, carboxin, fludioxonil, and blasticidin for the selection of transformants also has been suggested for genetic engineering of filamentous fungi [3,5,10].
Here, we have tested different bacterial resistance markers for the DNA-mediated transformation of the filamentous fungus Penicillium brevicompactum. In 1893, this filamentous ascomycete was discovered to be the first-known producer with antibacterial activity [11]. Since then, diverse P. brevicompactum strains have been described as producing a large range of bioactive compounds. These products, including secondary metabolites, are of significant biotechnological and medical importance because of their antiviral, antifungal, antibacterial, antitumor, and antipsoriasis activities [12]. Currently, the pharmaceutical industry uses P. brevicompactum for the large-scale biosynthesis of mycophenolic acid, an immunosuppressant agent with derivatives that are applied for autoimmune conditions as well as for kidney, heart, and liver transplantation patients [13].
Because of the limited number of selection markers, few studies have demonstrated genetic engineering of P. brevicompactum. In one report, the nitrate reductase gene from Fusarium oxysporum was used as a selection maker in an appropriate nitrate reductase-deficient recipient [14]. Successful A. tumefaciens–mediated transformation of P. brevicompactum has been described, as well, in which hph was used as a selectable marker under the control of the gpdA promoter from Aspergillus nidulans [15]. Here, we used ble and the nourseothricin-resistance marker nat1 for gene deletion analysis, followed by complementation studies. Although the previously applied ble [16,17] worked properly in the complementation studies, the commercially available nat1 could not successfully be used for selection of antibiotic-resistant P. brevicompactum transformants, although other studies have applied this resistance marker successfully in filamentous fungi [18,19]. To circumvent this difficulty, we developed a novel codon-adapted nat1 selection marker that is suitable for high-frequency transformation of P. brevicompactum. Additionally, we demonstrated that this newly constructed gene together with ble is suitable for site-specific gene deletion experiments. Our findings extend the options for genetic manipulation of this important biotechnological fungus, which is the subject of metabolic engineering experiments, to optimize the production of mycophenolic acid and its analogues [13].
2. Material and Methods
2.1. Strains, Plasmids and Culture Conditions
All wild type and recombinant strains from Penicillium brevicompactum used in this investigation are shown in Table 1. The strains Dutch Centraalbureau voor Schimmelcultures CBS 257.29 and CBS 110068 were chosen for transformation and isolation of genomic DNA. To prepare the spore suspension, all P. brevicompactum strains were grown on Czapek Yeast Agar (CYA) [20] for 7 days at 27 °C. Medium M334, containing 2% glucose monohydrate, 2% tryptone, 0.3% potassium dihydrogen phosphate and 0.1% magnesium sulphate heptahydrate, was used as a minimal liquid growth medium. For sensitivity tests against antibiotics, we used a complete culture medium (CCM) [21].

Table 1.
Fungal strains used in this investigation.
For plasmid construction we used Escherichia coli (E. coli), K12 XL1-Blue. A standard cloning protocol was carried out for electroporation of E. coli cells [22].
2.2. Construction of Transformation Vectors
The custom-synthetized Pbnat1 gene is codon-adapted and carries restriction sites for NcoI and BamHI that flank the gene. Prior to ligation, both Pbnat1 and the vector pPtrpCnat1 were digested with NcoI and BamHI. The resulting plasmid, pPtrpC-Pbnat1, served for the construction of deletion vectors. To ensure high homologous recombination efficiency, we used about 1 kb of the 5’ and 3’ flanking regions of MAT1-2-1 and flbA. The MAT1-2-1- 5’ flanking region was amplified by PCR using the oligonucleotides 5´-Pb-MAT1-2-1-MluI-for and 5’-Pb-MAT1-2-1-EcoRI-rv. Restriction with corresponding enzymes and N-terminal fusion to PtrpC-Pbnat1 were followed by C-terminal fusion of the 3’ flanking regions, which had been amplified by PCR using oligonucleotides 3’-Pb-MAT1-2-1-NotI-for/ 3’-Pb-MAT1-2-1-NotI-rv. For ligation, the amplicon was digested with NotI. We used the same strategy for the construction of the plasmid pPb-flbA-KO. The downstream fragment was generated by PCR using 5’-Pb-flbA-PstI-for/ 5´-Pb-flbA-PstI-rv as primers, restricted with PstI, and fused N terminally to PtrpC-Pbnat vector. It then was amplified by PCR with primer pairs 3’-PbflbA-NotI-for/ 3’-flbA-NotI-rv and ligated into the NotI restriction site, which terminates the 3´end of Pbnat1.
For complementation of the corresponding deletion strains, MAT1-2-1 was amplified from genomic DNA of CBS 110068 using oligonucleotides MAT1-2-1-OE-EcoRI-for/ MAT1-2-1_EcoRI-OE-rv. To amplify flbA, we used as template genomic DNA from CBS 257.29, using oligonucleotides Pb-flbA-BglII-OE-for/ Pb-flbA-BamHI-OE-rv. Both genes were fused to the 3´-end of Pgpd-egfp and subsequently introduced separately into pN-EGFP [23]. nat1 in pN-EGFP was further substituted by ble. The sequences of all oligonucleotides are given in Table S3, and plasmids in Table S4.
2.3. Bioinformatics and Programs
We obtained the genome sequences of AgRF18 strains from online database Joint Genome Institute (JGI) (https://genome.jgi.doe.gov/portal/). The JGI local blast tool was used to receive the sequences of MAT1-2-1 and flbA genes. To generate the codon usage table, we used the Bioperl based fascodon program. The sequence of the synthetic Pbnat1 gene was designed, using the online tool GENEius-The Tuning Tool (https://www.eurofinsgenomics.eu/en/gene-synthesis-molecular-biology/geneius/). Custom gene synthesis was done by GenScript Corporation, 120 Centennial Ave., Piscataway, NJ 08854, USA (https://www.genscript.com/). For calculation of CAI, we used the online CAIcal algorithm local version 1.3 (http://genomes.urv.es/CAIcal/) [24]. For in silico cloning strategies, we used SnapGene version 5.0, GSL Biotech LLC program (https://www.snapgene.com/).
2.4. Transformation of P. brevicompactum Strains
DNA-mediated transformation of P. brevicompactum strains was performed with some slight modifications, as previously described by Bull et al. (1988) [25]. Briefly, strains were grown in M344 shaking liquid medium (230 rpm) at 25 °C for 24 h. Three grams of filtered mycelia were used for protoplast preparation. Protoplast preparation involved shaking the fungal mycelia at 110 rpm and 25 °C for 2h in 35 mL 0.9 M NaCl buffer containing 40 mg/mL Vinotaste®Pro enzyme. A total of 1 × 107 protoplasts in 100 µL transformation buffer (0.9M NaCl + 0.9M CaCl2) was transformed with 10 µg linear or circular DNA by means of a second transformation buffer (50% PEG 6000, 50 mM CaCl2, 10 mM Tris; pH 5.0). For selection of transformants on solid complete culture medium supplemented with 2% glucose, we used 200 µg/mL nourseothricin or 100 µg/mL phleomycin. To obtain purified transformants from single colonies, we streaked spores on CCM medium containing selection antibiotics. Molecular analysis of transgenic fungal strains by PCR and Southern blotting and hybridization was done as described previously [19,22].
3. Results and Discussion
The aim of this study was to develop an alternative transformation system for targeted gene disruption in different strains of the biotechnologically relevant fungus P. brevicompactum.
3.1. Test for Sensitivity Against Antibiotics and DNA Transformation
To use bacterial resistance markers for DNA-mediated transformation, we tested the sensitivity of six type culture collection strains from P. brevicompactum against different concentrations of nourseothricin and phleomycin. Our results indicated that all wild-type strains (Table 1) are sensitive to even low concentrations of nourseothricin (Figure 1). Indeed, we found a rather high sensitivity in all strains tested, and even a low antibiotic concentration of 10 µg/mL resulted in growth reduction. This exposure is much lower than the 150 µg/mL previously used to select transformants of the related P. chrysogenum [26]. However, a higher phleomycin concentration is required for growth inhibition of CBS 110068 and IBT 23078. Thus, a concentration of 100 µg/mL is necessary for reliable selection of transformants on phleomycin-containing medium.
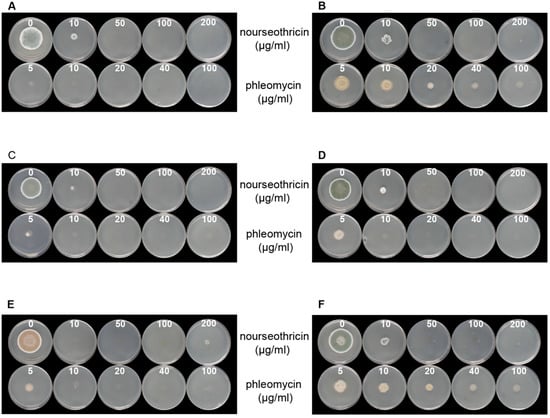
Figure 1.
Test for sensitivity against different concentrations of antibiotics as given for six P. brevicompactum wild type strains. (A) CBS 257.29; (B) CBS 110068; (C) CBS 110070; (D) CBS110071; (E) CBS 317.59; (F) IBT 27083. Antibiotic concentrations (µg/mL) are indicated in white on each plate. For further transformation experiments, CBS 257.29 and CBS 110068 were selected.
To examine the suitability of ble as well as nat1 for P. brevicompactum transformation, we used two transformation vectors that previously had been successfully applied for Penicillium chrysogenum: p1783-1 and pDrive/PtrpC-Tn5Phleo [21,27]. For hosts, we applied two strains carrying MAT1-1-1 (CBS 257.29) or MAT1-2-1 (CBS 110068). Both show a high production of 3.1 and 1.8 g mycophenolic acid per kilogram of mycelia, respectively, and thus can be considered as high producers of this immunosuppressant [28].
Using the vector pDrive/PtrpC-Tn5Phleo for transformation, we found a rather good frequency of transformants (about 6 per 10 µg DNA) for both recipient strains on phleomycin-containing plates. In contrast, the nat1 gene (p1783-1) yielded no transformants in 15 individual transformation experiments. This gene originates from the prokaryote Streptomyces nourseii, and its codon usage is not adapted to the expression machinery of eukaryotic host systems. To optimize expression of nat1, we decided to synthesize it with a codon bias adapted to P. brevicompactum.
3.2. Construction of a Codon-Adapted nat1 Gene
The successful expression of codon-optimized genes in diverse hosts from filamentous fungi has previously been reported. For example, a completely synthetized codon-optimized flp recombinase gene from yeast Saccharomyces cerevisiae allowed for successful establishment of a marker recycling system in P. chrysogenum and Sordaria macrospora [19]. Moreover, construction of a codon-adapted luciferase gene for expression in Neurospora crassa provided a novel reporter assay system for this fungus [29]. Furthermore, the efficient expression of a codon-optimized Derf7 gene encoding a mite allergen in Aspergillus oryzae suggested that fungi can serve as hosts for the synthesis of recombinant allergen used in immunotherapy [30].
To improve the expression efficiency of nat1 in P. brevicompactum, we determined the codon bias for the fungus. For this purpose, we accessed the available genome sequences of the P. brevicompactum strain AgRF18 (online database of the Joint Genome Institute: https://genome.jgi.doe.gov/portal/) to generate a codon usage table (Supplementary Table S1). Table 2 gives the corresponding frequencies of codon usages. We selected 12,343 protein-coding sequences (CDS) and 5,661,200 codons from P. brevicompactum and compared them with the codon usage of native nat1 from S. noursei. This comparison yielded significant differences between the codon biases. For instance, AGT is the preferential codon for the amino acid aspartate in P. brevicompactum (51.8%) but is used at a frequency of only 4.4% in the native nat1 gene, which shows a bias toward using AGC to encode aspartate (Table 2). Moreover, the GC content is quite different between nat1 (71.2%) and the P. brevicompactum genome (52.8%). Because of these differences, we designed a novel nat1 gene with a codon bias adapted to the codon-preferred bias of P. brevicompactum.

Table 2.
Comparison of the codon usage of nat1 and Pbnat1 with the genomic codon usage of P. brevicompactum (Pb) (AgRF18), (https://genome.jgi.doe.gov/portal/). Numbers give the usage bias of each codon for each amino acid in percent. Preferred amino acid codons are labelled in red. First column in the left indicate the first base of triplets, upper row the second base and last column the third base.
To obtain the most suitable codon, we generated the codon-optimized gene in silico, as described in the materials and methods section. This modified sequence is given in Figure 2, and served for the customized gene synthesis. We designated the corresponding gene as Pbnat1. This novel Pbnat1 gene with 88.4% DNA homology to nat1 has a GC content of 63.7%, which is more similar to the overall GC content of the P. brevicompactum genome (52.8%). This modification is important because GC content is a main mediator of codon and amino acid usage and thus the most significant factor determining codon bias [31]. In total, in Pbnat1, we changed 7.5% of the first (GC1) and 23.2% of the third (GC3) codon positions (Supplementary Table S2).

Figure 2.
Comparative sequence alignment of codon adapted Pbnat1 gene with the commercially available nat1 gene. 82 nucleotide changes are marked in light blue.
To examine the compatibility Pbnat1 with P. brevicompactum codon bias, we calculated the codon adaptation index (CAI) for both nat1 and Pbnat1. For this purpose, we used the online CAIcal algorithm (http://genomes.urv.es/CAIcal/) [24]. The CAI for a specific gene is defined by comparing its codon usage rate and frequency in a reference set of genes. Assigned values are between 0 and 1. With CAI values closer to 1, the expression level of the targeted gene is expected to be better in a heterologous host system [19,32,33]. Changing about 43% of all codons shifted the CAI value for Pbnat1 from 0.83 to 0.91. This result led us to predict that Pbnat1 compared with nat1 would have an optimized and more accurate expression in P. brevicompactum.
3.3. Use of the Codon-Optimized Pbnat1 Gene for Site-Specific Deletion of Two Nuclear Genes
To improve the transformation efficiency in P. brevicompactum, we developed a reliable procedure based on the use of protoplasts. For this purpose, we tested two buffers: potassium phosphate [14] and 0.9 M sodium chloride [25] for CBS 257.29 and CBS 110068. Based on our results, we concluded that 0.9 M sodium chloride containing 40 mg/mL Vinotaste®Pro digestive enzyme was the only effective buffer for protoplasting of P. brevicompactum strains.
After establishing the transformation method (see the material and methods section), we investigated the efficiency of the newly constructed Pbnat1 gene in P. brevicompactum using the vector pPtrpC-Pbnat1. This plasmid contains the Pbnat1 gene under the control of the trpC promoter from A. nidulans. Application of Pbnat1 led to successful transformation events at a frequency of about 10 nourseothricin-resistant transformants per 10 µg of circular DNA. All transformants were genetically stable on selective media and were propagated for more than 12 months.
In the next step, we applied the Pbnat1 gene containing the vector pPtrpC-Pbnat1 for site-specific deletion of two genes inferred to be involved in fungal development: flbA and MAT1‐2‐1. flbA encodes a regulator of G-protein signaling that is involved in asexual sporulation and mycelial proliferation, and MAT1-2-1 encodes a transcription factor that controls sexual development [17,34,35]. To generate deletion mutants by homologous recombination, we transformed the strains with a linear deletion cassette containing 5‘-and 3’- flanking regions derived from the target gene and each about 1 kb long (Figures S1A and S2A). The corresponding cassettes were obtained from plasmids, shown in Figure 3A,B. CBS 257.29 and CBS 110068 served as the recipient strains for disruption of flbA and MAT1-2-1, respectively. With linear DNA for deletion of MAT1-2-1, we observed a frequency of 10–12 transformants per 10 µg of linear DNA. The transformation frequency was considerably less when we used the flbA deletion cassette (pPb-flbA-KO), with about 2–4 transformants per 10 µg of DNA.
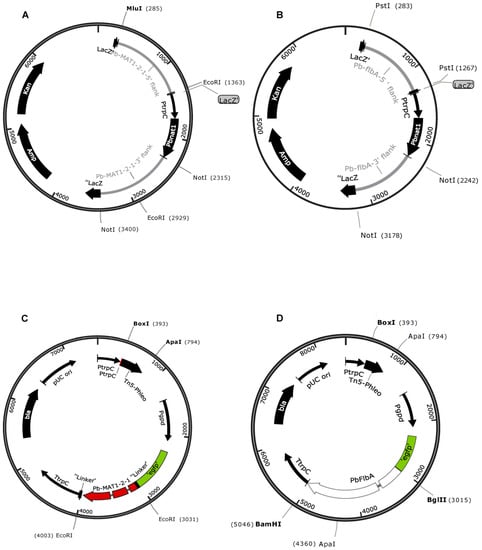
Figure 3.
Maps of vectors for DNA mediated transformation of P. brevicompactum. (A,B) Plasmids pPb-MAT1-2-1-KO (A) and pPb-flbA-KO (B) for site-specific deletion of MAT1-2-1 and flbA genes, carrying the codon adapted pbnat1 gene under the transcriptional control of the PtrpC promoter; (C,D) Plasmids pPb-MAT1-2-1-comp (C) and pPb-flbA-comp (D) for complementation of the corresponding deletion strains. Both plasmids carry the egfp-tagged gene of interest under the constitutive gpd promoter from A. nidulans.
To verify that homologous recombination had occurred, we performed PCR amplification and Southern hybridizations with a probe that was homologous to the 5’-flanking region of the target gene (Supplementary Figures S1B, S1C, S2B, S3F). Our data confirmed the presence of homokaryotic deletion strains. While the MAT1-2-1 deletion strains showed no detectable phenotype, the flbA deletion strains were distinct from the wild type because of a delayed and reduced sporulation phenotype (Figure 4). This developmental feature is similar to previously reported phenotypes that resulted when the homologous flbA gene was deleted in different Aspergillus species [36,37]. Verification of 60 putative MAT1-2-1 strains revealed a frequency of 10% for deletion of MAT1-2-1 gene. In contrast, only 1 of 24 putative flbA deletion strains was identified as a homokaryotic deletion strain.
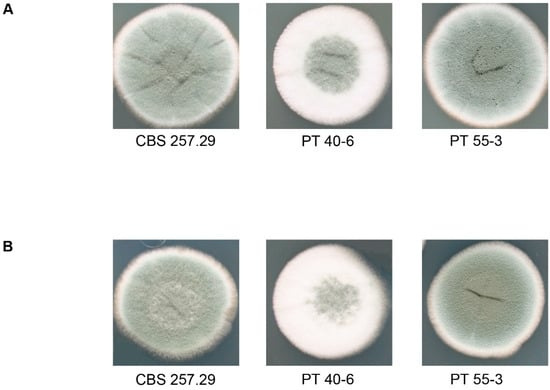
Figure 4.
Phenotypic analysis of flbA deletion and corresponding complementation strains after 7 days of growth on CCM medium at 27 °C, under different light condition. (A) light, (B) dark. CBS 257.29: wild type; PT 40-6: deletion strain; PT 55-3: complementation strain.
In the next step, we conducted complementation experiments to confirm the successful deletion of the target genes. For complementation, we used the plasmids pPb-MAT1-2-1-comp and pPb-flbA-comp to transform the appropriate deletion strains (Figure 3C, 3D). PCR analysis and Southern hybridization with a target gene–specific probe served as confirmation of the successful ectopic integration of the wild-type genes (Figure S3). The ectopic integration of flbA into the deletion strains activated a wild type–like sporulation phenotype (Figure 4).
In our experiments we observed a sufficiently high frequency of homologous recombination in P. brevicompactum. Therefore, it is not necessary to use mutants in which non-homologous end joining (NHEJ) is abolished by mutation of the NHEJ repair system. Neurospora crassa was the first filamentous ascomycete in which homologous recombination was enhanced by disrupting genes for the catalytic subunit (DNA-PKcs) or the regulatory DNA-binding subunits (Ku70/80 heterodimer) [38,39]. Later, strains from diverse ascomycetes were generated to serve as recipients for the targeted integration of foreign genes [6,40]. However, several reports showed that non-homologous end joining–deficient strains accumulate random mutations and thus are less suitable for long-term experiments. Furthermore, these recipient strains show an elevated sensitivity to different chemicals such as bleomycin, methyl methanesulfonate, and ethyl methanesulfonate (for review, see [6,40]). Thus, some investigators have avoided using specific recipients for homologous recombination because the natural homologous recombination frequency was similar to that described here [41,42,43].
In conclusion, we have extended the molecular tools for genetic manipulation of the biotechnologically relevant fungus P. brevicompactum. The opportunity to generate deletion and complementation strains by the use of different marker genes opens up future avenues to research to identify factors that control or regulate secondary metabolism in this mycophenolic acid–producing fungus.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/5/4/96/s1, Figure S1: Construction of a MAT1-2-1 deletion mutant; Figure S2: Construction of a flbA deletion strains; Figure S3: Complementation of MAT1-2-1 and flbA deletion strains; Table S1: Codon usage in P. brevicompactum. The genomic sequences of strain AgRF18 was used for calculating frequency of preferred amino acid codons; Table S2: Comparison of CAI and GC-content of nat1 and Pbnat1; Table S3: List of oligonucleotides used in this study; Table S4. List of plasmids, used in this study.
Author Contributions
Y.M., B.H., and U.K. designed the experimental strategy, Y.M. performed the experiments; Y.M. and U.K. analyzed data; Y.M., B.H., U.K. wrote the manuscript.
Funding
The work of the authors was funded by the Christian Doppler Society (Vienna, Austria) and a grand from the German Research Foundation (DFG KU 517/15-1).
Acknowledgement
The authors are thankful to Fabian Becker for conducting some of the experiments during his Bachelor thesis and T. Dahlmann for help in bioinformatics applications. We would like to acknowledge the technical advice of I. Godehardt, and the fruitful discussion with T. Dahlmann (RUB, Bochum), J. Finke (Janssen-Cilag GmbH, Neuss, D), H. Hecker (Novartis, Kundl, A), H. Kürnsteiner (Novartis, Kundl, A), and I. Zadra (Novartis, Kundl, A).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kluge, J.; Terfehr, D.; Kück, U. Inducible promoters and functional genomic approaches for the genetic engineering of filamentous fungi. Appl. Microbiol. Biotechnol. 2018, 102, 6357–6372. [Google Scholar] [CrossRef] [PubMed]
- He, Z.M.; Price, M.S.; Obrian, G.R.; Georgianna, D.R.; Payne, G.A. Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 2007, 7, 104. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Diez, B. Strategies for the transformation of filamentous fungi. J. Appl. Microbiol. 2002, 92, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Arentshorst, M.; Ram, A.F.; van den Hondel, C.A. Agrobacterium-mediated transformation leads to improved gene replacement efficiency in Aspergillus awamori. Fungal Genet. Biol. 2005, 42, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Weld, R.J.; Plummer, K.M.; Carpenter, M.A.; Ridgway, H.J. Approaches to functional genomics in filamentous fungi. Cell Res. 2006, 16, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Kück, U.; Hoff, B. New tools for the genetic manipulation of filamentous fungi. Appl. Microbiol. Biotechnol. 2010, 86, 51–62. [Google Scholar] [CrossRef]
- Krappmann, S. CRISPR-Cas9, the new kid on the block of fungal molecular biology. Med. Mycol. 2017, 55, 16–23. [Google Scholar] [CrossRef]
- Shi, T.Q.; Liu, G.N.; Ji, R.Y.; Shi, K.; Song, P.; Ren, L.J.; Huang, H.; Ji, X.J. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef]
- Austin, B.; Hall, R.M.; Tyler, B.M. Optimized vectors and selection for transformation of Neurospora crassa and Aspergillus nidulans to bleomycin and phleomycin resistance. Gene 1990, 93, 157–162. [Google Scholar] [CrossRef]
- Jiang, D.; Zhu, W.; Wang, Y.; Sun, C.; Zhang, K.Q.; Yang, J. Molecular tools for functional genomics in filamentous fungi: Recent advances and new strategies. Biotechnol. Adv. 2013, 31, 1562–1574. [Google Scholar] [CrossRef]
- Bentley, R. Mycophenolic Acid: A one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000, 100, 3801–3826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Cao, S.; Qiu, L.; Qi, F.; Li, Z.; Yang, Y.; Huang, S.; Bai, F.; Liu, C.; Wan, X.; et al. Functional characterization of MpaG’, the O-methyltransferase involved in the biosynthesis of mycophenolic acid. Chembiochem 2015, 16, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Regueira, T.B.; Kildegaard, K.R.; Hansen, B.G.; Mortensen, U.H.; Hertweck, C.; Nielsen, J. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl. Environ. Microbiol. 2011, 77, 3035–3043. [Google Scholar] [CrossRef] [PubMed]
- Varavallo, M.A.; de Queiroz, M.V.; Pereira, J.F.; Ribeiro, R.A.; Soares, M.A.; Ribeiro, J.B.; de Araujo, E.F. Development of a transformation system for Penicillium brevicompactum based on the Fusarium oxysporum nitrate reductase gene. Braz. J. Microbiol. 2005, 36, 184–189. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, J.; Xu, R.; Lv, X.; Wang, L.; Sun, A.; Wei, D. Insertion mutation in HMG-CoA lyase increases the production yield of MPA through Agrobacterium tumefaciens-mediated transformation. J. Microbiol. Biotechnol. 2016, 26, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- Kolar, M.; Punt, P.J.; Vandenhondel, C.A.M.J.J.; Schwab, H. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli LacZ fusion gene. Gene 1988, 62, 127–134. [Google Scholar] [CrossRef]
- Böhm, J.; Dahlmann, T.A.; Gümüser, H.; Kück, U. A MAT1-2 wild-type strain from Penicillium chrysogenum: Functional mating-type locus characterization, genome sequencing and mating with an industrial penicillin-producing strain. Mol. Microbiol. 2015, 95, 859–874. [Google Scholar] [CrossRef]
- Kück, U.; Hoff, B. Application of the nourseothricin acetyltransferase gene (nat1) as dominant marker for the transformation of filamentous fungi. Fungal Genet. Newsl. 2006, 53, 9–11. [Google Scholar] [CrossRef]
- Kopke, K.; Hoff, B.; Kück, U. Application of the Saccharomyces cerevisiae FLP/FRT recombination system in filamentous fungi for marker recycling and construction of knockout strains devoid of heterologous genes. Appl. Environ. Microbiol. 2010, 76, 4664–4674. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Böhm, J.; Hoff, B.; O’Gorman, C.M.; Wolfers, S.; Klix, V.; Binger, D.; Zadra, I.; Kurnsteiner, H.; Poggeler, S.; Dyer, P.S.; et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA 2013, 110, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.R.; Green, M.R. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Kück, U.; Pöggeler, S.; Nowrousian, M.; Nolting, N.; Engh, I. Sordaria macrospora, a model system for fungal development. In Physiology and Genetics: Selected Basic and Applied Aspects; Anke, T., Weber, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 17–39. [Google Scholar]
- Puigbò, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct 2008, 3, 38. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.H.; Smith, D.J.; Turner, G. Transformation of Penicillium chrysogenum with a dominant selectable marker. Curr. Genet. 1988, 13, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Hoff, B.; Kamerewerd, J.; Sigl, C.; Zadra, I.; Kück, U. Homologous recombination in the antibiotic producer Penicillium chrysogenum: Strain DeltaPcku70 shows up-regulation of genes from the HOG pathway. Appl. Microbiol. Biotechnol. 2010, 85, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Gesing, S.; Schindler, D.; Franzel, B.; Wolters, D.; Nowrousian, M. The histone chaperone ASF1 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 2012, 84, 748–765. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudjanlou, Y.; Dahlmann, T.; Kück, U. Molecular analysis of mating type genes in the mycophenolic acid producer Penicillium brevicompactum. 2019; in preparation. [Google Scholar]
- Gooch, V.D.; Mehra, A.; Larrondo, L.F.; Fox, J.; Touroutoutoudis, M.; Loros, J.J.; Dunlap, J.C. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot. Cell 2008, 7, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Tokuoka, M.; Tanaka, M.; Ono, K.; Takagi, S.; Shintani, T.; Gomi, K. Codon optimization increases steady-state mRNA levels in Aspergillus oryzae heterologous gene expression. Appl. Environ. Microbiol. 2008, 74, 6538–6546. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Dang, Y.; Zhou, M.; Li, L.; Yu, C.H.; Fu, J.; Chen, S.; Liu, Y. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl. Acad. Sci. USA 2016, 113, E6117–E6125. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Quax, T.E.; Claassens, N.J.; Soll, D.; van der Oost, J. Codon bias as a means to fine-tune gene expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef]
- Yu, J.H.; Wieser, J.; Adams, T.H. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996, 15, 5184–5190. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Beer, C.; Freitag, M.; Kück, U. Genome-wide identification of target genes of a mating-type-domain transcription factor reveals functions beyond sexual development. Mol. Microbiol. 2015, 96, 1002–1022. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.A.; Yu, J.H. The phosducin-like protein PhnA is required for Gbetagamma-mediated signaling for vegetative growth, developmental control, and toxin biosynthesis in Aspergillus nidulans. Eukaryot. Cell 2006, 5, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Aerts, D.; Hauer, E.E.; Ohm, R.A.; Arentshorst, M.; Teertstra, W.R.; Phippen, C.; Ram, A.F.J.; Frisvad, J.C.; Wosten, H.A.B. The FlbA-regulated predicted transcription factor Fum21 of Aspergillus niger is involved in fumonisin production. Antonie Van Leeuwenhoek 2018, 111, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, Y.; Suzuki, K.; Ishii, C.; Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2004, 101, 12248–12253. [Google Scholar] [CrossRef]
- Walker, J.R.; Corpina, R.A.; Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001, 412, 607–614. [Google Scholar] [CrossRef]
- Krappmann, S. Gene targeting in filamentous fungi: The benefits of impaired repair. Fungal Biol. Rev. 2007, 21, 25–29. [Google Scholar] [CrossRef]
- Siewers, V.; Kokkelink, L.; Smedsgaard, J.; Tudzynski, P. Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 2006, 72, 4619–4626. [Google Scholar] [CrossRef]
- Lin, C.H.; Chung, K.R. Specialized and shared functions of the histidine kinase- and HOG1 MAP kinase-mediated signaling pathways in Alternaria alternata, a filamentous fungal pathogen of citrus. Fungal Genet. Biol. 2010, 47, 818–827. [Google Scholar] [CrossRef]
- Lou, H.; Ye, Z.; Yun, F.; Lin, J.; Guo, L.; Chen, B.; Mu, Z. Targeted gene deletion in Cordyceps militaris using the split-marker approach. Mol. Biotechnol. 2018, 60, 380–385. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).