Abstract
The filamentous fungal genus Aspergillus represents an industrially significant group of eukaryotic microorganisms. For nearly a century, it has been widely utilized in the production of diverse high-value products, including organic acids, industrial enzymes, recombinant proteins, and various bioactive natural compounds. With the rapid advancement of synthetic biology, Aspergillus has been extensively exploited as a heterologous chassis for the production of heterologous proteins (e.g., sweet proteins and antibodies) and the synthesis of natural products (e.g., terpenoids and polyketides) due to its distinct advantages, such as superior protein secretion capacity, robust precursor supply, and efficient eukaryotic post-translational modifications. In this review, we provide a comprehensive summary of the advancements in the successful expression of heterologous proteins and the biosynthesis of natural products using Aspergillus platforms (including Aspergillus niger, Aspergillus nidulans, and Aspergillus oryzae) in recent years. Emphasis is placed on the applications of A. oryzae in the heterologous biosynthesis of terpenoids. More importantly, we thoroughly examine the current state of the art in utilizing CRISPR-Cas9 for genetic modifications in A. oryzae and A. niger. In addition, future perspectives on developing Aspergillus expression systems are discussed in this article, along with an exploration of their potential applications in natural product biosynthesis.
1. Introduction
Conventional microorganism chassis such as Escherichia coli and Saccharomyces cerevisiae face inherent constraints, including limited biosynthetic capacity, metabolic burden, and insufficient precursor supply, which collectively impair their ability to meet the escalating societal requirements for sustainable and eco-friendly chemical production [1,2,3]. As an essential group of industrial microorganisms in filamentous fungi, Aspergillus has been extensively utilized in various fields, including pharmaceuticals, food processing, agriculture, enzyme production, cosmetics, and chemical materials [4,5]. Over the past few years, the advancement of synthetic biology has further promoted the application of Aspergillus in the synthesis of critical pharmaceuticals and agrochemicals, such as penicillin/cephalosporin (antibiotics), statins (cholesterol control), echinocandin (antifungals), ergot alkaloids (α-blocking agent), citric acid (food additives), kojic acid (skin whitening), itaconic acid (antitumor), and gibberellin (as a plant growth stimulant) [6,7,8,9]. Compared to model organisms, such as E. coli and S. cerevisiae, Aspergillus species possess an expansive repertoire of secondary metabolic gene clusters, including polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS), and terpenoid synthases, enabling direct biosynthesis of complex natural products (e.g., lovastatin, penicillin) [10,11]. Moreover, Aspergillus provides an abundant supply of key precursors and coenzymes required for the biosynthesis of bioactive natural products, complemented by compatible transcription, translation, and post-translational modification machineries, including glycosylation, phosphorylation, and acetylation [12,13]. Thus, Aspergillus has become increasingly indispensable in synthetic biology research, especially as a heterologous host for proteins and natural products. For example, industrial fungal strains for organic acid production, such as Aspergillus terreus and A. niger, not only possess robust central carbon metabolism and highly efficient organic acid secretion capacity but also demonstrate excellent stress-resistant characteristics including acid tolerance and thermotolerance during industrial fermentation processes [14,15]. The edible strain A. oryzae has been successfully applied for the production of heterologous proteins, including bovine chymosin (CHY) and human lysozyme (HLY) [16].
Although many heterologous proteins have been successfully expressed in Aspergillus (e.g., human cytokine interleukin 6 (IL-6) in A. niger [17], adalimumab in A. oryzae [18]), there are still many challenges that need to be addressed, including improving production and optimizing expression systems. With the aid of Aspergillus expression platforms, the biosynthetic pathways of numerous natural products, including terpenoids, polyketides, and non-ribosomal peptides, have been elucidated [19]. For example, A. nidulans and A. oryzae have emerged as model organisms for filamentous fungal biology, which are currently recognized as preferred fungal chassis for heterologous production of bioactive natural products [19,20]. This has greatly facilitated the activation of silent gene clusters and the discovery of new compounds. Notably, among the Aspergillus expression platforms, A. oryzae exhibits exceptional advantages for terpenoid biosynthesis, making it an outstanding microbial host for the production of these compounds. For example, it has been efficiently engineered for the production of pleuromutilin (a diterpenoid antibiotic) and cephalosporin P1 (a triterpenoid antibiotic) [21,22].
In this review, we concentrate on the A. niger, A. nidulans and A. oryzae expression system and summarize recent advances in successfully expressing proteins and bioactive natural products through these species, emphasizing the biosynthesis of terpenoids through A. oryzae. Furthermore, we provide a comprehensive review of the latest advancements in the application of CRISPR-Cas9 for genetic engineering in A. oryzae, aiming to offer valuable insights for future exploration of Aspergillus chassis and the construction of high-efficiency fungal platforms.
2. Aspergillus Is an Expression Host for Heterologous Protein Production
2.1. A. niger
A. niger, as an indispensable industrial strain, has been extensively exploited for homologous and heterologous expressions of many protein products used in food, washing, textile, paper, and other industries [23] due to its high protein secretion efficiency, unique safety characteristics, and low–medium costs [24]. On the one hand, the genome of A. niger contains an extensive repertoire of genes encoding carbohydrate active enzymes (CAZymes) [25], making it an ideal host for CAZyme production and homologous expression. For example, Fiedler et al. used the tunable Tet-on system to control the glaA gene encoding glucoamylase (GlaA) and generated the ΔracA strain via racA gene deletion, leading to a 4-fold increase in GlaA secretion compared to the parental strain [26]. Moreover, Zhang et al. constructed gpdA promoters with one to four copies of the gpd box (PgpdA, PgpdA2B, PgpdA3B, PgpdA4B) and used them to regulate xynB expression in A. niger. The strain containing three copies of gpd box (PgpdA3B) demonstrated the highest xylanase activity (3588.38 U/mL), protein expression, and transcription efficiency [27]. Furthermore, Cai et al. cloned two β-glucosidase (BGL) genes from marine A. niger ZJUBE-1 and then inserted them into the host genome for constitutive homologous expression. By leveraging the gpdA promoter to drive stable gene expression independent of inducers, they achieved directed expression of BGLs [28]. Similarly, Alazi et al. used the strong constitutive gpdA promoter to overexpress the transcription factor gaaR in A. niger, enabling constitutive transcription of pectinase-encoding genes, D-galacturonic acid (GA) transporter genes, and catabolic pathway enzymes even under non-inducing conditions [29]. Liu et al. cloned the tannase gene tan7 from A. niger SH-2 and overexpressed it in the low-background secretory strain A. niger Bdel4 using the glaA promoter, achieving a peak tannase activity of 111.5 U/mL at 168 h with over 70% purity in the supernatant [30]. In addition, A. niger has successfully enabled the homologous expression of amylases, cellulases, and other CAZymes, facilitating their large-scale industrial production [31,32]. On the other hand, A. niger has also been used as a cell factory to express some heterologous proteins, including lysozyme [33], human lactoferrin [34], chymosin [35], thaumatin [36], lipase [37], and nuclease P1 [38]. However, achieving the efficient expression of most heterologous proteins is still difficult [39]. In recent years, driven by the rapid advancement and widespread application of gene editing technology, the CRISPR/Cas9 system has been effectively employed for genetic manipulation in A. niger [40]. For example, this technology enabled a significant enhancement in trehalase MthT production from Thermothelomyces thermophilus, with activity reaching up to 1698.83 U/mL [41]. Moreover, a CRISPR/Cas9-mediated multi-copy expression system for an alkaline serine protease from A. oryzae was successfully established in A. niger, yielding a protease activity of 11,023.2 U/mL and a protein concentration of 10.8 mg/mL [42].
2.2. A. oryzae
A. oryzae is identified as a GRAS (generally recognized as safe) organism by the U.S. Food and Drug Administration and has been used by the fermentation and food processing industries to produce sake, miso, soy sauce, and douchi for centuries [5]. Due to its strong protein secretion capabilities and reliable safety, A. oryzae is a suitable host for heterologous protein production [20]. Using A. oryzae as a host to express heterologous proteins has become increasingly well-established. For example, recombinant human lactoferrin [43], human lysozyme [44], calf chymosin [45], and other heterologous proteins have been successfully expressed to date. Moreover, in recent years, recombinant antibodies (such as adalimumab) with certain biological activity were also successfully expressed and purified in A. oryzae [18]. Additionally, cordycepin, a natural antibiotic with numerous pharmacological activities, was efficiently expressed by overexpressing two metabolic genes (cns1 and cns2) involved in cordycepin biosynthesis [43].
2.3. A. nidulans
A. nidulans has long been recognized as a model organism for eukaryotic research, owing to its well-characterized genetics, physiology, and amenability to molecular manipulation [44,45]. Although A. nidulans is not a predominant industrial species, it serves as a promising microbial platform for industrial enzyme production. To date, A. nidulans has synthesized several industrial enzymes, including endoglucanase, xylanase, cellulases, β-glucosidases, laccases, and lipases [46]. Most of them were produced under submerged fermentation rather than solid-state fermentation due to the availability of fermentation parameters and reduced fermentation time of submerged fermentation [47]. In addition, A. nidulans is also a versatile fungal cell factory for the heterologous production of different carbohydrate active enzymes (CAZymes), which are industrially relevant biocatalysts responsible for the degradation of plant cell walls, with a promising result for the production of bio-based compounds from lignocellulosic feedstock, such as biofuel and nutraceuticals [48]. However, due to its relatively low protein secretion capacity, A. nidulans has not been fully developed as a cell factory for heterologous protein expression. Therefore, Yan et al. successfully developed a high-efficiency A. nidulans strain (ΔagsB-derA) by performing stepwise modification of mycelial morphology and protein secretory pathway to alleviate this limitation. Consequently, a higher yield of secretory and expression of human interleukin-6 (HuIL-6) was achieved by further disruption of extracellular proteases, indicating that A. nidulans is a promising platform for efficient heterologous protein expression [49]. Very recently, Gerhardt et al. employed CRISPR/Cas9 technology to examine how deleting genes related to N-glycan assembly and endoplasmic reticulum protein quality control affects recombinant β-xylosidase secretion in A. nidulans. Their findings demonstrated that combined algC and algI deletion enhances the secretion and alters the secretome, which may be a promising strategy for boosting recombinant protein secretion [50].
Although the application of heterologous protein expression using A. niger, A. nidulans, and A. oryzae as hosts has reached a relatively mature stage, in-depth studies on the regulation of gene expression and protein secretory pathways are still required to further optimize expression platforms, enhance protein yields, and ultimately establish highly efficient expression systems for heterologous proteins [20,51,52].
3. Aspergillus Is a Microbial Chassis for Natural Product Synthesis
In addition to being exploited as excellent hosts for heterologous protein expression, filamentous fungi have also been developed as ideal platforms for the production of natural products due to their exceptional capacity for secondary metabolite production, their inherent pre-mRNA splicing mechanisms, and their abundant supply of biosynthetic precursors and coenzymes [19,53]. Compared to typical heterologous hosts such as E. coli and yeast, the most notable advantage of filamentous fungi is their capability to express entire gene clusters responsible for the biosynthesis of fungal natural products [54,55]. Moreover, during the cloning of these gene clusters, there is no requirement to eliminate introns, as most filamentous fungal hosts can accurately execute the splicing of introns from secondary metabolite genes derived from other fungi, thereby successfully yielding the desired products [56,57]. Several filamentous fungi, such as A. oryzae, A. nidulans, and A. niger, have been successfully utilized for the study of biosynthetic pathways and heterologous production of natural products.
3.1. A. oryzae Is a Chassis for Natural Product Production
A. oryzae is a robust platform for the heterologous production of natural products, which are still abundant in modern drug discovery [58]. This platform not only facilitates the elucidation of the biosynthetic pathway of a given natural product but also allows the activation of silent or cryptic biosynthetic gene clusters, making a great contribution to simplifying the biosynthetic pathway, discovering new natural products and their derivatives, and genome mining. Thus, the A. oryzae host has been widely utilized for genome mining and the biosynthesis of fungal natural products, including polyketides, non-ribosomal peptides, and terpenoids (e.g., sesquiterpenoids, diterpenoids, sesterterpenoids) [59,60], which have always been considered as important ingredients in therapeutic agents.
3.1.1. Transformation Methods in A. oryzae
Currently, the heterologous expression of biosynthetic genes encoding natural products in A. oryzae primarily relies on the PEG/CaCl2-mediated protoplast transformation method [61], and the process is shown in Figure 1. Using molecular biology, the exogenous genes were loaded onto the vectors of nutrient-deficient screening markers and resistance markers, and the target gene vectors were transformed into protoplasts with the help of the high-sugar and -salt osmotic pressure of the transformation reagents. The target gene can be inserted into the chromosome of A. oryzae after homologous recombination between the vector and the host genome [62]. Transformed single genes or gene clusters can be inserted individually or as a whole. For example, Sakai et al. [58] successfully cloned the entire gene clusters (40 kb for monacolin K biosynthesis in Monascus ruber and 12 kb for terrequinone A biosynthesis in A. nidulans) into vectors via cosmid library construction, achieving heterologous expression of both gene clusters in the A. oryzae host. Single-gene insertion is a process in which the 5′ and 3′ ends of the target gene are added to an amylase (amyB) [63] or amylase-enhanced [64] promoter and terminator, respectively, and then induced by starch or dextrin in the culture medium, which enables the efficient expression of the target gene [21,65].
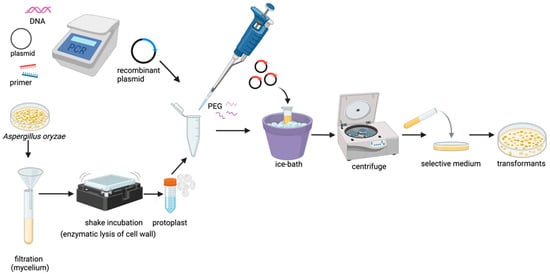
Figure 1.
The process of protoplast-mediated transformation in A. oryzae. (Figure 1 was created with BioRender.com).
Although protoplast transformation enables efficient selection of transformants with successful target gene integration and facilitates isolation of high-expression strains, transgenerational instability in fungal transformants may result in either physical loss of the transgene or its transcriptional silencing [66]. Therefore, optimizing the A. oryzae expression system is significant for achieving high-efficiency transformation. For instance, Liu et al. and Wei et al. successively achieved targeted gene knock-in in A. oryzae using optimized CRISPR-Cas9 genome editing systems [67,68]. This approach enables precise genomic integration of foreign genes with high efficiency, significantly reducing screening time. Notably, the selectable marker can be automatically excised after integration, eliminating marker-associated limitations and facilitating sequential multigene insertions [67]. Recently, Li et al. improved the transformation and editing efficiency of the CRISPR/Cas9 system by optimizing the preparation conditions of protoplasts in A. oryzae [69].
3.1.2. The Production of Terpenoids in A. oryzae
Terpenoids, the largest family and the most diverse group of natural products with over 90,000 structures reported, are derived from C5 of isopentenyl diphosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP). They are typically formed by terpene cyclases and further modified through oxidation, hydroxylation, glycosylation, or other enzymatic reactions [70]. According to the carbon numbers of the terpenoid molecule, they are categorized as monoterpenoids (C10), sesquiterpenoids (C15), diterpenoids (C20), sesterterpenoids (C25), or triterpenoids (C30) [71]. Due to their beneficial therapeutic properties including anti-cancer, anti-oxidation, antibacterial, antiviral, antimalarial, and potent anti-inflammatory activities, terpenoids are significant sources of natural product drugs, including menthol (monoterpenes), the well-known antimalarial drug artemisinin (sesquiterpenes), vitamin A, the anti-cancer drug paclitaxel (diterpenes), ginsenosides (triterpenoids), the cardiovascular drug tanshinone and so on [72,73,74,75,76]. In nature, terpenoid biosynthesis mainly occurs through two distinct pathways: the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway, predominantly in bacteria and plant plastids, and the mevalonate (MVA) pathway, primarily in eukaryotes and archaea [77]. As shown in Figure 2, A. oryzae possesses almost all precursor compounds for terpenoid synthesis, including IPP, DMAPP, geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), and geranylgeranyl pyrophosphate (GGPP) generated through the MVA pathway [78]. Moreover, A. oryzae exhibits robust protein secretion machinery and a superior post-translational modification capacity, enabling the precise recognition and splicing of introns. These attributes allow for the efficient heterologous expression of nearly all terpenoid cyclases in this fungal system [79]. Therefore, A. oryzae is an excellent heterologous expression chassis for terpenoids, particularly suitable for expressing fungal or plant-derived terpenoid biosynthetic gene(s)/clusters [80]. In recent years, A. oryzae has been widely applied in research on terpenoid biosynthesis, genomic mining, and synthetic biology. Here, we systematically review recent advances in terpenoid production using A. oryzae as a heterologous host, organized according to terpenoid classification systems. Table 1 presents the representative terpenoids heterologously produced in A. oryzae.
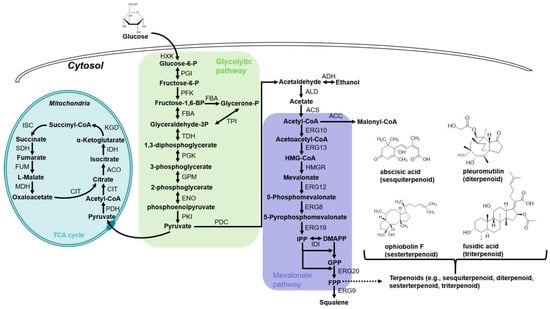
Figure 2.
Metabolism pathway for terpenoid production in A. oryzae. HXK, hexokinase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; FBA, fructose-bisphosphate aldolase; TPI, triose phosphate isomerase; TDH, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; GPM, phosphoglycerate mutase; ENO, enolase; PKI, pyruvate kinase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase; ALD, aldehyde dehydrogenase; ACS, acetyl-CoA synthase; ACC, acetyl-CoA carboxylase; ERG10, acetyl-CoA C-acetyltransferase; ERG13, hydroxymethylglutaryl-CoA synthase; HMGR, HMG-CoA reductase; ERG12, mevalonate kinase; ERG8, phosphomevalonate kinase; ERG19, diphosphomevalonate decarboxylase; IDI, Isopentenyl-diphosphate delta-isomerase; ERG20, FPP synthase; ERG9, squalene synthase; PDH, pyruvate dehydrogenase; CIT, citrate synthase; ACO, aconitase; IDH, isocitrate dehydrogenase; KGD, multifunctional 2-oxoglutarate metabolism enzyme; ISC, succinyl coenzyme A synthetase; SDH, succinatedehydrogenase; FUM, fumarase; MDH, malate dehydrogenase; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl diphosphate; FPP, farnesyl pyrophosphate.
Sesquiterpenoids
Sesquiterpenoids are ubiquitously distributed in the tissues of plants, marine organisms, microorganisms, and insects. Although they comprise a conserved C15 carbon skeleton, sesquiterpenoids exhibit remarkable skeletal diversity through multienzymatic secondary metabolic processes, generating over 180 skeletal frameworks and complex derivatives [81,82]. Thus, sesquiterpenoids represent one of the largest and most structurally diverse classes of terpenoids in terms of both quantitative abundance and chemical complexity. Takino et al. identified a novel type of sesquiterpenoid synthase gene cluster from Botrytis cinerea and achieved the biosynthesis of abscisic acid using A. oryzae as a heterologous expression host [83]. Murai et al. utilized A. oryzae as a heterologous expression host to biosynthesize a sesquiterpene alcohol with a novel skeleton, trichobrasilenol [84]. Moreover, Feng et al. heterologously expressed braA (terpene cyclase), braB (N-acetylglucosaminidase), and braC (P450 oxidase) from Annulohypoxylon truncatum in A. oryzae, resulting in the biosynthesis of novel compounds brasilane A, brasilane D, and brasilane E [85]. Furthermore, Fukaya et al. applied the hot-spot knock-in method and incorporated 13 melleolide biosynthetic genes into A. oryzae through stepwise reconstitution. Consequently, they successfully isolated 1α-hydroxymelleolide, a representative sesquiterpenoid and natural melleolide derivative, and elucidated its biosynthetic pathway [86]. Additionally, using A. oryzae as a heterologous expression system, Han et al. characterized 12 putative sesquiterpene synthases and one P450 enzyme in basidiomycete Flammulina velutipes, ultimately leading to the biosynthesis of 16 distinct sesquiterpenoid compounds [87].
Diterpenoids
Diterpenoids, composed of four isoprene units, are a diverse class of natural products biosynthesized from the C20 precursor GGPP. They have high value in the pharmaceutical industry and exhibit diverse biological activities, including antitumor, anti-inflammatory, and immunosuppressive effects [88,89]. Fujii et al. successfully introduced the whole gene cluster into A. oryzae by using four different vectors and demonstrated the total biosynthesis of a typical diterpene aphidicolin [90]. Qin et al. identified an unusual chimeric terpene synthase (EvVS) by genome mining of the fungus Emericella variecolor. Through heterologous expression in A. oryzae, a novel tricyclic diterpene hydrocarbon, variediene, was successfully obtained [91]. Furthermore, Bailey et al. achieved the biosynthesis of the biologically active compound pleuromutilin using A. oryzae as a heterologous host [21]. In 2017, based on the previous research, Alberti et al. gradually expressed the biosynthetic gene of pleuromutilin in A. oryzae and finally revealed its complete biosynthetic pathway [92]. In recent years, employing heterologous hosts of A. oryzae, integrated with pathway engineering methodologies, researchers have obtained new pleuromutilin congeners with antimicrobial activities, which provide more options for the expansion of this family of drugs [93,94,95]. Notably, Saito et al. obtained an A. oryzae strain generated by engineering multiple metabolic pathways, containing 13 metabolic modifications, which showed excellent productivities of pleuromutilin (161.6 mg/L), an 8.5-fold increase compared to the control strains without any metabolic modifications [96]. Additionally, Tazawa et al. employed an A. oryzae expression system to reconstruct the biosynthetic pathway of brassicicenes, which are a series of diterpenes, and the generated transformant successfully biosynthesized novel brassicicene derivatives (brassicicene I) [97]. Moreover, by using a newly developed hot-spot knock-in and plasmid recycling genome editing method, Liu et al. reconstituted the biosynthetic gene cluster of erinacine (a bioactive diterpene) in A. oryzae, which generated erinacine Q and its intermediates [67]. Very recently, Xu et al. identified a novel biosynthetic gene cluster (tdn) by using a genome mining method in Talaromyces adpressus and achieved heterologous expression of tdn genes in A. oryzae. As a result, a new diterpenoid, cycloaraneosene-9-ol-8-one, and three known diterpenoids were determined [98].
Sesterterpenoids
Sesterterpenoids are a relatively small group among terpenoids and are predominantly synthesized through cyclization of the linear precursor geranylfarnesyl diphosphate (GFPP) [99]. Although sesterterpenoids are the rarest of all isoprenoids, they are widely distributed in higher plants, microorganisms, insects, and marine invertebrate organisms, especially sponges, and exhibit diverse biological properties including anti-cancer, anti-inflammatory, antiprotozoal, antitubercular, and antifeedant activities [100,101]. During a screening of putative diterpene synthase genes, Oikawa’s group identified the first sesterterpene synthase (AcOS) from Aspergillus clavatus, which simultaneously possesses the functions of class I sesterterpene synthase and prenyltransferase. They used the A. oryzae heterologous expression system and obtained sesterterpene ophiobolin F [102]. On this basis, Quan et al. discovered a new ophiobolin F synthase (AcldOS) in Aspergillus calidoustus, which is a homologous gene of AcOS. They achieved the heterologous expression of AcldOS in A. oryzae and elucidated the absolute structure of ophiobolin F and its potential biosynthesis pathway [103]. Similarly, Okada et al. identified and characterized the bifunctional sesterterpene synthase EvQS by the genome mining approach; they introduced it into A. oryzae for heterologous expression and successfully obtained quiannulatene and quiannulatic acid [99]. Additionally, Quan et al. achieved heterologous expression in A. oryzae of a small biosynthetic gene cluster from Aspergillus calidoustus. This gene cluster, composed of two genes, acldAS for a chimeric sesterterpene synthase and acldA-P450 for a cytochrome P450 monooxygenase, enabled the biosynthesis of asperterpenol A and asperterpenol B [104]. Moreover, Qiao et al. discovered a unique sesterterpenoid gene cluster (consisting of pstA and pstB) and heterologously expressed both genes in A. oryzae, resulting in the production of two new sesterterpenoids, penisentenol and penisentone [105]. Notably, Liu’s group designed a further optimized A. oryzae chassis through an automated and high-throughput (auto-HTP) biofoundry workflow. Using this platform, they reconstituted 39 terpenoid BGCs into 208 A. oryzae strains and successfully elucidated the biosynthetic pathway for the sesterterpenoid mangicol J [80]. Recently, Yan et al. successfully achieved the heterologous production of bioactive fungal sesterterpenoids variecolin and variecolactone in A. oryzae by co-expressing the cytochrome P450 monooxygenase VrcB along with the sesterterpene synthase gene vrcA. Subsequently, they employed a series of vrcB homologues that were individually co-expressed with vrcA in the A. oryzae system, yielding three novel variecolin analogs [106].
Triterpenoids and Steroids
Triterpenoids possess versatile biological activities and are extensively utilized in the pharmaceutical, food, cosmetic, and chemical industries [107]. As a structurally diverse class of terpenoids, triterpenoids are ubiquitously distributed across natural sources and serve as a significant resource for drug discovery from natural products due to their anti-inflammatory, antibacterial, antiviral, anti-oxidant, anti-cardiovascular, anti-tumor, and immunomodulatory properties [108,109]. Lv et al. designated the hel gene cluster and individually introduced it into A. oryzae. Heterologous expression of this nine-gene cluster in A. oryzae enables the isolation of the fungi-derived triterpenoid antibiotic helvolic acid (~20 mg L−1) and the elucidation of its complete biosynthetic pathway [110]. Based on this, Cao et al. first identified the biosynthetic gene cluster of fusidic acid, which is the only fusidane-type antibiotic that has been clinically used. Consequently, they obtained fusidic acid and unraveled its full biosynthetic pathway through the introduction of the two uncharacterized genes (fusC1 and fusB1) into the previously established six-gene-expression A. oryzae strain [111]. After that, the biosynthetic gene cluster of cephalosporin P1, which is also a fusidane-type antibiotic, was identified by the same group and stepwise reconstitution in A. oryzae enabled them to obtain cephalosporin P1 and characterize its complete biosynthetic pathway [22]. Similarly, Li et al. individually co-expressed the biosynthetic gene cluster of the antifungal fernane-type triterpenoid polytolypin in A. oryzae, leading to the elucidation of its biosynthetic pathway and the generation of 13 fernane-type triterpenoids [112]. Very recently, Cao et al. introduced fsoA, fsoD, fsoE, and fsoF individually into A. oryzae and demonstrated that this biosynthetic gene cluster comprising the four genes is sufficient for producing fuscoatroside, the first enfumafungin-type antibiotics that belong to triterpenoids [113]. Through biosynthetic reconstitution in A. oryzae, the complete biosynthetic pathway of fuscoatroside has been revealed, and 11 novel derivatives have finally been discovered.

Table 1.
Heterologously produced representative terpenoids in A. oryzae, sources of the biosynthetic genes, and hosts used for the heterologous expression.
Table 1.
Heterologously produced representative terpenoids in A. oryzae, sources of the biosynthetic genes, and hosts used for the heterologous expression.
| Natural Products | Structure Type | Gene Source | Heterologous Hosts | References |
|---|---|---|---|---|
| abscisic acid | sesquiterpenoid | Botrytis cinerea SAS56 | A. oryzae NSAR1 | [83] |
| trichobrasilenol | sesquiterpenoid | T. atroviride FKI-3849 | A. oryzae NSAR1 | [84] |
| brasilane A, D and E | sesquiterpenoid | Annulohypoxylon truncatum CBS 140778 | A. oryzae NSAR1 | [85] |
| 1α-hydroxymelleolide | sesquiterpenoid | Armillaria mellea FMC 543 | A. oryzae NSPlD1 | [86] |
| pleuromutilin | diterpenoid | Clitopilus passeckerianus ATCC 34646 | A. oryzae NSAR1 | [21] |
| aphidicolin | diterpenoid | Phoma betae PS-13 | A. oryzae NSAR1 | [90] |
| brassicicene I | diterpenoid | Pseudocercospora fijiensis 10CR-1-24 | A. oryzae NSAR1 | [97] |
| erinacine Q | diterpenoid | Hericiumerinaceusyamabushitake Y2 | A. oryzae NSPlD1 | [67] |
| 20-prenylpaxilline 22-prenylpaxilline | diterpenoid | Tolypocladium inflatum | A. oryzae NSAR1 | [114] |
| cotylenin C, F, I and E | diterpenoid | Talaromyces adpressus | A. oryzae NSAR1 | [115] |
| cycloaraneosene-9-ol-8-one | diterpenoid | Talaromyces adpressus | A. oryzae NSAR1 | [98] |
| ophiobolin F | sesterterpenoid | Aspergillus clavatus NRRL | A. oryzae NSAR1 | [102] |
| astellifadiene | sesterterpenoid | Emericella variecolor NBRC32302 | A. oryzae NSAR1 | [116] |
| quiannulatene quiannulatic acid | sesterterpenoid | Emericella variecolor NBRC32302 | A. oryzae NSAR1 | [99] |
| asperterpenoid A | sesterterpenoid | Talaromyces wortmannii ATCC 26942 | A. oryzae NSAR | [117] |
| asperterpenol A asperterpenol B | sesterterpenoid | Aspergillus calidoustus CBS121601 | A. oryzae NSAR | [104] |
| mangicol J | sesterterpenoid | Fusarium graminearum J1-012 | A. oryzae NSAR1 | [80] |
| variecolin | sesterterpenoid | Aspergillus aculeatus ATCC 16872 | A. oryzae NSAR1 | [106] |
| helvolic acid | triterpenoid | Aspergillus fumigatus Af293 | A. oryzae NSAR1 | [110] |
| fusidic acid | triterpenoid | Acremonium fusidioides ATCC 14700 | A. oryzae NSAR1 | [111] |
| cephalosporin P1 | triterpenoid | Acremonium chrysogenum ATCC 11550 | A. oryzae NSAR1 | [22] |
| polytolypin | triterpenoid | Polytolypa hystricis UAMH7299 | A. oryzae NSAR1 | [112] |
| fuscoatroside | triterpenoid | Humicola fuscoatra NRRL 22980 | A. oryzae NSAR1 | [113] |
3.1.3. A. oryzae as a Chassis for Synthesis of Other Natural Products
Polyketides
Polyketides represent a structurally diverse class of natural products, characterized by their intricately assembled carbon skeletons derived from simple acyl building blocks [118]. More than 10,000 polyketide compounds have been discovered so far, among which clinically important representatives—including lovastatin (anti-cholesterol), erythromycin (antibiotic), azithromycin (antibiotic), and rapamycin (immunosuppressant)—serve as indispensable therapeutic agents in modern medicine [119,120]. With the development of synthetic biology and the rise of green manufacturing, an increasing number of researchers are employing A. oryzae as a chassis to modify and optimize the synthesis methods of polyketide compounds continuously. For example, He et al. heterologously expressed the citrinin biosynthetic genes in A. oryzae, with the non-reducing polyketide synthase gene (pksCT, annotated as citS) as the core component. Through heterologous co-expression of citS and downstream tailoring of enzyme genes (citA–citE) in A. oryzae, they achieved high-titer production of citrinin, validating its biosynthetic pathway and highlighting A. oryzae as a robust host for fungal polyketide biosynthesis [121]. Moreover, Sakai et al. successfully overexpressed the monacolin K (MK) gene cluster from Monascus pilosus in A. oryzae, resulting in the production of the corresponding metabolite, MK [58]. Furthermore, Yamamoto et al. reconstituted the biosynthetic gene cluster of phomoidride B in A. oryzae by using a genome editing-based hot-spot knock-in method, thereby elucidating the late-stage biosynthesis of phomoidrides [122]. Additionally, Han et al. identified a polyketide synthase (PKS, herA) in the basidiomycete Hericium erinaceus through genome mining. They cloned the herA gene and subsequently heterologously expressed it in A. oryzae, successfully achieving the production of orsellinic acid (OA) [123]. Similarly, Huang et al. identified the Phy biosynthetic gene cluster from Aspergillus chevalieri BYST01, which is responsible for the biosynthesis of physcion, a bioactive polyketide natural product derived from both plants and microorganisms. Through heterologous expression of key genes (PhyF, PhyG, and PhyL) in A. oryzae, they successfully reconstructed the biosynthetic pathway of physcion, thereby elucidating the molecular mechanisms underlying its production [124].
Non-Ribosomal Peptides
Non-ribosomal peptides (NRPs) represent a structurally diverse class of polypeptide secondary metabolites produced by various microorganisms. In contrast to polypeptides synthesized via ribosomal mechanisms, these compounds are assembled by multimodular enzyme complexes, such as non-ribosomal peptide synthetases (NRPSs) [125,126]. de Mattos-Shipley et al. identified the Pscy gene cluster from Penicillium species and heterologously expressed it in A. oryzae. This gene cluster, comprising a non-ribosomal peptide synthetase (NRPS) and a novel trans-N-methyltransferase (N-MeT), enabled the biosynthesis of cycloaspeptide A and cycloaspeptide E [127]. Moreover, Qi et al. identified a putative BGC (cri) from Eurotium cristatum NWAFU-1. They cloned the criC gene (encoding a non-ribosomal peptide synthetase, NRPS) and heterologously expressed it in A. oryzae, enabling the efficient production of the cyclic dipeptide compound cyclo-TA [128]. Recently, Katayama et al. utilized A. oryzae as a heterologous expression platform for cyclochlorotine, a fungal mycotoxin synthesized by NRPS. Through cyclochlorotine production in A. oryzae, they investigated the subcellular localization of five transmembrane proteins and confirmed the normal function of UstYa family proteins and transporters [129].
3.2. A. nidulans as a Chassis for Natural Product Synthesis
Similar to A. oryzae, A. nidulans has also been widely used in synthetic biology to study gene clusters from other species. As one of the most popular fungal hosts for the reconstitution of fungal biosynthesis pathways, A. nidulans has abundant secondary metabolite production capacity and an efficient gene targeting system with simple nutritional requirements and rapid growth and reproduction rates, so it is the most advanced and efficient molecular genetic system and a well-established model organism among filamentous fungi [57,130]. Furthermore, due to the successful expression of many types of natural products in A. nidulans, the application of A. nidulans as a versatile chassis has greatly facilitated the complete characterization and reconstruction of biosynthetic pathways for diverse natural products, as well as the discovery of new compounds [10,131]. In recent years, the A. nidulans expression system has successfully produced many fungal natural products. For example, by heterologously reconstructing the biosynthetic pathways in A. nidulans, the Chooi group successfully biosynthesized a set of natural products, including two polyketides ((M)-viriditoxin from Paecilomyces variotii [132] and stemphyloxin II from Parastagonospora nodorum [133]) and a benzazepine (alkaloid nanangelenin A from Aspergillus nanangensis [134]). Yuan et al. achieved the biosynthesis of cosmosporaside C, a fungal hybrid terpenoid saccharide, and elucidated the assembly line, which contained several unusual steps [135]; a group of dimeric indole piperazine alkaloids was also heterologously synthesized in A. nidulans by the combination of the non-ribosomal peptide synthetase (NRPS)-like gene cpsA and other genes, and one of them, campesine G, has been shown to have insecticidal activity [136]. Yang et al. identified a biosynthetic gene cluster (bgt) in the genome of Trichoderma erinaceum F1-1, a marine-derived fungus, which encodes the UbiA terpene synthase BgtA; then, they achieved the transcriptional activation of the silent bgt gene cluster through successful heterologous expression in A. nidulans LO8030, resulting in the isolation of eight previously unreported bergamotene-derived sesquiterpenoids [137].
3.3. A. niger Is a Chassis for Natural Product Synthesis
One of the earliest demonstrations of heterologous expression for a complete fungal biosynthetic pathway is the reconstitution of the BGC from Penicillium chrysogenum in A. niger [138]. Although it is not as widely utilized as A. oryzae and A. nidulans in the field of heterologous natural product biosynthesis, it is still a strong candidate host, especially for the production of non-ribosomal peptides (NRPs), due to its substantial industrial advantages [139]. For instance, Yeh et al. introduced mica, an NRPS-like gene from A. nidulans into A. niger, and their findings confirmed that only micA is necessary and sufficient for the synthesis of microperfuranone [140]. Meyer et al. successfully expressed the non-ribosomal peptide synthetase gene esyn1, derived from Fusarium oxysporum, in the Tet-on system of A. niger, yielding approximately 5 g/L of the cyclic depsipeptide enniatin [141]. In addition, the polyketide synthase gene terA from A. terreus was successfully expressed in A. niger, yielding a mixture of three distinct compounds with varying sizes and structures: orsellinic acid, 4-hydroxy-6-methylpyranone, and 6,7-dihydroxymellein [142]. Löhr et al. introduced the mushroom polyketide synthase genes from Cortinarius odorifer CoPKS1–6 individually in A. niger; the result indicated that CoPKS1 and CoPKS4 are a new class of atrochrysone carboxylic acid synthases that either exclusively generate an octaketide or synthesize both hepta- and octaketides simultaneously [143].
4. Application of CRISPR/Cas9-Based Genome Editing Technology in A. oryzae and A. niger
The clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system is a highly efficient and revolutionary genome editing technology, originally derived from the adaptive immune system of bacteria and archaea [40]. This system is primarily composed of two core components: a single guide RNA (sgRNA) and the Cas9 nuclease. The sgRNA directs the system to target specific DNA sequences through complementary base pairing, while the Cas9 nuclease executes double-stranded DNA cleavage at the designated sites, enabling precise genome editing [40,144]. In recent years, CRISPR/Cas9-mediated genome editing technology has been effectively utilized in both fundamental research and industrial production of natural products and recombinant proteins in the genus Aspergillus, with A. niger and A. oryzae emerging as the most extensively utilized species [40,145,146].
4.1. CRISPR/Cas9-Based Genome Editing Technology for A. niger
A. niger possesses exceptional tolerance to extreme environmental conditions, high production economy, robust fermentation stability, and superior food safety; it has been granted generally recognized as safe (GRAS) status by the U.S. Food and Drug Administration (FDA) [32,147], so it is a suitable cell factory for diverse biotechnological applications [40]. In 2016, Kuivanen et al. first successfully established the CRISPR-Cas9 system for genetic engineering in A. niger [148]. To disrupt the catabolism of galactaric acid in A. niger and thereby generate an engineered strain capable of the efficiently producing galactaric acid, they identified seven genes—putatively encoding metabolic enzymes and transport proteins—through RNA sequencing, and then they deleted these genes by using CRISPR/Cas9 technology in conjunction with in vitro synthesized sgRNA. Subsequently, the same group successively achieved the effective deletion of gluD and gluF genes using the identical CRISPR/Cas9 strategy, which encode the NADPH-requiring 2-keto-L-gulonate reductase and NADPH-requiring 5-keto-d-gluconate reductase, respectively [149,150]. On this basis, Kuivanen et al. further developed an optimized CRISPR/Cas9 genome editing method for A. niger, based on in vitro-assembled Cas9/gRNA ribonucleoprotein (RNP) complexes, and achieved a 100% targeting efficiency for a single genomic target [151]. In terms of site-specific gene insertion, Sarkari et al. established an efficient genetic engineering toolbox for A. niger, which combines the GoldenMOCS cloning system (a Golden Gate-derived modular cloning strategy) with a CRISPR/Cas9-mediated integration approach to enable site-specific insertion of heterologous expression cassettes at the pyrG locus [152]. This system used a transiently expressed Cas9 (via a size-reduced AMA1 plasmid that is readily lost) and a split-pyrG marker for direct selection, achieving a high integration efficiency of up to 100%. Beyond targeted gene editing, transcriptional regulation of gene expression also serves as a pivotal approach for constructing and optimizing cell factories. The Tet-on system is an effective method for functional analysis of essential genes, allowing for precise modulation of gene expression levels in response to inducer concentrations [153,154]. In recent years, researchers have focused on integrating CRISPR/Cas9 genome editing technology with the Tet-on regulatory system in A. niger, aiming to quantitatively elucidate the impacts of differential expression levels of target genes on product biosynthesis. For example, Cairns et al. employed CRISPR-based genome editing to integrate the titratable Tet-on expression system upstream of ageB, secG, and geaB in A. niger. They utilized this strategy to investigate the functional associations between protein secretion, filamentous growth, and organic acid production [155]. Moreover, Zhang et al. replaced the native promoter of the pyrG gene in A. niger with a Tet-on inducible promoter through in situ promoter substitution mediated by CRISPR/Cas9 genome editing [156]. By titrating the concentration of the inducer doxycycline (Dox), they established a quantitative relationship between the differential expression levels of pyrG and the performance of citric acid fermentation, including titers and productivity. Very recently, a CRISPR/Cas9-based visual toolkit enabling multiplex integration at specific genomic loci has been established in A. niger [157], which allows simultaneous insertion of recombinant genes into multiple high-expression loci through the combined application of a visual multigene editing system (VMS) and the CRISPR-HDR system.
4.2. CRISPR/Cas9-Based Genome Editing Technology for A. oryzae
To achieve accurate and efficient genome editing in A. oryzae and better meet the demands of industrial production, the CRISPR-Cas9 system has been rapidly adopted in this filamentous fungus recently [144]. In 2016, Katayama et al. successfully established a CRISPR/Cas9-mediated genome editing method in A. oryzae, marking the first application of targeted mutagenesis in this filamentous fungus [146]. Using the CRISPR/Cas9 system, they knocked out the uracil synthesis gene pyrG (encoding orotidine 5′-phosphate decarboxylase) to construct a stable uridine/uracil auxotrophic A. oryzae strain, achieving a 10% mutagenesis efficiency with a characteristic 1 bp deletion at the target site 3–4 bp upstream of the PAM sequence. On this basis, the same group further applied this system in 2017 to delete the ligD gene—a DNA ligase gene participating in non-homologous end joining (NHEJ) in A. oryzae [158]. This study demonstrated that introducing the ligD mutation by genome editing is an effective strategy to improve gene targeting efficiency in A. oryzae industrial strains. Furthermore, Katayama et al. established an optimized CRISPR/Cas9 method using AMA1-based autonomous replicating genome editing plasmids harboring a drug resistance marker (ptrA) [159]. Notably, this system enabled highly efficient marker-free multiple gene modifications, with mutation efficiency significantly improved to 50–100% in both wild-type and industrial A. oryzae strains. Additionally, Li et al. developed a CRISPR/Cas9 genome editing method by optimizing protoplast preparation conditions, which further enhanced the transformation and multiplexed genome editing efficiency in A. oryzae (37.6% single-gene and 19.8% dual-gene editing efficiency) [69]. In this study, the morphological gene yA was identified as a promising selection marker for the rapid and precise isolation of positive transformants during genetic manipulation in A. oryzae. Very recently, a novel modular synthetic biology toolkit for A. oryzae was developed, featuring a recyclable ribonucleoprotein (RNP)-based CRISPR-Cas9 method [160]. Unlike traditional plasmid-based systems, this approach achieved gene editing through direct transformation of pre-assembled CRISPR-Cas9 RNP complexes, eliminating the need for plasmid-based encoding of Cas9 and sgRNAs. Notably, this RNP-based method demonstrates comparable efficiency and versatility to plasmid-based CRISPR-Cas9 systems for genetic engineering in A. oryzae [159].
5. Conclusions and Perspectives
With the rapid development of synthetic biology and molecular biology techniques, Aspergillus species exhibit substantial potential as expression hosts and play an increasingly vital role in the heterologous production of proteins and the synthesis of bioactive natural products. On the one hand, the engineered strains of Aspergillus enable the efficient production of heterologous proteins, significantly increasing their yields to meet industrial standards [24]. On the other hand, as an extraordinary chassis, Aspergillus can activate silent biosynthetic gene clusters (BGCs), facilitating the discovery of novel natural products, as well as promoting the elucidation of complete biosynthetic pathways, thus simplifying and optimizing their intricate synthesis routes [161]. For instance, A. oryzae, A. niger, and A. nidulans are being widely developed as research tools in synthetic biology [162,163]. Notably, A. oryzae serves as a powerful platform that not only enables rapid and efficient functional identification of natural product biosynthetic gene clusters but also facilitates genome-guided mining of bioactive natural products. Moreover, this system can be exploited for targeted engineering and optimization of functional genes to enhance natural product biosynthesis [58,80]. To date, A. oryzae has successfully enabled the heterologous expression of diverse natural product classes, particularly sesquiterpenoids, diterpenoids, and polyketides (PKs) [79,164,165]. Remarkably, this platform exhibits outstanding adaptability, efficiently integrating biosynthetic genes not only from filamentous fungi but also from basidiomycetes, bacteria, and even higher plants [166,167]. Such versatility has significantly accelerated the rapid elucidation and reconstruction of biosynthetic pathways for complex natural products, establishing A. oryzae as an ideal platform for synthetic biology research on natural products [5,62]. However, compared to A. niger, A. oryzae remains less amenable to complex genetic manipulation. The active non-homologous end-joining (NHEJ) pathway in A. oryzae inherently results in low homologous recombination rates, making targeted gene knockouts or integrations challenging [168,169]. Moreover, the rigid cell wall of A. oryzae significantly impairs the efficiency of conventional transformation approaches, including electroporation and biolistics [166]. Consequently, genetic manipulation of A. oryzae is predominantly dependent on polyethylene glycol (PEG)-mediated protoplast transformation, a laborious and time-consuming procedure that demands technical expertise [170]. In addition, despite the application of CRISPR/Cas9 technology in A. oryzae, its efficiency is constrained by multiple factors. The Cas9 protein exhibits low nuclear localization efficiency, and although partial improvement can be achieved through codon optimization and fusion with nuclear localization signals (NLS) as reported in previous studies [146], this remains a limiting factor. Furthermore, A. oryzae displays intrinsic resistance to most antifungal antibiotics. This characteristic leads to a scarcity of viable dominant selectable markers, compelling researchers to predominantly rely on auxotrophic markers such as pyrG and argB for genetic selection procedures [16,166].
Since 2015, the CRISPR/Cas system with its diverse engineering efficiencies has been progressively applied to genome editing in various filamentous fungi [171,172,173]; this system guides the Cas protein to target specific DNA sequences via the guide RNA (gRNA), enabling efficient and precise editing of the genome, including gene knockout, insertion, and point mutation, thereby contributing to the construction of a highly efficient filamentous fungal expression platform [174]. Although the CRISPR/Cas9 system has indeed enhanced the genome editing efficiency of Aspergillus species, it still faces several challenges. For example, in most non-model strains, the gene editing efficiency of the CRISPR/Cas system remains relatively low, which is determined by multiple factors (e.g., host cellular barriers and nucleic acid degradation mechanisms) [175]; another major limitation is unexpected off-target effects, which means the recognition sequence of the gRNA may bind to non-target DNA, resulting in non-specific editing and uncontrollable genomic variations [176]. In the future, the CRISPR/Cas9 system can be further optimized in multiple dimensions, including reducing the off-target effect, overcoming PAM sequence constraints of Cas proteins, and enhancing precise on-target editing efficiency [177].
This article provides a comprehensive overview of the latest research advances in the Aspergillus expression system for the heterologous expression of proteins and the biosynthesis of bioactive natural products, with a detailed focus on the progress in the heterologous biosynthesis of terpenoid compounds. Additionally, this article highlights the recent advancements in the application of CRISPR/Cas9-based genome editing technology in A. oryzae over the past few years. In the genus Aspergillus, A. oryzae, A. nidulans and A. niger are widely utilized for the efficient production of heterologous proteins and recognized as excellent chassis for the biosynthesis of natural products. These species have become a robust and versatile platform for synthetic biology research [62,80,178]. Heterologous expression hosts of the genus Aspergillus have achieved significant success in both heterologous protein expression and natural product biosynthesis. However, further refinement and optimization of this system remain imperative, including but not limited to developing novel transformation methods, enhancing transformation efficiency, identifying and designing more efficient enhanced promoters, and optimizing host metabolic pathways [170,179]. These optimization needs are not only critical for advancing fundamental research but also lay the groundwork for the broader application of Aspergillus in synthetic biology.
Author Contributions
Conceptualization, Y.S.; investigation, Y.S.; visualization, S.Q.; resources, Y.D.; writing—original draft preparation, Y.S.; writing—review and editing, S.Q.; project administration, B.Z.; funding acquisition, B.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The Key Special Projects of the National Key Research and Development Plan (2021YFA1301302), Project of the Department of Science and Technology of Jiangxi Province (20213AAG02020), the National Natural Science Foundation of China (32200606), Self-made Experimental Instruments and Equipment Project of Shenzhen Technology University (JSZZ202301021), Project of Shenzhen Science and Technology Innovation Commission (KJZD20230923114408018), 2024 Off-Campus Practice Base Project for Professional Degree Postgraduate Students of Shenzhen Technology University (SJJD2024003).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pan, R.; Yang, X.; Qiu, M.; Jiang, W.; Zhang, W.; Jiang, Y.; Xin, F.; Jiang, M. Construction of Coculture System Containing Escherichia coli with Different Microbial Species for Biochemical Production. ACS Synth. Biol. 2023, 12, 2208–2216. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, X.; Yao, M.; Xiao, W.; Zha, J. Chassis engineering for microbial production of chemicals: From natural microbes to synthetic organisms. Curr. Opin. Biotechnol. 2020, 66, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, Y.; Du, G.; Ledesma-Amaro, R.; Liu, L. Microbial Chassis Development for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 779–796. [Google Scholar] [CrossRef]
- Knuf, C.; Nielsen, J. Aspergilli: Systems biology and industrial applications. Biotechnol. J. 2012, 7, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wu, Y.; Long, S.; Feng, S.; Jia, X.; Hu, Y.; Ma, M.; Liu, J.; Zeng, B. Aspergillus oryzae as a Cell Factory: Research and Applications in Industrial Production. J. Fungi 2024, 10, 248. [Google Scholar] [CrossRef]
- Alberti, F.; Foster, G.D.; Bailey, A.M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 2017, 101, 493–500. [Google Scholar] [CrossRef]
- Van Den Berg, M.; Gidijala, L.; Kiela, J.; Bovenberg, R.; Vander Keli, I. Biosynthesis of active pharmaceuticals: β-lactam biosynthesis in filamentous fungi. Biotechnol. Genet. Eng. Rev. 2010, 27, 1–32. [Google Scholar] [CrossRef]
- Ding, Q.; Ye, C. Microbial cell factories based on filamentous bacteria, yeasts, and fungi. Microb. Cell Fact. 2023, 22, 20. [Google Scholar] [CrossRef]
- Felipe, M.T.D.; Barbosa, R.D.; Bezerra, J.D.P.; de Souza-Motta, C.M. Production of kojic acid by Aspergillus species: Trends and applications. Fungal Biol. Rev. 2023, 45, 100313. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Lin, T.S.; Wang, C.C.C. Total Heterologous Biosynthesis of Fungal Natural Products in Aspergillus nidulans. J. Nat. Prod. 2022, 85, 2484–2518. [Google Scholar] [CrossRef]
- Guo, C.J.; Wang, C.C. Recent advances in genome mining of secondary metabolites in Aspergillus terreus. Front. Microbiol. 2014, 5, 717. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Fang, Y.; Ding, M.; Zhang, Y.; Jia, K.; Li, Z.; Collemare, J.; Liu, W. Developing fungal heterologous expression platforms to explore and improve the production of natural products from fungal biodiversity. Biotechnol. Adv. 2022, 54, 107866. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.J. Engineered and total biosynthesis of fungal specialized metabolites. Nat. Rev. Chem. 2024, 8, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Men, P.; Tang, S.; Lu, X. Aspergillus terreus as an industrial filamentous fungus for pharmaceutical biotechnology. Curr. Opin. Biotechnol. 2021, 69, 273–280. [Google Scholar] [CrossRef]
- West, T.P. Citric Acid Production by Aspergillus niger Using Solid-State Fermentation of Agricultural Processing Coproducts. Appl. Biosci. 2023, 2, 1–13. [Google Scholar] [CrossRef]
- Daba, G.M.; Mostafa, F.A.; Elkhateeb, W.A. The ancient koji mold (Aspergillus oryzae) as a modern biotechnological tool. Bioresour. Bioprocess. 2021, 8, 52. [Google Scholar] [CrossRef]
- Punt, P.J.; van Biezen, N.; Conesa, A.; Albers, A.; Mangnus, J.; van den Hondel, C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002, 20, 200–206. [Google Scholar] [CrossRef]
- Huynh, H.H.; Morita, N.; Sakamoto, T.; Katayama, T.; Miyakawa, T.; Tanokura, M.; Chiba, Y.; Shinkura, R.; Maruyama, J.I. Functional production of human antibody by the filamentous fungus Aspergillus oryzae. Fungal Biol. Biotechnol. 2020, 7, 7. [Google Scholar] [CrossRef]
- Fan, J.; Wei, P.L.; Li, Y.; Zhang, S.; Ren, Z.; Li, W.; Yin, W.B. Developing filamentous fungal chassis for natural product production. Bioresour. Technol. 2025, 415, 131703. [Google Scholar] [CrossRef]
- Ntana, F.; Mortensen, U.H.; Sarazin, C.; Figge, R. Aspergillus: A Powerful Protein Production Platform. Catalysts 2020, 10, 1064. [Google Scholar] [CrossRef]
- Bailey, A.M.; Alberti, F.; Kilaru, S.; Collins, C.M.; de Mattos-Shipley, K.; Hartley, A.J.; Hayes, P.; Griffin, A.; Lazarus, C.M.; Cox, R.J.; et al. Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production. Sci. Rep. 2016, 6, 25202. [Google Scholar] [CrossRef]
- Cao, Z.Q.; Lv, J.M.; Liu, Q.; Qin, S.Y.; Chen, G.D.; Dai, P.; Zhong, Y.; Gao, H.; Yao, X.S.; Hu, D. Biosynthetic Study of Cephalosporin P(1) Reveals a Multifunctional P450 Enzyme and a Site-Selective Acetyltransferase. ACS Chem. Biol. 2020, 15, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Lübeck, M.; Lübeck, P.S. Fungal Cell Factories for Efficient and Sustainable Production of Proteins and Peptides. Microorganisms 2022, 10, 753. [Google Scholar] [CrossRef]
- Gruben, B.S.; Mäkelä, M.R.; Kowalczyk, J.E.; Zhou, M.; Benoit-Gelber, I.; De Vries, R.P. Expression-based clustering of CAZyme-encoding genes of Aspergillus niger. BMC Genom. 2017, 18, 900. [Google Scholar] [CrossRef]
- Fiedler, M.R.M.; Barthel, L.; Kubisch, C.; Nai, C.; Meyer, V. Construction of an improved Aspergillus niger platform for enhanced glucoamylase secretion. Microb. Cell Fact. 2018, 17, 95. [Google Scholar] [CrossRef]
- Zhang, H.; Yan, J.N.; Zhang, H.; Liu, T.Q.; Xu, Y.; Zhang, Y.Y.; Li, J. Effect of gpd box copy numbers in the gpdA promoter of Aspergillus nidulans on its transcription efficiency in Aspergillus niger. FEMS Microbiol. Lett. 2018, 365, fny154. [Google Scholar] [CrossRef]
- Cai, L.N.; Xu, S.N.; Lu, T.; Lin, D.Q.; Yao, S.J. Directed expression of halophilic and acidophilic β-glucosidases by introducing homologous constitutive expression cassettes in marine Aspergillus niger. J. Biotechnol. 2019, 292, 12–22. [Google Scholar] [CrossRef]
- Alazi, E.; Knetsch, T.; Di Falco, M.; Reid, I.D.; Arentshorst, M.; Visser, J.; Tsang, A.; Ram, A.F.J. Inducer-independent production of pectinases in Aspergillus niger by overexpression of the D-galacturonic acid-responsive transcription factor gaaR. Appl. Microbiol. Biotechnol. 2018, 102, 2723–2736. [Google Scholar] [CrossRef]
- Liu, F.; Wang, B.; Ye, Y.; Pan, L. High level expression and characterization of tannase tan7 using Aspergillus niger SH-2 with low-background endogenous secretory proteins as the host. Protein Expr. Purif. 2018, 144, 71–75. [Google Scholar] [CrossRef]
- Zhu, S.Y.; Xu, Y.; Yu, X.W. Improved Homologous Expression of the Acidic Lipase from Aspergillus niger. J. Microbiol. Biotechnol. 2020, 30, 196–205. [Google Scholar] [CrossRef]
- Li, C.; Zhou, J.; Du, G.; Chen, J.; Takahashi, S.; Liu, S. Developing Aspergillus niger as a cell factory for food enzyme production. Biotechnol. Adv. 2020, 44, 107630. [Google Scholar] [CrossRef]
- Archer, D.B.; Jeenes, D.J.; MacKenzie, D.A.; Brightwell, G.; Lambert, N.; Lowe, G.; Radford, S.E.; Dobson, C.M. Hen egg white lysozyme expressed in, and secreted from, Aspergillus niger is correctly processed and folded. Biotechnology 1990, 8, 741–745. [Google Scholar] [CrossRef]
- Ward, P.P.; Piddington, C.S.; Cunningham, G.A.; Zhou, X.; Wyatt, R.D.; Conneely, O.M. A system for production of commercial quantities of human lactoferrin: A broad spectrum natural antibiotic. Biotechnology 1995, 13, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Dunn-Coleman, N.S.; Bloebaum, P.; Berka, R.M.; Bodie, E.; Robinson, N.; Armstrong, G.; Ward, M.; Przetak, M.; Carter, G.L.; LaCost, R.; et al. Commercial levels of chymosin production by Aspergillus. Biotechnology 1991, 9, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Faus, I.; del Moral, C.; Adroer, N.; del Río, J.L.; Patiño, C.; Sisniega, H.; Casas, C.; Bladé, J.; Rubio, V. Secretion of the sweet-tasting protein thaumatin by recombinant strains of Aspergillus niger var. awamori. Appl. Microbiol. Biotechnol. 1998, 49, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Prathumpai, W.; Flitter, S.J.; McIntyre, M.; Nielsen, J. Lipase production by recombinant strains of Aspergillus niger expressing a lipase-encoding gene from Thermomyces lanuginosus. Appl. Microbiol. Biotechnol. 2004, 65, 714–719. [Google Scholar] [CrossRef]
- Chen, X.; Wang, B.; Pan, L. Heterologous expression and characterization of Penicillium citrinum nuclease P1 in Aspergillus niger and its application in the production of nucleotides. Protein Expr. Purif. 2019, 156, 36–43. [Google Scholar] [CrossRef]
- Kamaruddin, N.; Storms, R.; Mahadi, N.M.; Illias, R.M.; Abu Bakar, F.D.; Murad, A.M.A. Reduction of Extracellular Proteases Increased Activity and Stability of Heterologous Protein in Aspergillus niger. Arab. J. Sci. Eng. 2018, 43, 3327–3338. [Google Scholar] [CrossRef]
- Jin, F.J.; Wang, B.T.; Wang, Z.D.; Jin, L.; Han, P. CRISPR/Cas9-Based Genome Editing and Its Application in Aspergillus Species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef]
- Dong, L.; Lin, X.; Yu, D.; Huang, L.; Wang, B.; Pan, L. High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation. J. Ind. Microbiol. Biotechnol. 2020, 47, 133–144. [Google Scholar] [CrossRef]
- Wang, S.; Xue, Y.; Zhang, P.; Yan, Q.; Li, Y.; Jiang, Z. CRISPR/Cas9 System-Mediated Multi-copy Expression of an Alkaline Serine Protease in Aspergillus niger for the Production of XOD-Inhibitory Peptides. J. Agric. Food Chem. 2023, 71, 15194–15203. [Google Scholar] [CrossRef] [PubMed]
- Jeennor, S.; Anantayanon, J.; Panchanawaporn, S.; Chutrakul, C.; Vongsangnak, W.; Laoteng, K. Efficient de novo production of bioactive cordycepin by Aspergillus oryzae using a food-grade expression platform. Microb. Cell Fact. 2023, 22, 253. [Google Scholar] [CrossRef] [PubMed]
- Goldman, G.H.; Kafer, E. Aspergillus nidulans as a model system to characterize the DNA damage response in eukaryotes. Fungal Genet. Biol. 2004, 41, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2007, 2, 811–821. [Google Scholar] [CrossRef]
- Kumar, A. Aspergillus nidulans: A Potential Resource of the Production of the Native and Heterologous Enzymes for Industrial Applications. Int. J. Microbiol. 2020, 2020, 8894215. [Google Scholar] [CrossRef]
- Liu, E.; Li, M.; Abdella, A.; Wilkins, M.R. Development of a cost-effective medium for submerged production of fungal aryl alcohol oxidase using a genetically modified Aspergillus nidulans strain. Bioresour. Technol. 2020, 305, 123038. [Google Scholar] [CrossRef]
- Lopes, A.M.M.; Martins, M.; Goldbeck, R. Heterologous Expression of Lignocellulose-Modifying Enzymes in Microorganisms: Current Status. Mol. Biotechnol. 2021, 63, 184–199. [Google Scholar] [CrossRef]
- Yan, Q.; Han, L.; Liu, Z.; Zhou, S.; Zhou, Z. Stepwise genetic modification for efficient expression of heterologous proteins in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2023, 107, 6923–6935. [Google Scholar] [CrossRef]
- Gerhardt, J.A.; Rubio, M.V.; Terrasan, C.R.F.; Wassano, N.S.; Rodrigues, A.; Figueiredo, F.L.; Antoniel, E.P.; Contesini, F.J.; Dias, A.H.S.; Mortensen, U.H.; et al. Improving recombinant protein secretion in Aspergillus nidulans by targeting the N-glycosylation machinery. Metab. Eng. Commun. 2025, 20, e00264. [Google Scholar] [CrossRef]
- Fleissner, A.; Dersch, P. Expression and export: Recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 2010, 87, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Garrigues, S.; de Vries, R.P. Heterologous protein production in filamentous fungi. Appl. Microbiol. Biotechnol. 2023, 107, 5019–5033. [Google Scholar] [CrossRef]
- Lubertozzi, D.; Keasling, J.D. Developing Aspergillus as a host for heterologous expression. Biotechnol. Adv. 2009, 27, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, C.M.; Williams, K.; Bailey, A.M. Reconstructing fungal natural product biosynthetic pathways. Nat. Prod. Rep. 2014, 31, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Blank, L.M. Recent Advances in Yeast Recombinant Biosynthesis of the Triterpenoid Protopanaxadiol and Glycosylated Derivatives Thereof. J. Agric. Food Chem. 2023, 71, 2197–2210. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Oakley, C.E.; Ahuja, M.; Entwistle, R.; Schultz, A.; Chang, S.L.; Sung, C.T.; Wang, C.C.; Oakley, B.R. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J. Am. Chem. Soc. 2013, 135, 7720–7731. [Google Scholar] [CrossRef]
- Yaegashi, J.; Oakley, B.R.; Wang, C.C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J. Ind. Microbiol. Biotechnol. 2014, 41, 433–442. [Google Scholar] [CrossRef]
- Sakai, K.; Kinoshita, H.; Nihira, T. Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 2012, 93, 2011–2022. [Google Scholar] [CrossRef]
- Oikawa, H. Reconstitution of biosynthetic machinery of fungal natural products in heterologous hosts. Biosci. Biotechnol. Biochem. 2020, 84, 433–444. [Google Scholar] [CrossRef]
- Oikawa, H. Heterologous production of fungal natural products: Reconstitution of biosynthetic gene clusters in model host Aspergillus oryzae. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 420–430. [Google Scholar] [CrossRef]
- Gallagher, R.R.; Patel, J.R.; Interiano, A.L.; Rovner, A.J.; Isaacs, F.J. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 2015, 43, 1945–1954. [Google Scholar] [CrossRef]
- Yang, H.; Song, C.; Liu, C.; Wang, P. Synthetic Biology Tools for Engineering Aspergillus oryzae. J. Fungi 2024, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Wakimoto, T.; Mori, T.; Awakawa, T.; Abe, I. Complete biosynthetic pathway of anditomin: Nature’s sophisticated synthetic route to a complex fungal meroterpenoid. J. Am. Chem. Soc. 2014, 136, 15326–15336. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, X.; Chen, L.; Yan, D.; Wang, W.G.; Matsuda, Y. Heterologous Biosynthesis of Tetrahydroxanthone Dimers: Determination of Key Factors for Selective or Divergent Synthesis. J. Nat. Prod. 2021, 84, 1544–1549. [Google Scholar] [CrossRef] [PubMed]
- Yamane, M.; Minami, A.; Liu, C.; Ozaki, T.; Takeuchi, I.; Tsukagoshi, T.; Tokiwano, T.; Gomi, K.; Oikawa, H. Biosynthetic Machinery of Diterpene Pleuromutilin Isolated from Basidiomycete Fungi. Chembiochem 2017, 18, 2317–2322. [Google Scholar] [CrossRef]
- Zhang, S.L.; Ban, A.; Ebara, N.; Mizutani, O.; Tanaka, M.; Shintani, T.; Gomi, K. Self-excising Cre/mutant lox marker recycling system for multiple gene integrations and consecutive gene deletions in Aspergillus oryzae. J. Biosci. Bioeng. 2017, 123, 403–411. [Google Scholar] [CrossRef]
- Liu, C.; Minami, A.; Ozaki, T.; Wu, J.; Kawagishi, H.; Maruyama, J.I.; Oikawa, H. Efficient Reconstitution of Basidiomycota Diterpene Erinacine Gene Cluster in Ascomycota Host Aspergillus oryzae Based on Genomic DNA Sequences. J. Am. Chem. Soc. 2019, 141, 15519–15523. [Google Scholar] [CrossRef]
- Wei, X.; Matsuyama, T.; Sato, H.; Yan, D.; Chan, P.M.; Miyamoto, K.; Uchiyama, M.; Matsuda, Y. Molecular and Computational Bases for Spirofuranone Formation in Setosusin Biosynthesis. J. Am. Chem. Soc. 2021, 143, 17708–17715. [Google Scholar] [CrossRef]
- Li, Q.H.; Lu, J.C.; Zhang, G.Q.; Zhou, J.W.; Li, J.H.; Du, G.C.; Chen, J. CRISPR/Cas9-Mediated Multiplexed Genome Editing in Aspergillus oryzae. J. Fungi 2023, 9, 109. [Google Scholar] [CrossRef]
- Shataer, D.; Chang, Y.H.; Obul, M.; Aierken, K.; Liu, H.P. An Up-to-date Review on the Classification, Pharmacology, and Production of Terpenes and Terpenoids. Curr. Org. Chem. 2025, 29, 1508–1522. [Google Scholar] [CrossRef]
- Zhang, Y.; Nielsen, J.; Liu, Z. Engineering yeast metabolism for production of terpenoids for use as perfume ingredients, pharmaceuticals and biofuels. FEMS Yeast Res. 2017, 17, fox080. [Google Scholar] [CrossRef]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, H.; Liu, W.; Liu, E.; Pang, Y.; Gao, H.; He, Q.; Liao, W.; Yao, Y.; Zeng, J.; et al. Menthol: An underestimated anticancer agent. Front. Pharmacol. 2023, 14, 1148790. [Google Scholar] [CrossRef]
- Abu Samaan, T.M.; Samec, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Paclitaxel’s Mechanistic and Clinical Effects on Breast Cancer. Biomolecules 2019, 9, 789. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, Z.; Liao, F.; Jiang, T.; Tu, Y. The birth of artemisinin. Pharmacol. Ther. 2020, 216, 107658. [Google Scholar] [CrossRef]
- Gao, S.; Liu, Z.; Li, H.; Little, P.J.; Liu, P.; Xu, S. Cardiovascular actions and therapeutic potential of tanshinone IIA. Atherosclerosis 2012, 220, 3–10. [Google Scholar] [CrossRef]
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 2016, 34, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.Y.; Yi, J.; Chang, Y.B.; Sun, C.P.; Ma, X.C. Recent studies on terpenoids in Aspergillus fungi: Chemical diversity, biosynthesis, and bioactivity. Phytochemistry 2022, 193, 113011. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, S.; Liu, C.; Nishishita, J.; Kozaki, T.; Sogahata, K.; Sato, Y.; Minami, A.; Ozaki, T.; Schmidt-Dannert, C.; Maruyama, J.I.; et al. Ascomycete Aspergillus oryzae Is an Efficient Expression Host for Production of Basidiomycete Terpenes by Using Genomic DNA Sequences. Appl. Environ. Microbiol. 2019, 85, e00409-19. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.J.; Cheng, S.; Bian, G.K.; Yan, P.; Ma, Z.N.; Dai, W.; Chen, R.; Fu, S.; Huang, H.W.; Chi, H.M.; et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi. Nat. Catal. 2022, 5, 277. [Google Scholar] [CrossRef]
- Dai, Q.; Zhang, F.L.; Feng, T. Sesquiterpenoids Specially Produced by Fungi: Structures, Biological Activities, Chemical and Biosynthesis (2015–2020). J. Fungi 2021, 7, 1026. [Google Scholar] [CrossRef]
- Chen, D.L.; Wang, B.W.; Sun, Z.C.; Yang, J.S.; Xu, X.D.; Ma, G.X. Natural Nitrogenous Sesquiterpenoids and Their Bioactivity: A Review. Molecules 2020, 25, 2485. [Google Scholar] [CrossRef]
- Takino, J.; Kozaki, T.; Sato, Y.; Liu, C.; Ozaki, T.; Minami, A.; Oikawa, H. Unveiling Biosynthesis of the Phytohormone Abscisic Acid in Fungi: Unprecedented Mechanism of Core Scaffold Formation Catalyzed by an Unusual Sesquiterpene Synthase. J. Am. Chem. Soc. 2018, 140, 12392–12395. [Google Scholar] [CrossRef]
- Murai, K.; Lauterbach, L.; Teramoto, K.; Quan, Z.; Barra, L.; Yamamoto, T.; Nonaka, K.; Shiomi, K.; Nishiyama, M.; Kuzuyama, T.; et al. An Unusual Skeletal Rearrangement in the Biosynthesis of the Sesquiterpene Trichobrasilenol from Trichoderma. Angew. Chem. Int. Ed. Engl. 2019, 58, 15046–15050. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Surup, F.; Hauser, M.; Miller, A.; Wennrich, J.P.; Stadler, M.; Cox, R.J.; Kuhnert, E. Biosynthesis of oxygenated brasilane terpene glycosides involves a promiscuous N-acetylglucosamine transferase. Chem. Commun. 2020, 56, 12419–12422. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, M.; Nagamine, S.; Ozaki, T.; Liu, Y.; Ozeki, M.; Matsuyama, T.; Miyamoto, K.; Kawagishi, H.; Uchiyama, M.; Oikawa, H.; et al. Total Biosynthesis of Melleolides from Basidiomycota Fungi: Mechanistic Analysis of the Multifunctional GMC Oxidase Mld7. Angew. Chem. Int. Ed. Engl. 2023, 62, e202308881. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Zhang, P.; Xie, Z.; Qi, J.; Wang, P.; Li, C.; Xue, Z.; Wu, R.; Liu, C. Functional Characterization of Sesquiterpene Synthases and P450 Enzymes in Flammulina velutipes for Biosynthesis of Spiro [4.5] Decane Terpene. J. Agric. Food Chem. 2024, 72, 9227–9235. [Google Scholar] [CrossRef]
- Rico-Martínez, M.; Medina, F.G.; Marrero, J.G.; Osegueda-Robles, S. Biotransformation of diterpenes. Rsc Adv. 2014, 4, 10627–10647. [Google Scholar] [CrossRef]
- Hu, Z.M.; Liu, X.Y.; Tian, M.; Ma, Y.; Jin, B.L.; Gao, W.; Cui, G.H.; Guo, J.; Huang, L.Q. Recent progress and new perspectives for diterpenoid biosynthesis in medicinal plants. Med. Res. Rev. 2021, 41, 2971–2997. [Google Scholar] [CrossRef]
- Fujii, R.; Minami, A.; Tsukagoshi, T.; Sato, N.; Sahara, T.; Ohgiya, S.; Gomi, K.; Oikawa, H. Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase α: Heterologous expression of four biosynthetic genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2011, 75, 1813–1817. [Google Scholar] [CrossRef]
- Qin, B.; Matsuda, Y.; Mori, T.; Okada, M.; Quan, Z.; Mitsuhashi, T.; Wakimoto, T.; Abe, I. An Unusual Chimeric Diterpene Synthase from Emericella variecolor and Its Functional Conversion into a Sesterterpene Synthase by Domain Swapping. Angew. Chem. Int. Ed. Engl. 2016, 55, 1658–1661. [Google Scholar] [CrossRef]
- Alberti, F.; Khairudin, K.; Venegas, E.R.; Davies, J.A.; Hayes, P.M.; Willis, C.L.; Bailey, A.M.; Foster, G.D. Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives. Nat. Commun. 2017, 8, 1831. [Google Scholar] [CrossRef]
- Alberti, F.; Khairudin, K.; Davies, J.A.; Sangmalee, S.; Willis, C.L.; Foster, G.D.; Bailey, A.M. Biosynthesis of pleuromutilin congeners using an Aspergillus oryzae expression platform. Chem. Sci. 2023, 14, 3826–3833. [Google Scholar] [CrossRef] [PubMed]
- Schafhauser, T.; Wibberg, D.; Binder, A.; Rückert, C.; Busche, T.; Wohlleben, W.; Kalinowski, J. Genome Assembly and Genetic Traits of the Pleuromutilin-Producer Clitopilus passeckerianus DSM1602. J. Fungi 2022, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Dai, H.; Zhang, M.; Liao, H.; Zhang, R.; Chen, B.; Han, J.; Liu, H. Molecular networking assisted discovery and combinatorial biosynthesis of new antimicrobial pleuromutilins. Eur. J. Med. Chem. 2022, 243, 114713. [Google Scholar] [CrossRef]
- Saito, N.; Katayama, T.; Minami, A.; Oikawa, H.; Maruyama, J.I. Versatile filamentous fungal host highly-producing heterologous natural products developed by genome editing-mediated engineering of multiple metabolic pathways. Commun. Biol. 2024, 7, 1263. [Google Scholar] [CrossRef] [PubMed]
- Tazawa, A.; Ye, Y.; Ozaki, T.; Liu, C.; Ogasawara, Y.; Dairi, T.; Higuchi, Y.; Kato, N.; Gomi, K.; Minami, A.; et al. Total Biosynthesis of Brassicicenes: Identification of a Key Enzyme for Skeletal Diversification. Org. Lett. 2018, 20, 6178–6182. [Google Scholar] [CrossRef]
- Xu, Q.; Ren, X.; Hu, L.; Xu, Q.; Zhang, X.; Deng, M.; Ye, Y.; Zhang, Y.; Lu, Y.; Qiao, Y. Uncovering a novel biosynthetic gene cluster for sordarin through genome mining in the fungus Talaromyces adpressus. Bioresour. Bioprocess. 2025, 12, 35. [Google Scholar] [CrossRef]
- Okada, M.; Matsuda, Y.; Mitsuhashi, T.; Hoshino, S.; Mori, T.; Nakagawa, K.; Quan, Z.; Qin, B.; Zhang, H.; Hayashi, F.; et al. Genome-Based Discovery of an Unprecedented Cyclization Mode in Fungal Sesterterpenoid Biosynthesis. J. Am. Chem. Soc. 2016, 138, 10011–10018. [Google Scholar] [CrossRef]
- Li, K.; Gustafson, K.R. Sesterterpenoids: Chemistry, biology, and biosynthesis. Nat. Prod. Rep. 2021, 38, 1251–1281. [Google Scholar] [CrossRef]
- Guo, K.; Liu, Y.; Li, S.H. The untapped potential of plant sesterterpenoids: Chemistry, biological activities and biosynthesis. Nat. Prod. Rep. 2021, 38, 2293–2314. [Google Scholar] [CrossRef] [PubMed]
- Chiba, R.; Minami, A.; Gomi, K.; Oikawa, H. Identification of ophiobolin F synthase by a genome mining approach: A sesterterpene synthase from Aspergillus clavatus. Org. Lett. 2013, 15, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Quan, Z.; Dickschat, J.S. On the mechanism of ophiobolin F synthase and the absolute configuration of its product by isotopic labelling experiments. Org. Biomol. Chem. 2020, 18, 6072–6076. [Google Scholar] [CrossRef]
- Quan, Z.; Dickschat, J.S. Biosynthetic Gene Cluster for Asperterpenols A and B and the Cyclization Mechanism of Asperterpenol A Synthase. Org. Lett. 2020, 22, 7552–7555. [Google Scholar] [CrossRef]
- Qiao, Y.B.; Xu, Q.X.; Huang, Z.J.; Chen, X.; Ren, X.M.; Yuan, W.L.; Guan, Z.H.; Li, P.K.; Li, F.L.; Xiong, C.M.; et al. Genome mining reveals a new cyclopentane-forming sesterterpene synthase with unprecedented stereo-control. Org. Chem. Front. 2022, 9, 5808–5818. [Google Scholar] [CrossRef]
- Yan, D.X.; Arakelyan, J.; Wan, T.; Raina, R.; Chan, T.K.; Ahn, D.; Kushnarev, V.; Cheung, T.K.; Chan, H.C.; Choi, I.; et al. Genomics-driven derivatization of the bioactive fungal sesterterpenoid variecolin: Creation of an unnatural analogue with improved anticancer properties. Acta Pharm. Sin. B 2024, 14, 421–432. [Google Scholar] [CrossRef]
- Qiu, S.; Gilani, M.D.S.; Müller, C.; Zarazua-Navarro, R.M.; Liebal, U.; Eerlings, R.; Blank, L.M. Cultivation optimization promotes ginsenoside and universal triterpenoid production by engineered yeast. New Biotechnol. 2024, 83, 219–230. [Google Scholar] [CrossRef]
- Wang, S.; Meng, D.; Feng, M.; Li, C.; Wang, Y. Efficient Plant Triterpenoids Synthesis in Saccharomyces cerevisiae: From Mechanisms to Engineering Strategies. ACS Synth. Biol. 2024, 13, 1059–1076. [Google Scholar] [CrossRef]
- Li, Y.L.; Wang, J.; Li, L.Y.; Song, W.H.; Li, M.; Hua, X.; Wang, Y.; Yuan, J.F.; Xue, Z.Y. Natural products of pentacyclic triterpenoids: From discovery to heterologous biosynthesis. Nat. Product. Rep. 2023, 40, 1303–1353. [Google Scholar] [CrossRef]
- Lv, J.M.; Hu, D.; Gao, H.; Kushiro, T.; Awakawa, T.; Chen, G.D.; Wang, C.X.; Abe, I.; Yao, X.S. Biosynthesis of helvolic acid and identification of an unusual C-4-demethylation process distinct from sterol biosynthesis. Nat. Commun. 2017, 8, 1644. [Google Scholar] [CrossRef]
- Cao, Z.; Li, S.; Lv, J.; Gao, H.; Chen, G.; Awakawa, T.; Abe, I.; Yao, X.; Hu, D. Biosynthesis of clinically used antibiotic fusidic acid and identification of two short-chain dehydrogenase/reductases with converse stereoselectivity. Acta Pharm. Sin. B 2019, 9, 433–442. [Google Scholar] [CrossRef]
- Li, X.Y.; Lv, J.M.; Cao, Z.Q.; Wang, G.Q.; Lin, F.L.; Chen, G.D.; Qin, S.Y.; Hu, D.; Gao, H.; Yao, X.S. Biosynthetic characterization of the antifungal fernane-type triterpenoid polytolypin for generation of new analogues via combinatorial biosynthesis. Org. Biomol. Chem. 2023, 21, 851–857. [Google Scholar] [CrossRef]
- Cao, Z.Q.; Wang, G.Q.; Luo, R.; Gao, Y.H.; Lv, J.M.; Qin, S.Y.; Chen, G.D.; Awakawa, T.; Bao, X.F.; Mei, Q.H.; et al. Biosynthesis of Enfumafungin-type Antibiotic Reveals an Unusual Enzymatic Fusion Pattern and Unprecedented C-C Bond Cleavage. J. Am. Chem. Soc. 2024, 146, 12723–12733. [Google Scholar] [CrossRef]
- Han, H.; Peng, S.; Wang, Q.; Wang, H.; Wang, P.; Li, C.; Qi, J.; Liu, C. Biochemical characterization of a multiple prenyltransferase from Tolypocladium inflatum. Appl. Microbiol. Biotechnol. 2024, 108, 275. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Yao, N.; Yuan, W.; Li, F.; Xiao, Y.; Rehmutulla, M.; Xie, Y.; Chen, C.; Zhu, H.; Zhou, Y.; et al. Total biosynthesis of cotylenin diterpene glycosides as 14-3-3 protein-protein interaction stabilizers. Chem. Sci. 2025, 16, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Mitsuhashi, T.; Lee, S.; Hoshino, M.; Mori, T.; Okada, M.; Zhang, H.; Hayashi, F.; Fujita, M.; Abe, I. Astellifadiene: Structure Determination by NMR Spectroscopy and Crystalline Sponge Method, and Elucidation of its Biosynthesis. Angew. Chem. Int. Ed. Engl. 2016, 55, 5785–5788. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.H.; Lv, J.M.; Wang, Q.Z.; Zou, J.; Lu, Y.J.; Wang, Q.L.; Chen, D.N.; Yao, X.S.; Gao, H.; Hu, D. Biosynthesis of an anti-tuberculosis sesterterpenoid asperterpenoid A. Org. Biomol. Chem. 2019, 17, 248–251. [Google Scholar] [CrossRef]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009, 48, 4688–4716. [Google Scholar] [CrossRef]
- Nivina, A.; Yuet, K.P.; Hsu, J.; Khosla, C. Evolution and Diversity of Assembly-Line Polyketide Synthases. Chem. Rev. 2019, 119, 12524–12547. [Google Scholar] [CrossRef]
- Paulsel, T.Q.; Williams, G.J. Current State-of-the-Art Toward Chemoenzymatic Synthesis of Polyketide Natural Products. Chembiochem 2023, 24, e202300386. [Google Scholar] [CrossRef]
- He, Y.; Cox, R.J. The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 2016, 7, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Matsuyama, T.; Ozaki, T.; Takino, J.; Sato, H.; Uchiyama, M.; Minami, A.; Oikawa, H. Elucidation of Late-Stage Biosynthesis of Phomoidride: Proposal of Cyclization Mechanism Affording Characteristic Nine-Membered Ring of Fungal Dimeric Anhydride. J. Am. Chem. Soc. 2022, 144, 20998–21004. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Yu, C.; Qi, J.; Wang, P.; Zhao, P.; Gong, W.; Xie, C.; Xia, X.; Liu, C. High-efficient production of mushroom polyketide compounds in a platform host Aspergillus oryzae. Microb. Cell Fact. 2023, 22, 60. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.D.; Zhang, S.X.; Wang, Y.; Song, Z.W.; Wang, W.Y.; Yin, C.P.; Zhang, Y.L. Biosynthesis of Physcion and Identification of an O-Methyltransferase with C6-OH Selectivity in Aspergillus chevalieri BYST01. ACS Chem. Biol. 2025, 20, 1048–1058. [Google Scholar] [CrossRef]
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 2003, 20, 275–287. [Google Scholar] [CrossRef]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. Engl. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- de Mattos-Shipley, K.M.J.; Greco, C.; Heard, D.M.; Hough, G.; Mulholland, N.P.; Vincent, J.L.; Micklefield, J.; Simpson, T.J.; Willis, C.L.; Cox, R.J.; et al. The cycloaspeptides: Uncovering a new model for methylated nonribosomal peptide biosynthesis. Chem. Sci. 2018, 9, 4109–4117. [Google Scholar] [CrossRef]
- Qi, J.; Han, H.; Sui, D.; Tan, S.; Liu, C.; Wang, P.; Xie, C.; Xia, X.; Gao, J.M.; Liu, C. Efficient production of a cyclic dipeptide (cyclo-TA) using heterologous expression system of filamentous fungus Aspergillus oryzae. Microb. Cell Fact. 2022, 21, 146. [Google Scholar] [CrossRef]
- Katayama, T.; Jiang, Y.; Ozaki, T.; Oikawa, H.; Minami, A.; Maruyama, J.I. Subcellular compartmentalized localization of transmembrane proteins essential for production of fungal cyclic peptide cyclochlorotine. Biosci. Biotechnol. Biochem. 2024, 88, 1279–1288. [Google Scholar] [CrossRef]
- Pinheiro, Â.; Piontkivska, D.; Sequeira, P.; Martins, T.M.; Silva Pereira, C. Aspergillus nidulans . Trends Microbiol. 2023, 31, 212–213. [Google Scholar] [CrossRef]
- Lin, S.Y.; Oakley, C.E.; Jenkinson, C.B.; Chiang, Y.M.; Lee, C.K.; Jones, C.G.; Seidler, P.M.; Nelson, H.M.; Todd, R.B.; Wang, C.C.C.; et al. A heterologous expression platform in Aspergillus nidulans for the elucidation of cryptic secondary metabolism biosynthetic gene clusters: Discovery of the Aspergillus fumigatus sartorypyrone biosynthetic pathway. Chem. Sci. 2023, 14, 11022–11032. [Google Scholar] [CrossRef]
- Hu, J.; Li, H.; Chooi, Y.H. Fungal Dirigent Protein Controls the Stereoselectivity of Multicopper Oxidase-Catalyzed Phenol Coupling in Viriditoxin Biosynthesis. J. Am. Chem. Soc. 2019, 141, 8068–8072. [Google Scholar] [CrossRef]
- Li, H.; Hu, J.; Wei, H.; Solomon, P.S.; Stubbs, K.A.; Chooi, Y.H. Biosynthesis of a Tricyclo [6.2.2.0(2,7)] dodecane System by a Berberine Bridge Enzyme-Like Aldolase. Chemistry 2019, 25, 15062–15066. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gilchrist, C.L.M.; Phan, C.S.; Lacey, H.J.; Vuong, D.; Moggach, S.A.; Lacey, E.; Piggott, A.M.; Chooi, Y.H. Biosynthesis of a New Benzazepine Alkaloid Nanangelenin A from Aspergillus nanangensis Involves an Unusual l-Kynurenine-Incorporating NRPS Catalyzing Regioselective Lactamization. J. Am. Chem. Soc. 2020, 142, 7145–7152. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.Y.; Zhang, J.M.; Xu, Q.D.; Zhang, H.R.; Hu, C.; Zou, Y. Biosynthesis of Cosmosporasides Reveals the Assembly Line for Fungal Hybrid Terpenoid Saccharides. Angew. Chem. Int. Ed. Engl. 2023, 62, e202308887. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhang, H.R.; Zou, Y. A Cytochrome P450 Catalyzes Oxidative Coupling Formation of Insecticidal Dimeric Indole Piperazine Alkaloids. Angew. Chem. Int. Ed. Engl. 2024, 63, e202404000. [Google Scholar] [CrossRef]
- Yang, W.; Tian, S.; Du, Y.F.; Zeng, X.L.; Liang, J.J.; Lan, W.J.; Li, H. Genome Mining of the Marine-Derived Fungus Trichoderma erinaceum F1-1 Unearths Bergamotene-Type Sesquiterpenoids. J. Nat. Prod. 2024, 87, 2746–2756. [Google Scholar] [CrossRef]
- Smith, D.J.; Burnham, M.K.; Edwards, J.; Earl, A.J.; Turner, G. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillum chrysogenum. Biotechnology 1990, 8, 39–41. [Google Scholar] [CrossRef]
- Park, D.; Swayambhu, G.; Pfeifer, B.A. Heterologous biosynthesis as a platform for producing new generation natural products. Curr. Opin. Biotechnol. 2020, 66, 123–130. [Google Scholar] [CrossRef]
- Yeh, H.H.; Chiang, Y.M.; Entwistle, R.; Ahuja, M.; Lee, K.H.; Bruno, K.S.; Wu, T.K.; Oakley, B.R.; Wang, C.C. Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl. Microbiol. Biotechnol. 2012, 96, 739–748. [Google Scholar] [CrossRef]
- Meyer, V.; Fiedler, M.; Nitsche, B.; King, R. The Cell Factory Aspergillus Enters the Big Data Era: Opportunities and Challenges for Optimising Product Formation. Adv. Biochem. Eng. Biotechnol. 2015, 149, 91–132. [Google Scholar] [CrossRef]
- Zaehle, C.; Gressler, M.; Shelest, E.; Geib, E.; Hertweck, C.; Brock, M. Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity. Chem. Biol. 2014, 21, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Löhr, N.A.; Eisen, F.; Thiele, W.; Platz, L.; Motter, J.; Hüttel, W.; Gressler, M.; Müller, M.; Hoffmeister, D. Unprecedented Mushroom Polyketide Synthases Produce the Universal Anthraquinone Precursor. Angew. Chem. Int. Ed. Engl. 2022, 61, e202116142. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, J.I. Genome Editing Technology and Its Application Potentials in the Industrial Filamentous Fungus Aspergillus oryzae. J. Fungi 2021, 7, 638. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Deng, S.; Czajka, J.J.; Dai, Z.; Hofstad, B.A.; Kim, J.; Pomraning, K.R. CRISPR-Cas9/Cas12a systems for efficient genome editing and large genomic fragment deletions in Aspergillus niger. Front. Bioeng. Biotechnol. 2024, 12, 1452496. [Google Scholar] [CrossRef]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Møller, L.L.H.; Larsen, T.O.; Kumar, R.; Arnau, J. Safety of the fungal workhorses of industrial biotechnology: Update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 2018, 102, 9481–9515. [Google Scholar] [CrossRef]
- Kuivanen, J.; Wang, Y.J.; Richard, P. Engineering Aspergillus niger for galactaric acid production: Elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Fact. 2016, 15, 210. [Google Scholar] [CrossRef]
- Kuivanen, J.; Arvas, M.; Richard, P. Clustered Genes Encoding 2-Keto-l-Gulonate Reductase and l-Idonate 5-Dehydrogenase in the Novel Fungal d-Glucuronic Acid Pathway. Front. Microbiol. 2017, 8, 225. [Google Scholar] [CrossRef]
- Kuivanen, J.; Richard, P. NADPH-dependent 5-keto-D-gluconate reductase is a part of the fungal pathway for D-glucuronate catabolism. FEBS Lett. 2018, 592, 71–77. [Google Scholar] [CrossRef]
- Kuivanen, J.; Korja, V.; Holmström, S.; Richard, P. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes. Fungal Biol. Biotechnol. 2019, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Sarkari, P.; Marx, H.; Blumhoff, M.L.; Mattanovich, D.; Sauer, M.; Steiger, M.G. An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger. Bioresour. Technol. 2017, 245, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Wanka, F.; van Gent, J.; Arentshorst, M.; van den Hondel, C.A.; Ram, A.F. Fungal gene expression on demand: An inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 2011, 77, 2975–2983. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Cairns, T.; Zheng, P.; Meyer, V.; Sun, J. Protocol for gene characterization in Aspergillus niger using 5S rRNA-CRISPR-Cas9-mediated Tet-on inducible promoter exchange. STAR Protoc. 2022, 3, 101838. [Google Scholar] [CrossRef]
- Cairns, T.C.; Feurstein, C.; Zheng, X.; Zhang, L.H.; Zheng, P.; Sun, J.; Meyer, V. Functional exploration of co-expression networks identifies a nexus for modulating protein and citric acid titres in Aspergillus niger submerged culture. Fungal Biol. Biotechnol. 2019, 6, 18. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, X.; Cairns, T.C.; Zhang, Z.; Wang, D.; Zheng, P.; Sun, J. Disruption or reduced expression of the orotidine-5′-decarboxylase gene pyrG increases citric acid production: A new discovery during recyclable genome editing in Aspergillus niger. Microb. Cell Fact. 2020, 19, 76. [Google Scholar] [CrossRef]
- Li, Y.; Li, C.; Fu, Y.; Zhang, Q.; Ma, J.; Zhou, J.; Li, J.; Du, G.; Liu, S. A CRISPR/Cas9-based visual toolkit enabling multiplex integration at specific genomic loci in Aspergillus niger. Synth. Syst. Biotechnol. 2024, 9, 209–216. [Google Scholar] [CrossRef]
- Nakamura, H.; Katayama, T.; Okabe, T.; Iwashita, K.; Fujii, W.; Kitamoto, K.; Maruyama, J.I. Highly efficient gene targeting in Aspergillus oryzae industrial strains under ligD mutation introduced by genome editing: Strain-specific differences in the effects of deleting EcdR, the negative regulator of sclerotia formation. J. Gen. Appl. Microbiol. 2017, 63, 172–178. [Google Scholar] [CrossRef]
- Katayama, T.; Nakamura, H.; Zhang, Y.; Pascal, A.; Fujii, W.; Maruyama, J.I. Forced Recycling of an AMA1-Based Genome-Editing Plasmid Allows for Efficient Multiple Gene Deletion/Integration in the Industrial Filamentous Fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2019, 85, e01896-18. [Google Scholar] [CrossRef]
- Maini Rekdal, V.; van der Luijt, C.R.B.; Chen, Y.; Kakumanu, R.; Baidoo, E.E.K.; Petzold, C.J.; Cruz-Morales, P.; Keasling, J.D. Edible mycelium bioengineered for enhanced nutritional value and sensory appeal using a modular synthetic biology toolkit. Nat. Commun. 2024, 15, 2099. [Google Scholar] [CrossRef]
- Mózsik, L.; Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Transcriptional Activation of Biosynthetic Gene Clusters in Filamentous Fungi. Front. Bioeng. Biotechnol. 2022, 10, 901037. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A.B. Filamentous fungi for the production of enzymes, chemicals and materials. Curr. Opin. Biotechnol. 2019, 59, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Jun, S.C.; Han, K.H.; Hong, S.B.; Yu, J.H. Diversity, Application, and Synthetic Biology of Industrially Important Aspergillus Fungi. Adv. Appl. Microbiol. 2017, 100, 161–202. [Google Scholar] [CrossRef]
- Feng, J.; Hauser, M.; Cox, R.J.; Skellam, E. Engineering Aspergillus oryzae for the Heterologous Expression of a Bacterial Modular Polyketide Synthase. J. Fungi 2021, 7, 1085. [Google Scholar] [CrossRef] [PubMed]
- Kotani, A.; Ozaki, T.; Takino, J.; Mochizuki, S.; Akimitsu, K.; Minami, A.; Oikawa, H. Heterologous expression of a polyketide synthase ACRTS2 in Aspergillus oryzae produces host-selective ACR toxins: Coproduction of minor metabolites. Biosci. Biotechnol. Biochem. 2022, 86, 287–293. [Google Scholar] [CrossRef]
- Jin, F.J.; Hu, S.; Wang, B.T.; Jin, L. Advances in Genetic Engineering Technology and Its Application in the Industrial Fungus Aspergillus oryzae. Front. Microbiol. 2021, 12, 644404. [Google Scholar] [CrossRef]
- Panchanawaporn, S.; Chutrakul, C.; Jeennor, S.; Anantayanon, J.; Rattanaphan, N.; Laoteng, K. Potential of Aspergillus oryzae as a biosynthetic platform for indigoidine, a non-ribosomal peptide pigment with antioxidant activity. PLoS ONE 2022, 17, e0270359. [Google Scholar] [CrossRef]
- Takahashi, T.; Masuda, T.; Koyama, Y. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol. Genet. Genom. 2006, 275, 460–470. [Google Scholar] [CrossRef]
- Mizutani, O.; Arazoe, T.; Toshida, K.; Hayashi, R.; Ohsato, S.; Sakuma, T.; Yamamoto, T.; Kuwata, S.; Yamada, O. Detailed analysis of targeted gene mutations caused by the Platinum-Fungal TALENs in Aspergillus oryzae RIB40 strain and a ligD disruptant. J. Biosci. Bioeng. 2017, 123, 287–293. [Google Scholar] [CrossRef]
- Son, Y.E.; Park, H.S. Genetic Manipulation and Transformation Methods for Aspergillus spp. Mycobiology 2021, 49, 95–104. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Nodvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Matsu-Ura, T.; Baek, M.; Kwon, J.; Hong, C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015, 2, 4. [Google Scholar] [CrossRef]
- Jiang, C.M.; Lv, G.B.; Tu, Y.Y.; Cheng, X.J.; Duan, Y.T.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Ruan, H.; Zhang, H.; Wu, T.; Zhu, K.; Han, W.; Dong, R.; Ming, T.; Qi, H.; Zhang, Y. Utilization of CRISPR-Cas genome editing technology in filamentous fungi: Function and advancement potentiality. Front. Microbiol. 2024, 15, 1375120. [Google Scholar] [CrossRef]
- Wang, D.; Jin, S.; Lu, Q.; Chen, Y. Advances and Challenges in CRISPR/Cas-Based Fungal Genome Engineering for Secondary Metabolite Production: A Review. J. Fungi 2023, 9, 362. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yang, D.; Zhang, J.; Xu, J.; Chen, Y.E. Recent Advances in Improving Gene-Editing Specificity through CRISPR-Cas9 Nuclease Engineering. Cells 2022, 11, 2186. [Google Scholar] [CrossRef] [PubMed]
- Sakekar, A.A.; Gaikwad, S.R.; Punekar, N.S. Protein expression and secretion by filamentous fungi. J. Biosci. 2021, 46, 5. [Google Scholar] [CrossRef]
- He, Y.; Wang, B.; Chen, W.P.; Cox, R.J.; He, J.R.; Chen, F.S. Recent advances in reconstructing microbial secondary metabolites biosynthesis in Aspergillus spp. Biotechnol. Adv. 2018, 36, 739–783. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).