Abstract
Tar spot is a prevalent fungal disease characterized by shiny black spots on the leaves, stems, and fruits of various plants. It is typically caused by members of the family Phyllachoraceae, which consists of biotrophic and obligate plant parasitic fungi. During field investigations of tar spot fungi in Sichuan Province, China, 70 fungal samples associated with tar spots belonging to the family Poaceae were collected from 13 different locations. Through morphological studies and multi-locus phylogenetic analysis of combined ITS, SSU, and LSU datasets, the collected samples were classified into eight Phyllachora species. Among these, five were identified as new species (Phyllachora cylindricae, P. festucae, P. luzhouensis, P. palmifoliae, and P. wenchuanensis), and two represented new host records (P. chongzhouensis, P. panicicola). The new species are accompanied by descriptions and illustrations, while their characteristics are discussed in relation to comparable taxa. Additionally, P. yuanjiangensis is synonymized under P. xinpingensis. These findings enhance our understanding of tar spot fungi in Sichuan and, given that Phyllachora species are important pathogens of plants in the Poaceae family, establish a foundation for further research to better understand their potential impacts on agriculture and the local ecology.
1. Introduction
The genus Phyllachora (Phyllachoraceae, Phyllachorales) comprises biotrophic, obligate fungi that are widely distributed in tropical and subtropical regions, primarily parasitising the leaves and stems of Poaceae plants [1,2,3]. As a major plant pathogen, most Phyllachora species infect plant hosts and produce raised, melanized structures known as stromata, commonly referred to as tar spots [4]. These species are important plant pathogens, leading to diseases that severely affect the health and productivity of crops, ornamental plants, and forage grasses [5,6]. For instance, Phyllachora maydis, the pathogen responsible for tar spot disease in maize, was first identified in North America in 2015 [7]. Since its discovery, it has spread across the United States and Canada, resulting in reduced grain yield and diminished quality of silage, stover, and husks. In 2021, the economic losses in the United States attributed to tar spot were estimated at 5.97 million metric tons (235 million bushels) in yield reductions, equating to approximately 1.2 billion U.S. dollars (Crop Protection Network Disease Loss Calculator; https://loss.cropprotectionnetwork.org/, accessed on 5 December 2024) [8].
The family Phyllachoraceae was introduced by Theissen and Sydow [9] in 1915 and currently includes 54 genera, with Phyllachora as the type genus [10,11,12]. Phyllachora is the largest genus within the family Phyllachoraceae and has presently 1520 epithets in Index Fungorum (http://www.indexfungorum.org/ (accessed on 18 January 2025)). After excluding species that have been synonymized or transferred to other genera, approximately 990 species remain, although many still exhibit cases of synonymy. Species of this genus are morphologically characterized by conspicuous black clypeate and stromata on the surface or within the leaf tissue, typically appearing oblong to elongated, fusiform, or irregular, surrounded by yellow or pale brown necrotic lesions [13,14]. The ascomata are typically globose, oval, or irregular and are situated subcuticular or immersed within the host tissue, accompanied by numerous branched paraphyses that are slightly longer than the asci [15,16]. The asci are cylindrical, hyaline, short to medium pedicellate, containing four or eight ascospores [13,17]. The ascospores are arranged in monostichous, diplostichous, or irregular patterns [14]. They are round, ovoid, elliptical, or fusiform, hyaline or pale yellow, with smooth or verrucose and may sometimes possess a gelatinous sheath [13,16].
Sichuan Province is recognized as a biodiversity hotspot, showcasing an impressive variety of fungal species [18,19]. Numerous studies have documented the region’s abundance of tar spots, particularly impacting Poaceae crops, with both newly identified and previously known species of Phyllachora being recorded [14,16,20]. Additionally, Sichuan Province, especially the Yangtze River Delta, is a crucial area in China for cultivating Poaceae crops, including significant quantities of rice, maize, wheat, barley, foxtail millet, and broomcorn millet [21,22]. Also, numerous reports indicate the persistent threat of pathogenic fungi to crop production globally. These pathogens have shown a remarkable ability to evolve genetically, increasing their virulence under diverse selection pressures, often transitioning from natural environments to agricultural contexts [23]. Therefore, given the significant prevalence of tar spots, Sichuan presents a considerable challenge for local agriculture. This situation highlights the significance of investigating the diversity and effects of Phyllachora species in Poaceae plants within natural environments in Sichuan province. Therefore, in this study, we present findings from an ongoing survey of tar spot fungi in Sichuan Province, China, conducted from October 2021 to September 2023. A total of 70 specimens were collected from 13 sampling sites across a diverse range of Poaceae plants. Combining morphological studies with multi-gene phylogenetic analysis, we identified eight species, comprising five new species and two new host records. Detailed morphological descriptions, illustrations, and sequence data for these species are provided, offering valuable insights into the diversity within the genus Phyllachora.
2. Materials and Methods
2.1. Sample Collection, Morphological Examination, and Preservation
A survey of tar spot fungi was conducted in Sichuan Province, China, from October 2021 to September 2023. Diseased graminaceous plants were collected from 13 sampling sites across seven regions, including Chengdu City, Meishan City, Luzhou City, Mianyang City, Guangyuan City, Aba Tibetan and Qiang Autonomous Prefecture, and Ganzi Tibetan Autonomous Prefecture. More detailed information is provided in Supporting Information Supplementary Table S1. These fresh specimens were carefully stored in paper envelopes and subsequently transported to the laboratory for further examination. All specimens were preserved in a refrigerator at 4 °C for extended periods. The macromorphological observations and recordings were conducted using a Nikon SMZ800N stereomicroscope with a Nikon DS-Fi3 camera (Nikon Corporation, Tokyo, Japan). Hand sections were prepared under the stereomicroscope and mounted on slides with sterile water. The fungal microstructures were then photographed using the Nikon DS-Ri2 microscope camera (Nikon Corporation, Tokyo, Japan) attached to a Nikon Eclipse Ni-U microscope (Nikon Corporation, Tokyo, Japan). All measurements were recorded using Nikon NIS-Elements D 5.21 (Nikon Corporation, Tokyo, Japan) software, and the images were processed with Adobe Photoshop version 22.0 (Adobe Inc., San Jose, CA, USA). The holotypes were deposited in the Herbarium of Cryptogams at the Kunming Institute of Botany, Academia Sinica (HKAS) in Kunming, China, or the Herbarium of the University of Electronic Science and Technology (HUEST) in Chengdu, China. The taxonomic descriptions of the new taxa are deposited in MycoBank.
2.2. DNA Extraction, PCR Amplification, and Sequencing
Fungal genomic DNA was extracted from ascomata using the SteadyPure Plant Genomic DNA Extraction Kit (AG21026, Accurate Biotechnology, Changsha, Hunan, China) according to the manufacturer’s protocol. The DNA was stored at −20 °C for long-term storage. The three barcodes, which include the nuclear ribosomal internal transcribed spacer (ITS: ITS1-5.8S-ITS2), the partial nuclear ribosomal small subunit rRNA (SSU) and the partial nuclear ribosomal large subunit rRNA (LSU) were amplified by polymerase chain reaction (PCR). The pairwise primers were ITS9mun and ITS4_KYO1 [24] for ITS, LR0R [25] and LR5 [26] for LSU, and PNS1 [27] and NS41 [28] for SSU. The final PCR system was 30 µL, containing 15 μL PCR Master Mix (CoWin Biosciences, Taizhou, China), 11 µL of double-distilled water (ddH2O), 1 μL each forward and reverse primers (10 μM), and 2 μL DNA template. The PCR products were analyzed by electrophoresis in 1% agarose gels. Sanger sequencing was conducted by Sangon Biotech (Shanghai, China).
2.3. Phylogenetic Analyses
The consensus sequences were generated by manual editing, trimming, and assembling raw Sanger sequencing chromatograms using SeqMan Pro version 11.1.0 (DNASTAR, Inc., Madison, WI, USA). Barcode sequences of the family Phyllachoraceae species currently available in GenBank, along with the outgroup taxon Telimena bicincta (MM 133), were downloaded from the NCBI nucleotide database using the R package Analysis of Phylogenetics and Evolution (APE) version 5.8-1 [29]. Multiple sequence alignments were performed using MAFFT version 7.310 [30] with options “--maxiterate 1000 --genafpair --adjustdirectionaccurately” and the alignment results were further trimmed using trimAl version 1.4 [31] with the option “-gapthreshold 0.5”, which only allows 50% of taxa with a gap in each site. The best-fit nucleotide substitution models for each alignment dataset were selected using ModelFinder based on the Corrected Akaike Information Criterion (AICc) [32].
DNA barcode sequence datasets from the trimmed alignments were concatenated using an in-house Python (version 3.11.10) script. Maximum Likelihood (ML) and Bayesian Inference (BI) analyses were performed using the combined datasets. The ML phylogenetic tree was generated using IQ-TREE version 2.2.2.6 [33], with its topology evaluated through 1000 ultrafast bootstrap replicates. BI analysis was conducted using parallel MrBayes version 3.2.7a [34]. Two independent analyses were conducted with 20 million generations and four chains. The initial 25% of sampled trees were discarded as burn-in. Convergence of the Markov Chain Monte Carlo (MCMC) runs was confirmed using Tracer version 1.7.1 [35], ensuring all adequate sample size (ESS) values exceeded 200. The ML tree was annotated using TreeAnnotator version 2.7.3, which was implemented in BEAST version 2.7.3 [36] based on MrBayes MCMC trees without discarding burn-in and applying no posterior probability limit. Finally, the ML tree was visualized with ggtree [37] and further refined using Adobe Illustrator version 22.0.0 (Adobe Inc., CA, USA).
3. Results
3.1. Molecular Phylogeny
The newly generated sequences were deposited in GenBank, and the accession numbers are listed in Table 1. The concatenated dataset consists of three loci, namely LSU, SSU, and ITS, obtained from 157 specimens (including 70 specimens collected in this study) belonging to the family Phyllachoraceae, with Telimena bicincta (MM 133) as the outgroup. The dataset comprises a total of 2369 characters (LSU: 1–819; SSU: 820–1825; ITS: 1826–2369), including gaps, and consists of 1542 distinct patterns, 995 parsimony-informative sites, 424 singleton sites, and 950 constant sites. The best-fit evolution models for ML and MrBayes phylogenetic analysis were GTR+F+I+G4 for the LSU, K2P+G4 for the SSU, and SYM+I+G4 for the ITS. The best-scoring ML tree (lnL = –28,490.650), with ultrafast bootstrap values from ML analyses and posterior probabilities from the MrBayes analysis, is depicted at the node in Figure 1. Phylogenetic analyses demonstrated that our newly collected 70 specimens clustered into eight distinct clades within the genus Phyllachora. These comprise five new species (Phyllachora cylindricae, P. festucae, P. luzhouensis, P. palmifoliae, and P. wenchuanensis) and three known species (P. chongzhouensis, P. graminis, and P panicicola).

Table 1.
Species information and corresponding GenBank accession numbers used in this study. The newly generated sequences are indicated in bold. Type collections are marked with “T”. “-” means sequence unavailable.
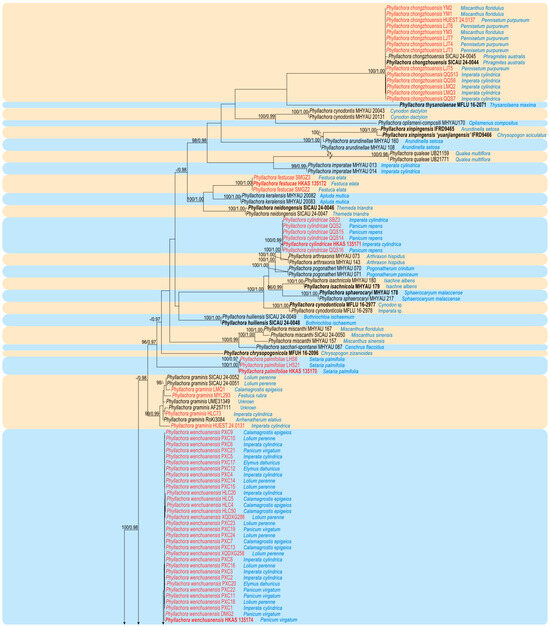
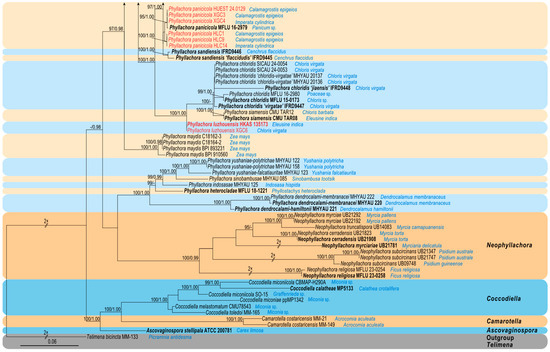
Figure 1.
A phylogram of the best-scoring ML consensus tree based on the concatenated dataset (LSU-SSU-ITS). The type specimens are indicated in bold, while the specimens collected in this study and synonymized taxa are highlighted in red, accompanied by the host species corresponding to their specimen number. The ML ultrafast bootstrap values/Bayesian PP greater than 95%/0.95 are displayed at the respective nodes. The tree is rooted with Telimena bicincta MM-113.
3.2. Taxonomy
3.2.1. Phyllachora chongzhouensis Q.R. Sun, X.L. Xu & C.L. Yang. J. Fungi 10 (No. 588): 8 (2024). Figure 2
MycoBank: MB 902115
Parasitic on leaves and stems of Poaceae. Tar spots 2–4 mm wide, oval, oblong or elongated, black, shiny, carbonaceous. Sexual morph: Ascomata 295–370 μm high, 250–300 μm diam ( = 320 × 280 μm, n = 15), permeating the leaf tissue, covering its entire thickness, like black nevus, domed above the leaf surface, subglobose, ellipsoidal, ostiole inconspicuous, scattered, solitary to gregarious, black, unicellular or multilocular. Peridium 40–120 μm wide ( = 83 μm, n = 20), composed of dark brown to black cells of textura angularis with slender, darker cells of outer layer, and large, slightly paler cells of interlayer. Paraphyses 1.5–5 μm wide ( = 3 μm, n = 20), hyaline, filament-like, and without any branches, septate, not constricted at septate, possessing lengths that surpass the asci, with apically narrowing ends, originating from the innermost basal and lateral walls. Asci 92–163 × 17–31 μm ( = 125 × 23 μm, n = 35), 8-spored, cylindrical, straight or curved, short to medium pedicellate, apex obtuse to rounded, hyaline. Ascospores 17–25 × 10–15 μm ( = 20 × 12 μm, n = 40), uniseriate, oblique, sometimes irregularly arranged, subglobose to ellipsoidal, rounded at the ends, aseptate, verrucous, hyaline, with a gelatinous sheath. Asexual morph: Undetermined.
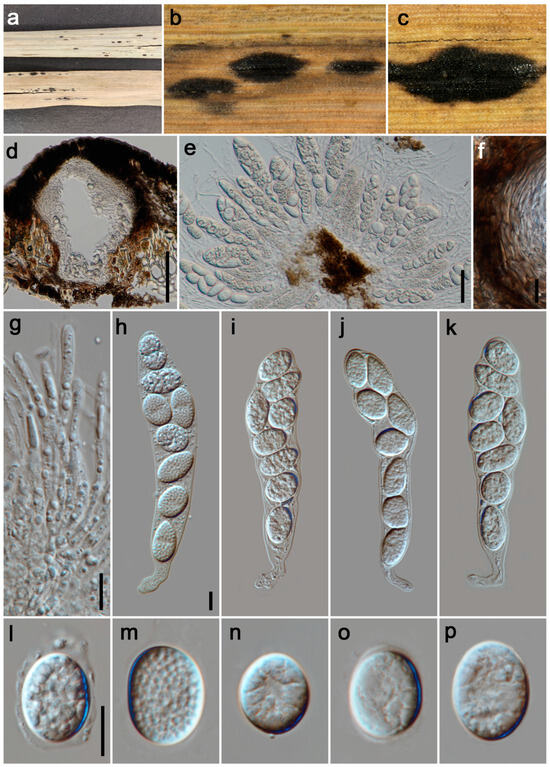
Figure 2.
Phyllachora chongzhouensis (HUEST 24.0137). (a) Black spots on Pennisetum purpureum (Poaceae); (b,c) Stromata; (d) Vertical section of ascomata; (e) Asci and paraphyses; (f) Peridium; (g) Paraphyses; (h–k) Asci; (l–p) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,l). Scale bar of (h) applies to (i–k); Scale bar of (l) applies to (m–p).
Host distribution: Imperata cylindrica, Miscanthus floridulus, Pennisetum purpureum, Phragmites australis [16].
Material examined: China, Sichuan Province: Chengdu City, Wenjiang District, Lujiatan Wetland (30°41′38″ N, 103°46′5″ E, elevation 495 m), on stems and leaves of Pennisetum purpureum (Poaceae), 3 September 2023, P.W. Su, LJT2 (HUEST 24.0137); ibid., LJT3, LJT4, LJT5, LJT6, LJT7. Guangyuan City, Jiange County, Lanmaqiang (32°0′42″ N, 105°24′37″ E, elevation 655 m), on stems and leaves of Imperata cylindrica (Poaceae), 24 May 2023, P.W. Su, LMQ2; ibid., LMQ3. Mianyang City, Zitong County, Qiqu Mountain (31°41′37″ N, 105°11′8″ E, elevation 725 m), on stems and leaves of Imperata cylindrica (Poaceae), 25 May 2023, P.W. Su, QQS6; ibid., QQS7, QQS13. Chengdu City, Chongzhou City, Yangma Wetland Park (30°39′18″ N, 103°45′3″ E, elevation 492 m), on stems and leaves of Miscanthus floridulus (Poaceae), 27 May 2023, P.W. Su, YM1; ibid., YM2, YM3.
Notes: In the phylogenetic tree (Figure 1), our 14 specimens (HUEST 24.0137, LJT3–LJT7, LMQ2, LMQ3, QQS6, QQS7, QQS13, YM1–YM3) clustered together with the known species P. chongzhouensi (SICAU 24-0044 and SICAU 24-0045). Phyllachora chongzhouensis was first identified parasitizing the leaves of Phragmites australis in China [16]. Morphologically (Table 2), there are no significant differences between our specimens and the type species of P. chongzhouensis. Based on morphological diagnosis and phylogenetic analyses, our 14 collections are identified as P. chongzhouensis, marking the first records of this species from Imperata cylindrica, Miscanthus floridulus, and Pennisetum purpureum.
3.2.2. Phyllachora cylindricae P.W. Su & Maharachch., sp. Nov. Figure 3
MycoBank: MB 852909
Etymology: Name reflects the epithet of the host plant, Imperata cylindrica, from which the fungus was collected.
Parasitic on leaves and stems of Poaceae. Tar spots 1–2 mm wide, oblong, fusiform, black, carbonaceous. Sexual morph: Ascomata 200–270 μm high, 260–295 μm diam ( = 235 × 280 μm, n = 15), distributed throughout the leaf tissue, spanning its full thickness, domed above the leaf surface, subglobose or ellipsoidal, like black nevus, ostiole inconspicuous, scattered, solitary to gregarious, black, shiny, unicellular or multilocular. Peridium 55–70 μm wide ( = 61 μm, n = 15), an outer region composed of brown to black cells of textura angularis, inner layers consisting of multiple hyaline, thin-walled strata, flattened fungal cells. Paraphyses 1.5–3.5 μm wide ( = 2.5 μm, n = 20), hyaline, abundant, thread-like and unbranched, aseptate, longer than the asci, with tapering apices, originating from the inner basal and lateral walls. Asci 80–128 × 10–16 μm ( = 95 × 13 μm, n = 35), 8-spored, cylindrical, straight or curved, short to medium pedicellate, apex obtuse to rounded, hyaline. Ascospores 11–15 × 6–8 μm ( = 13 × 7 μm, n = 40), uniseriate, sometimes oblique, tear-shaped, ovoid to ellipsoidal, rounded at the base, tapered at the apex, aseptate, smooth, hyaline, with a gelatinous sheath. Asexual morph: Undetermined.
Host distributions: Imperata cylindrica, Panicum repens.
Material examined: China, Sichuan Province, Mianyang City, Zitong County, Shuangban Town, 31°49′40″ N, 105°3′11″ E, elevation 655 m, on stems and leaves of Imperata cylindrica (Poaceae), 25 May 2023, P.W. Su, SBZ2 (HKAS 135171, holotype; HUEST 24.0130, isotype); ibid., SBZ3. Mianyang City, Zitong County, Qiqu Mountain, 31°41′37″ N, 105°11′8″ E, elevation 725 m, on stems and leaves of Panicum repens (Poaceae), 25 May 2023, P.W. Su, QQS2, QQS14; ibid., QQS15, QQS16.
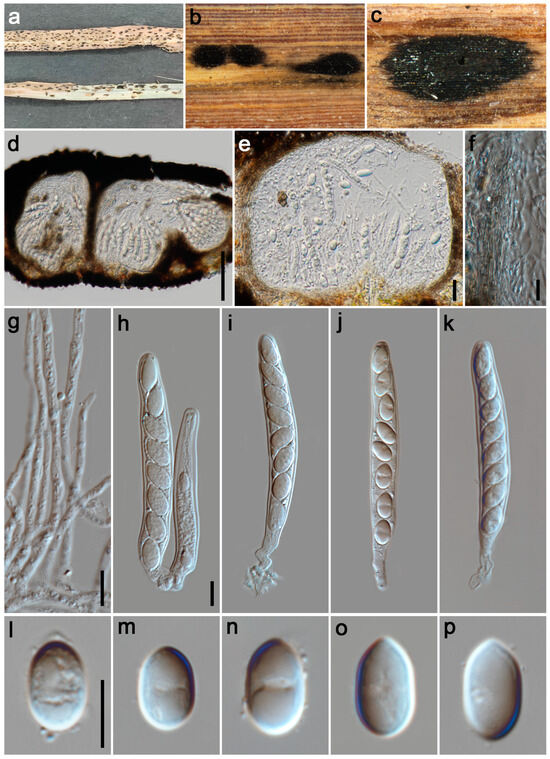
Figure 3.
Phyllachora cylindricae (HKAS 135171, holotype). (a) Black spots on Imperata cylindrica (Poaceae); (b,c) Stromata; (d,e) Vertical section of ascomata; (f) Peridium; (g) Paraphyses; (h–k) Asci; (l–p) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,l). Scale bar of (h) applies to (i–k); Scale bar of (l) applies to (m–p).
Notes: The phylogenetic tree (Figure 1) revealed that specimens HKAS 135171, SBZ3, QQS2, QQS14, QQS15, and QQS16 form a clade that is sister to Phyllachora arthraxonis (MHYAU 073 and MHYAU 143), with high statistical support values (100% ML, 1.00 PP). Phyllachora arthraxonis was first collected by Hennings et al. [38] in 1904 from the leaves of Arthraxon ciliaris, and its molecular data were subsequently deposited to the NCBI by Li et al. [15]. Morphologically, the asci (79.26–128.26 × 10.34–15.98 μm vs. 35–45 × 8–12 μm) and ascospores (10.67–15.27 × 6.45–8.27 μm vs. 8–11 × 4–5 μm) of HKAS 135171 are significantly larger than those of the type specimen of P. arthraxonis [38]. Additionally, the BLASTn analysis of HKAS 135171 and MHYAU 073 shows a 98% identity (525/534, 1 gap) using ITS. Therefore, based on morphological characteristics and phylogenetic analyses, we describe our collections as a new species of Phyllachora.
3.2.3. Phyllachora festucae P.W. Su & Maharachch., sp. Nov. Figure 4
MycoBank: MB 852908
Etymology: Name reflects the host genus, Festuca, from which the fungus was collected.
Parasitic on leaves and stems of Festuca elata. Tar spots 2–4 mm wide, on the upper leaf surface, oval, oblong or elongated, black, shiny, sometimes yellow to brown stripe at the edge of tar spots. Sexual morph: Ascomata 205–365 μm high, 205–720 μm diam ( = 310 × 550 μm, n = 15), permeating the leaf tissue, encompassing its total thickness, domed above the leaf surface, subglobose or ellipsoidal, like black nevus, ostiole inconspicuous, scattered, solitary to gregarious, black, shiny, irregularly multilocular. Peridium 55–95 μm wide ( = 75 μm, n = 20), an outer region composed of brown to dark brown cells of textura angularis, the inner layers are formed by multiple hyaline, thin-walled layers, flattened fungal cells. Paraphyses 2–3 μm wide ( = 2.5 μm, n = 20), hyaline, filiform, unbranched, aseptate, exceeding the length of the asci, featuring tapering apices that emerge from the inner basal and lateral walls. Asci 135–222 × 12–18 μm ( = 163 × 15 μm, n = 35), 8-spored, long and cylindrical, short pedicellate, apex obtuse to rounded, hyaline. Ascospores 17–27 × 8–12 μm ( = 23 × 10 μm, n = 40), uniseriate, sometimes overlapping and oblique, long ellipsoidal to fusiform, acute at the ends, hyaline, with a gelatinous sheath. Asexual morph: Undetermined.
Host distribution: Festuca elata.
Material examined: China, Sichuan Province, Aba Tibetan and Qiang Autonomous Prefecture, Wenchuan County, Shuimoguzhen, 30°56′4″ N, 103°25′19″ E, elevation 902 m, on stems and leaves of Festuca elata (Poaceae), 28 May 2023, P.W. Su, SMGZ1 (HKAS 135172, holotype; HUEST 24.0132, isotype); ibid., SMGZ2, SMGZ3.
Notes: Multi-gene phylogenetic analysis revealed that specimens HKAS 135172, SMGZ2, and SMGZ3 clustered together in a distinct clade, forming a sister group to Phyllachora keralensis (MHYAU 20082, MHYAU 20083). The BLASTn analysis of HKAS 135172 and MHYAU 20082 shows only 83% identity (426/515, 19 gaps) using ITS. Additionally, our specimens HKAS 135172 have significantly larger asci (135.54–222.89 × 12.02–17.82 μm vs. 52–68 × 7–9.5 μm) and ascospores (17.02–27.17 × 8.59–12.46 μm vs. 10–14 × 5–6.5 μm) compared to the type specimen of P. keralensis [39]. Based on differences in morphological features, molecular sequences, and multi-gene phylogenetic analysis, we describe the newly collected specimens HKAS 135172, SMGZ2, and SMGZ3 as a new species, Phyllachora festucae.
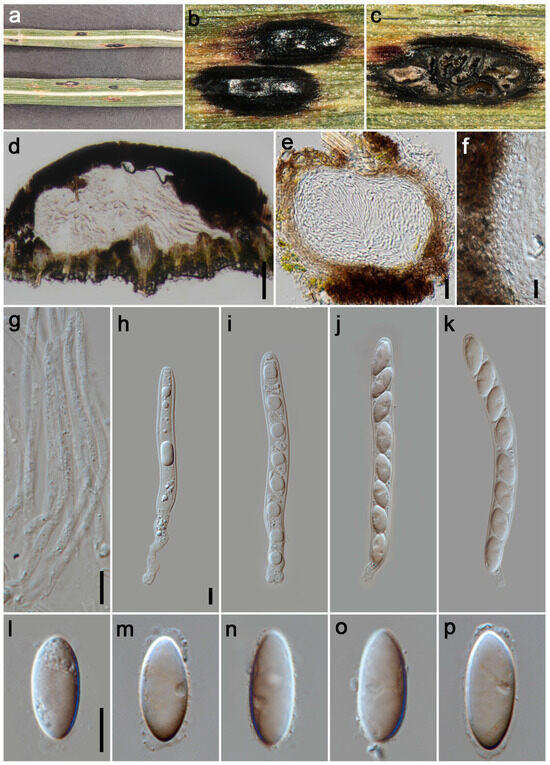
Figure 4.
Phyllachora festucae (HKAS 135171, holotype). (a) Black spots on Festuca elata (Poaceae); (b) Stromata; (c) Horizontal section of ascomata; (d,e) Vertical section of ascomata; (f) Peridium; (g) Paraphyses; (h–k) Asci; (l–p) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,l). Scale bar of (h) applies to (i–k); Scale bar of (l) applies to (m–p).
3.2.4. Phyllachora graminis (Pers.) Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 216 (1870). Figure 5
MycoBank: MB 200927
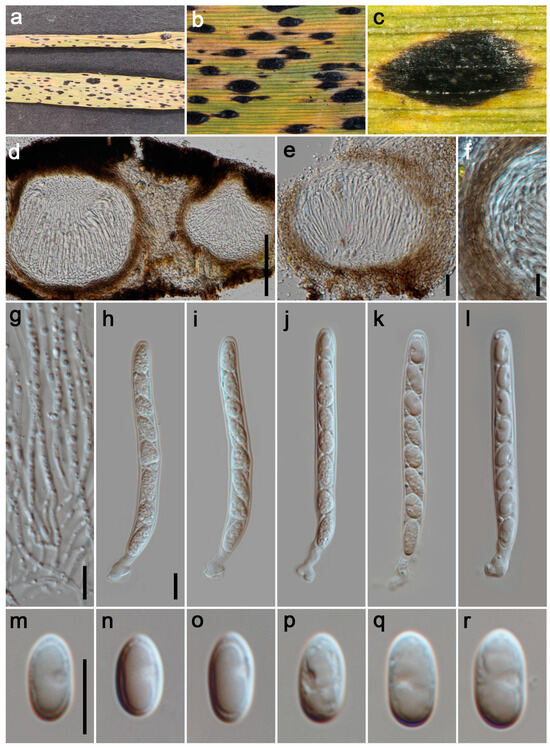
Figure 5.
Phyllachora graminis (HUEST 24.0131). (a) Black spots on Imperata cylindrica (Poaceae); (b,c) Stromata; (d,e) Vertical section of ascomata; (f) Peridium; (g) Paraphyses; (h–l) Asci; (m–r) Ascospores. The scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,m). Scale bar of (h) applies to (i–l); the scale bar of (m) applies to (n–r).
Parasitic on leaves and stems of Poaceae. Tar spots 2–4 mm wide, on both sides of the leaf, fusiform, oblong, black, shiny. Sexual morph: Ascomata 50–290 μm high, 180–570 μm diam ( = 195 × 260 μm, n = 15), subglobose, ellipsoidal, fusiform or irregular, like black nevus, domed above the leaf surface, scattered, sometimes gregarious, black and slightly shiny, unicellular or multilocular. Ostiole 50–55 μm high, 40–48 μm diam ( = 52 × 44 μm, n = 15), circular, brown, located in the center of the ascomata. Peridium 26–53 μm wide ( = 36 μm, n = 25), an outer region composed of brown to dark brown cells of textura angularis, the internal structure comprises multiple layers of hyaline, thin walls, flattened fungal cells. Paraphyses 2–3 μm wide ( = 2.5 μm, n = 25), hyaline, filiform, unbranched, septate, not constricted at septate, surpassing the asci in length, with apices that taper, emanating from the inner basal and lateral walls. Asci 83–105 × 7–9 μm ( = 93 × 8 μm, n = 35), 8-spored, cylindrical, straight or curved, short to medium pedicellate, apex obtuse to rounded, hyaline. Ascospores 10–13 × 5–7 μm ( = 12 × 6 μm, n = 40), uniseriate, sometimes overlapping and oblique, ovoid to ellipsoidal, aseptate, smooth, hyaline, with one large guttule. Asexual morph: Undetermined.
Host distribution: Various Poaceae plant genera, such as Agropyron, Brachypodium, Chloris, Eulalia, Festuca and Imperata [14,40].
Material examined: China, Sichuan Province, Aba Tibetan Qiang Autonomous Prefecture, Li County, Miyaluo, 31°43′7″ N, 102°48′19″ E, elevation 3500 m, on stems and leaves of Imperata cylindrica (Poaceae), 19 October 2021, P.W. Su, MYL295 (HUEST 24.0131); ibid., on stems and leaves of Festuca rubra (Poaceae), MYL293. Ganzi Tibetan Autonomous Prefecture, Luding County, Hualin Village, 29°43′44″ N, 102°17′38″ E, elevation 2065 m, on stems and leaves of Imperata cylindrica (Poaceae), 17 November 2022, P.W. Su, HLC73. Guangyuan City, Jiange County, Lanmaqiang, 32°0′42″ N, 105°24′37″ E, elevation 655 m, on stems and leaves of Calamagrostis epigeios (Poaceae), 24 May 2023, P.W. Su, LMQ1.
Notes: Phyllachora graminis was introduced by Fuckel in 1870 and recognized as the type of the genus, which is widely parasitic on various grasses [40]. The phylogenetic tree indicates that our four specimens (HUEST 24.0131, MYL293, HLC73, and LMQ1) are grouped with other P. graminis specimens (Figure 1). Morphological comparisons show no significant differences between our newly collected specimens and other previously reported P. graminis specimens (Table 2). Based on thorough analysis, HUEST 24.0131, MYL293, HLC73, and LMQ1 can be classified within P. graminis.
3.2.5. Phyllachora luzhouensis P.W. Su & Maharachch., sp. Nov. Figure 6
MycoBank: MB 852910
Etymology: Name reflects Luzhou, the city where the fungus was collected.
Parasitic on leaves and stems of Eleusine indica and Chloris virgata. Tar spots 1–2 mm wide, subcircular, rounded to oblong, black, carbonaceous. Sexual morph: Ascomata 140–360 μm high, 145–560 μm diam ( = 230 × 345 μm, n = 15), permeating the leaf tissue, encompassing its total thickness, domed above the leaf surface, subglobose, ellipsoidal to irregular, ostiole inconspicuous, scattered, solitary to gregarious, black, irregularly multilocular. Peridium 18–63 μm wide ( = 36 μm, n = 15), composed of brown to dark brown cells of textura angularis with slender, darker cells of outer layer, and large, slightly paler cells of interlayer. Paraphyses 1.5–3.5 μm wide ( = 2.5 μm, n = 20), hyaline, plentiful, filamentous, and lacking branches, extending beyond the asci with apices that narrow, originating from the inner basal and lateral walls. Asci 71–132 × 7–10 μm ( = 95 × 8 μm, n = 35), 8-spored, long and cylindrical, short to medium pedicellate, apex obtuse to rounded, hyaline. Ascospores 9–14 × 4–7 μm ( = 12 × 6 μm, n = 40), uniseriate, sometimes overlapping and oblique, ovoid, fusiform to ellipsoidal, acute at the ends, aseptate, hyaline, with one large guttule. Asexual morph: Undetermined.
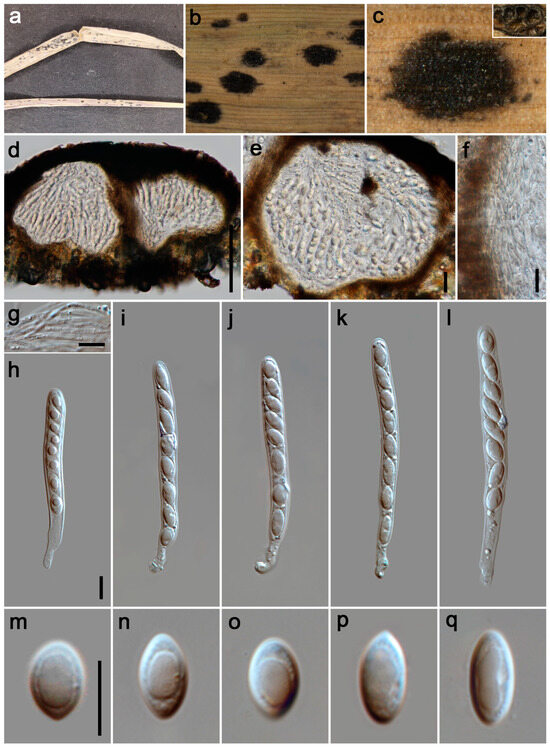
Figure 6.
Phyllachora luzhouensis (HKAS 135173, holotype). (a) Black spots on Eleusine indica (Poaceae); (b,c) Stromata and horizontal section of ascomata; (d,e) Vertical section of ascomata; (f) Peridium; (g) Paraphyses; (h–l) Asci; (m–q) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,m). Scale bar of (h) applies to (i–l); Scale bar of (m) applies to (n–q).
Host distribution: Chloris virgata, Eleusine indica.
Material examined: China, Sichuan Province, Luzhou City, Lu County, Xingguang Village, 29°5′55″ N, 105°27′38″ E, elevation 295 m, on stems and leaves of Eleusine indica (Poaceae), 30 March 2023, P.W. Su, XGC5 (HKAS 135173, holotype; HUEST 24.0133, isotype); ibid., on stems and leaves of Chloris virgata (Poaceae), XGC6.
Notes: In the multi-gene phylogenetic tree (Figure 1), our specimens HUEST 24.0133 and XGC6 form a distinct clade with high statistical support values (100% ML/1.00 PP), closely related to Phyllachora jiaensis (IFRD9448), P. virgatae (IFRD9447), P. chloridis (MFLU 15-0173), and P. siamensis (CMU TAR08). Detailed morphological comparisons reveal that HUEST 24.0133 differs from these species in the size of the asci as well as the shape and size of the ascospores (Table 2). Given the significant differences in morphology and molecular data, we propose the specimens HUEST 24.0133 and XGC6 as a new species, Phyllachora luzhouensis.
3.2.6. Phyllachora palmifoliae P.W. Su & Maharachch., sp. Nov. Figure 7
MycoBank: MB 852911
Etymology: Name reflects the epithet of the host plant, Setaria palmifolia, from where the fungus was collected.
Parasitic on leaves and stems of Setaria palmifolia. Tar spots 2–4 mm wide, subcircular, oblong or irregular, black, shiny, carbonaceous, sometimes yellow to brown stripe at the edge of tar spots. Sexual morph: Ascomata 155–345 μm high, 170–585 μm diam ( = 250 × 375 μm, n = 15), distributed throughout the leaf tissue, spanning its full thickness, like black nevus, domed above the leaf surface, subglobose or ellipsoidal, ostiole inconspicuous, scattered, solitary to gregarious, black, unicellular or multilocular. Peridium 31–58 μm wide ( = 45 μm, n = 15), an outer region composed of brown to black cells of textura angularis, internal layers made up of various hyaline, thin-walled strata, flattened fungal cells. Paraphyses 1.5–3 μm wide ( = 2 μm, n = 20), hyaline, a multitude of slender, non-branching filaments, exceeding the length of the asci, featuring tapering apices that emerge from the inner basal and lateral walls. Asci 110–175 × 9–19 μm ( = 142 × 13 μm, n = 35), 8-spored, elongated and cylindrical, either straight or curved, with a pedicel length ranging from short to medium, and an apex that is either obtuse or rounded, exhibiting hyaline. Ascospores 12–17 × 5–8 μm ( = 15 × 7 μm, n = 40), uniseriate, oblique, sometimes irregularly arranged, ovoid, fusiform to ellipsoidal, aseptate, hyaline, with a gelatinous sheath, one large guttule. Asexual morph: Undetermined.
Host distribution: Setaria palmifolia.
Material examined: China, Sichuan Province, Meishan City, Danling County, Longhu Mountain, 30°3′52″ N, 103°28′45″ E, elevation 623 m, on the leaves of Setaria palmifolia (Poaceae), 11 July 2022, P.W. Su, LHS13 (HKAS 135170, holotype; HUEST 24.0128, isotype); ibid., LHS6, LHS21.
Notes: The phylogenetic tree (Figure 1) shows that the specimens HKAS 135170, LHS6, and LHS21 cluster together, forming a distinct evolutionary branch with strong statistical support (96% ML/0.97 PP). The BLASTn analysis reveals that the ITS sequence of HKAS 135170 most closely matches Phyllachora sp. MHYAU 120, with only 96% identity (489/509, six gaps). MHYAU 120 has not yet been published, with only the sequence data available in the GenBank database. Furthermore, morphological comparisons with closely related species indicate significant differences in the size of the asci and ascospores between HUEST 24.0128 and these species. Therefore, considering the differences in morphology and phylogeny, we describe the newly collected specimens HKAS 135170, LHS6, and LHS21 as a new species, Phyllachora palmifoliae.
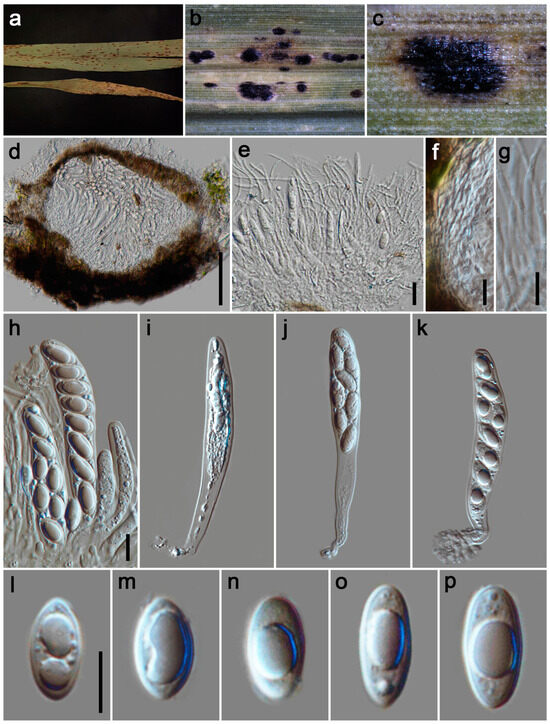
Figure 7.
Phyllachora palmifoliae (HKAS 135173). (a) Black spots on Setaria palmifolia (Poaceae); (b,c) Stromata; (d) Vertical section of ascomata; (e) Asci and paraphyses; (f) Peridium; (g) Paraphyses; (h–k) Asci; (l–p) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,l). Scale bar of (h) applies to (i–k); Scale bar of (l) applies to (m–p).
3.2.7. Phyllachora panicicola Dayar. & K.D. Hyde, Mycosphere 8(10): 1610 (2017). Figure 8
MycoBank: MB 552806
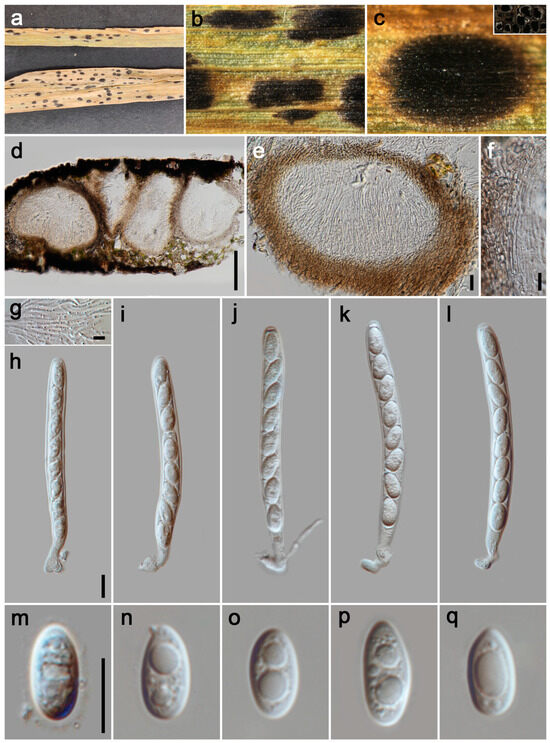
Figure 8.
Phyllachora panicicola (HUEST 24.0129). (a) Black spots on Calamagrostis epigeios (Poaceae); (b,c) Stromata and horizontal section of ascomata; (d,e) Vertical section of ascomata; (f) Peridium; (g) Paraphyses; (h–l) Asci; (m–q) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,m). Scale bar of (h) applies to (i–l); Scale bar of (m) applies to (n–q).
Parasitic on leaves and stems of Poaceae. Tar spots 1–3 mm wide, subcircular, rounded to oblong, black, carbonaceous, sometimes pale yellow to yellow stripe at the edge of tar spots. Sexual morph: Ascomata 175–330 μm high, 235–600 μm diam ( = 235 × 380 μm, n = 20), infused throughout the leaf tissue, covering its entire thickness, like black nevus, domed above the leaf surface, subglobose or ellipsoidal, ostiole inconspicuous, scattered, solitary to gregarious, black, multilocular. Peridium 29–67 μm wide ( = 40 μm, n = 15), an outer region composed of brown to black cells of textura angularis, internal configurations consist of numerous hyaline, thinly walled layers, flattened fungal cells. Paraphyses 1.5–3 μm wide ( = 2 μm, n = 20), hyaline, a multitude of slender, non-branching filaments, exhibiting a length greater than that of the asci, with apices that gradually narrow, stemming from the basal and lateral inner walls. Asci 86–145 × 8–11 μm ( = 105 × 9 μm, n = 35), 8-spored, cylindrical, short to medium pedicellate, apex obtuse to rounded, hyaline. Ascospores 10–14 × 5–7 μm ( = 12 × 6 μm, n = 40), uniseriate, sometimes overlapping and oblique, ovoid to ellipsoidal, aseptate, hyaline, with guttules and a gelatinous sheath. Asexual morph: Undetermined.
Host distribution: Calamagrostis epigeios, Imperata cylindrica, Panicum sp. [4].
Material examined: China, Sichuan Province, Luzhou City, Luxian County, Xingguang Village, 29°5′55″ N, 105°27′38″ E, elevation 295 m, on the stems and leaves of Calamagrostis epigeios and Imperata cylindrica (Poaceae), 30 March 2023, P.W. Su, XGC2 (HUEST 24.0129); ibid., XGC3, XGC4. Ganzi Tibetan Autonomous Prefecture, Luding County, Hualin Village, 29°43′44″ N, 102°17′38″ E, elevation 2065 m, on the stems and leaves of Calamagrostis epigeios and Imperata cylindrica (Poaceae), 17 November 2022, P.W. Su, HLC1; ibid., HLC9, HLC14.
Notes: In the phylogenetic tree (Figure 1), our six specimens (HUEST 24.0129, XGC3, XGC4, HLC1, HLC9, HLC14) are clustered with the known species P. panicicola (MFLU 16-2979). Morphological comparisons (Table 2) show no significant differences between our specimens and P. panicicola. Based on morphological diagnoses and phylogenetic analyses, our six specimens are identified as P. panicicola, representing the first records of this species from Calamagrostis epigeios and Imperata cylindrica.
3.2.8. Phyllachora xinpingensis H.X. Wu & J.C. Li, in Li, Wu & Song, Phytotaxa 578(3): 279 (2023)
- = Phyllachora yuanjiangensis H.X. Wu & J.C. Li, in Li, Wu & Song, Phytotaxa 578(3): 282 (2023)
Notes: Phyllachora yuanjiangensis and P. xinpingensis were introduced in the same study by Li et al. [41]. Here, we synonymized P. yuanjiangensis under P. xinpingensis based on morphological and molecular evidence, as well as the lack of host specificity within the genus. Morphologically, the differences highlighted in the original description, such as ascus length, are minor and fall within the natural variation observed in P. xinpingensis. The molecular phylogenetic analyses in the current study also demonstrate that the two taxa cluster as a single species (Figure 1), providing strong evidence of their conspecificity. Furthermore, the justification for P. yuanjiangensis as a distinct species relied on its association with a specific host plant, yet previous research [42] and this study confirm that species within Phyllachora are not strictly host-specific. These findings collectively support the conclusion that P. yuanjiangensis does not represent a separate taxon but is conspecific with P. xinpingensis.
3.2.9. Phyllachora wenchuanensis P.W. Su & Maharachch., sp. Nov. Figure 9
MycoBank: MB 852912
Etymology: Name reflects Wenchuan, the place where the fungus was collected.
Parasitic on leaves and stems of Poaceae. Tar spots 1–2 mm wide, fusiform, oblong, black, shiny. Sexual morph: Ascomata 160–225 μm high, 200–255 μm diam ( = 175 × 225 μm, n = 20), infused throughout the leaf tissue, covering its entire thickness, like black nevus, domed above the leaf surface, subglobose or ellipsoidal, ostiole inconspicuous, scattered, solitary to gregarious, black, unicellular or multilocular. Peridium 26–63 μm wide ( = 43 μm, n = 15), an outer region composed of brown cells of textura angularis, internal configurations consist of numerous hyaline, thinly walled layers, flattened fungal cells. Paraphyses 1.5–2.5 μm wide ( = 2 μm, n = 20), hyaline, a multitude of slender, non-branching filaments, exhibiting a length greater than that of the asci, with apices that gradually narrow, stemming from the basal and lateral inner walls. Asci 75–115 × 7–10 μm ( = 105 × 9 μm, n = 35), 8-spored, cylindrical, short to medium pedicellate, apex obtuse to rounded, hyaline. Ascospores 10–14 × 5–7 μm ( = 12 × 6 μm, n = 40), uniseriate, oblique, ovoid or ellipsoidal, aseptate, hyaline, one guttule. Asexual morph: Undetermined.
Host distribution: Calamagrostis epigeios, Elymus dahuricus, Imperata cylindrica, Lolium perenne, Panicum virgatum.
Material examined: China, Sichuan Province, Aba Tibetan and Qiang Autonomous Prefecture, Wenchuan County, Damengou, 31°34′2″ N, 103°31′21″ E, elevation 1873 m, on the stems and leaves of Panicum virgatum (Poaceae), 7 July 2023, P.W. Su, DMG1 (HKAS 135174, holotype; HUEST 24.0134, isotype); ibid., DMG2. Ganzi Tibetan Autonomous Prefecture, Luding County, Hualin Village, 29°43′44″ N, 102°17′38″ E, elevation 2065 m, on the stems and leaves of Calamagrostis epigeios and Imperata cylindrica (Poaceae), 17 November 2022, P.W. Su, HLC4; ibid., HLC5, HLC20, HLC50. Aba Tibetan and Qiang Autonomous Prefecture, Wenchuan County, Xiqiang Valley, 31°29′27″ N, 103°37′1″ E, elevation 1500 m, on the stems and leaves of Lolium perenne (Poaceae), 20 October 2021, P.W. Su, XQDXG258; ibid., XQDXG286. Aba Tibetan and Qiang Autonomous Prefecture, Li County, Puxi Village, 31°29′19″ N, 103°16′52″ E, elevation 2412 m, on the stems and leaves of various Poaceae species (Calamagrostis epigeios, Elymus dahuricus, Imperata cylindrica, Lolium perenne, Panicum virgatum), P.W. Su, PXC1–PXC24.
Note: The phylogenetic tree (Figure 1) indicates that our 32 specimens cluster together and are closely related to P. panicicola (MFLU 16-2979), P. sandiensis (IFRD9446), and P. flaccidudis (IFRD9445). Our collected specimens exhibit similar morphological characteristics in the shape of asci, as seen in P. panicicola, P. sandiensis, and P. flaccidudis. However, P. wenchuanensis differs from P. panicicola in having shorter asci (74.81–115.06 × 7.76–10.52 vs. 110–130 × 10–14) [4]. Additionally, the BLASTn analysis (Table 3) of the ITS sequence of HKAS 135174 reveals 97% identity (512/527, seven gaps) with P. sandiensis (IFRD9446) and 96% identity (506/526, 7 gaps) with P. flaccidudis (IFRD9445). Therefore, given the differences in morphology and phylogeny, we describe the newly collected specimens as a new species.

Figure 9.
Phyllachora wenchuanensis (HKAS 135174, holotype). (a) Black spots on Panicum virgatum (Poaceae); (b,c) Stromata; (d,e) Vertical section of ascomata; (f) Peridium; (g) Paraphyses; (h–k) Asci; (l–q) Ascospores. Scale bars: 100 μm (d), 20 μm (e), 10 μm (f–h,l). Scale bar of (h) applies to (i–k); Scale bar of (l) applies to (m–q).

Table 2.
Morphological comparison of new species and related species of Phyllachora.
Table 2.
Morphological comparison of new species and related species of Phyllachora.
| Species | Voucher | Hosts | Asci (µm) | Ascospores (µm) | Reference | |
|---|---|---|---|---|---|---|
| Size | Shape | |||||
| P. arthraxonis | HMAS 4306 | Arthraxon hispidus (Arthraxon) | 58–83 × 6–16 | 6–14 × 5–7 | ellipsoidal | [14] |
| P. arthraxonis T | - | Arthraxon ciliaris (Arthraxon) | 35–45 × 8–12 | 8–11 × 4–5 | ellipsoidal, ovoid | [38] |
| P. chloridis T | MFLU 15-0173 | Chloris sp. (Poaceae) | 50–72 × 6–8 | 8–12 × 3.5–4.8 | fusiform to oval | [4] |
| P. chongzhouensis T | SICAU 24-0044 | Phragmites australis (Poaceae) | 86–142 × 14–29 | 13–28 × 9–15 | ellipsoidal, ovoid | [16] |
| P. chongzhouensis | HUEST 24.0137 | Pennisetum purpureum (Poaceae) | 92–163 × 17–31 | 17–25 × 10–15 | subglobose to ellipsoidal | This study |
| P. chrysopogonicola T | MFLU 16-2096 | Chrysopogon zizanioides (Poaceae) | 70–122 × 12.5–15 | 20–26 (–30) × 6–8 | unicellular to papillate to turbinate, clavate, | [43] |
| P. cylindrica T | HKAS 135171 | Imperata cylindrica (Poaceae) | 80–128 × 10–16 | 11–15 × 6–8 | tear-shaped, ovoid to ellipsoidal | This study |
| P. festucae T | HKAS 135172 | Festuca elata (Poaceae) | 135–222 × 12–18 | 17–27 × 8–12 | long ellipsoidal to fusiform | This study |
| P. graminis | HUEST 24.0131 | Imperata cylindrica (Poaceae) | 83–105 × 7–9 | 10–13 × 5–7 | ovoid to ellipsoidal | This study |
| P. graminis | SICAU 24-005 | Lolium perenne (Poaceae) | 54–101 × 6–10 | 8–15 × 4–7 | ellipsoidal | [16] |
| P. graminis T | - | Elymus sp. (Poaceae) | 78–80 × 7–8 | 8–12 × 4–5 | oval to ovoid or ovoid | [14,40] |
| P. keralensis | MHYAU 13716 | Apluda mutica (Poaceae) | 48.7–79.1 × 10.7–13.6 | 9.2–12.8 × 5.6–6.9 | ellipsoid or oblong | [44] |
| P. keralensis T | - | Apluda mutica (Poaceae) | 52–68 × 7–9.5 | 10–14 × 5–6.5 | ellipsoid | [39] |
| P. luzhouensis T | HKAS 135173 | Eleusine indica (Poaceae) | 71–132 × 7–10 | 9–14 × 4–7 | ovoid, fusiform to ellipsoidal | This study |
| P. palmifoliae T | HKAS 135170 | Setaria palmifolia (Poaceae) | 110–175 × 9–19 | 12–17 × 5–8 | ovoid, fusiform to ellipsoidal | This study |
| P. panicicola | HUEST 24.0129 | Calamagrostis epigeios (Poaceae) | 86–145 × 7–11 | 10–14 × 5–7 | ovoid to ellipsoidal | This study |
| P. panicicola T | MFLU 16-2979 | Panicum sp. (Poaceae) | 110–130 × 10–14 | 14–16 × 6–8 | ellipsoidal, rounded at the ends | [4] |
| P. flaccidudis T | IFRD9445 | Cenchrus flaccidus (Poaceae) | 80–110 × 7–10 | 11–13 × 4–7 | drop shape, oval to ellipse, rounded at the ends | [13] |
| P. sandiensis T | IFRD9446 | Cenchrus flaccidus (Poaceae) | 92–126 × 7–10 | 10–14 × 6–7 | drop shape, oval to ellipse | [13] |
| P. siamensis T | CMU TAR08 | Eleusine indica (Poaceae) | 62–105 × 7–10 | 10–13 × 5–6 | unicellular to ovate | [42] |
| P. wenchuanensis T | HKAS 135174 | Panicum virgatum (Poaceae) | 75–115 × 7–10 | 10–14 × 5–7 | ovoid or ellipsoidal | This study |
T Type species. “-” not available.

Table 3.
Comparison of ITS sequence similarity among some species of Phyllachora.
Table 3.
Comparison of ITS sequence similarity among some species of Phyllachora.
| ITS (%) | P. panicicola MFLU 16-2979 | P. sandiensis IFRD9446 | P. flaccidudis IFRD9445 | P. panicicola HUEST 24.0129 | P. wenchuanensis HKAS 135174 |
|---|---|---|---|---|---|
| P. panicicola MFLU 16-2979 | 100 | ||||
| P. sandiensis IFRD9446 | 97.9 | 100 | |||
| P. flaccidudis IFRD9445 | 96.89 | 99.03 | 100 | ||
| P. panicicola HUEST 24.0129 | 99.81 | 97.7 | 96.57 | 100 | |
| P. wenchuanensis HKAS 135174 | 97.7 | 97.15 | 96.20 | 97.89 | 100 |
4. Discussion
Between 2019 and 2023, we conducted surveys on the fungal diversity of Phyllachora in Sichuan Province, collecting 70 specimens and identifying eight species, including five new ones. These findings underscore the significant diversity of Phyllachora in the region. However, due to the province’s vast geographical area, this study primarily concentrated on central Sichuan, leaving several crucial ecological zones unexplored, such as the mountainous regions of western Sichuan, the temperate forests in the northeast, and the subtropical areas in the south. To gain a more comprehensive understanding of the diversity of Phyllachora in Sichuan and to potentially uncover additional undiscovered species, future research should prioritize expanding the sampling range, with a particular focus on conducting thorough investigations in the southern regions, including the valleys of the Dadu River and the hills of Yunnan–Guizhou Plateau.
According to Flora Fungorum Sinicorum, Vol. 46: Phyllachora, as of 2009, 96 species of Phyllachora have been reported in China, including 26 new species, primarily found in the southern regions, namely Yunnan, Guangxi, Guangdong, and Sichuan provinces [14]. Since then, only 17 new species of Phyllachora have been identified in China, with molecular data available for 15 species [4,13,15,16,20,44,45,46,47]. Among these, Yunnan Province has been the most extensively studied, with the discovery of seven new species: P. dendrocalami-hamiltoniicola, P. dendrocalami-membranacei, P. isachnicola, P. panicicola, P. sphaerocaryi, P. xinpingensis, and P. yuanjiangensis. Additionally, four new species (P. chongzhouensis, P. heteroclada, P. huiliensis, P. neidongensis) were found in Sichuan Province, two in Hainan Province (P. hainanensis and P. jianfengensis), two in Shaanxi Province (P. jiaensis and P. sandiensis), and two in Shanxi Province (P. flaccidudis and P. virgatae).
Despite a significant number of species reported, the taxonomy of Phyllachora remains fraught with several unresolved issues that require urgent attention. One of the most pressing challenges is the long-standing assumption of host specificity, which has traditionally been used to define species within this genus [42]. Although Phyllachora species have often been regarded as host-specific, present results indicate this assumption is largely flawed. Furthermore, previous results showed that, despite being phylogenetically distinct, P. siamensis and P. chloridis were isolated from the same host genus (Chloris sp.) [43,48]. This lack of host specificity questions the accuracy of previous species delineations based solely on host–plant relationships. Thus, the traditional method of classifying species by their host associations has likely exaggerated the species count within the genus. For example, many Phyllachora species recorded in databases such as Index Fungorum and MycoBank rely on historical descriptions associated with host specificity rather than comprehensive taxonomic analysis evaluations. Reassessing species boundaries requires an approach that incorporates morphological comparisons and multi-locus phylogenetic analyses epitypification. Epitypification is particularly important for older species that were described solely based on morphology without accompanying sequence data [49]. By designating epitypes with molecular data, researchers can provide a stable taxonomic framework for these species. For instance, revisiting the type species, Phyllachora graminis, with modern sequencing techniques could clarify its phylogenetic position and aid in resolving species complexes within the genus. Similarly, Phyllachora maydis, a significant pathogen responsible for tar spot disease in maize, could benefit from epitypification to standardize its identity and facilitate comparisons with closely related species.
In addition to the issue of host specificity, the genus Phyllachora encounters notable taxonomic challenges due to its morphological similarities with other genera, such as Polystigma. These two genera are often indistinguishable based solely on their morphology, with stromatal pigmentation being one of the few distinguishing features [50]. However, these subtle morphological differences complicate the reliable differentiation of species without molecular analysis. The limited availability of type sequences for Phyllachora in GenBank exacerbates this issue, hindering the establishment of accurate species boundaries. The scarcity of type material, coupled with the morphological plasticity of the genus, highlights the necessity for a more comprehensive revision of its taxonomy.
Phyllachora is an ancient and species-rich genus comprising over 900 morphologically recognized species. However, molecular data are available for only 44 species in GenBank, including the five new species identified in this study. This limitation primarily arises because the taxa are biotrophic and unable to grow in culture, making obtaining sequence data from freshly collected specimens challenging. Consequently, the taxonomy of the genus remains largely understudied. Only a limited number of studies have explored the phylogeny of Phyllachora, predominantly based on ITS, LSU, and SSU sequence data, which has revealed that Phyllachora is polyphyletic [4,11]. In this study, we observed that while ITS provided some degree of species resolution, LSU and SSU sequences showed limited power in distinguishing Phyllachora species (Figure S1), with both newly described and previously reported taxa clustering together. Furthermore, the limited resolution of LSU and SSU markers in our study limits the application of the concept of Genealogical Concordance Phylogenetic Species Recognition, as these regions do not provide a sufficient phylogenetic signal to resolve species-level relationships. Although we attempted to include additional protein-coding markers, such as TEF1 and RPB2, PCR amplification was unsuccessful due to primer mismatches and weak amplification signals. We also further attempted whole-genome sequencing, which was complicated by contamination from the plant host DNA, highlighting the need for optimized DNA extraction techniques and novel primers tailored for Phyllachora phylogenetics. Therefore, we emphasize the need for further studies incorporating additional gene regions, particularly those derived from protein-coding genes, to better resolve the relationships among closely related species in the genus Phyllachora. Phylogenomic analysis can also be utilized to identify additional genetic markers for distinguishing taxa within species of Sordariomycetes [51], including those in the genus Phyllachora. Additionally, sequences from the type specimen of P. graminis should be generated to understand the species boundaries of Phyllachora sensu stricto.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof11030208/s1. Table S1: Detailed information of sampling sites. Figure S1: Single-gene trees. Novel isolates are indicated in red. Type isolates are in bold. The ML ultrafast bootstrap values greater than 95% are indicated at the respective nodes.
Author Contributions
Conceptualization, S.S.N.M.; methodology, P.S. and Y.J.; software, P.S. and Y.L.; validation, Y.C. and S.S.N.M.; formal analysis, P.S., Y.C., and S.S.N.M.; investigation, S.S.N.M. and J.-K.L.; resources, S.S.N.M.; data curation, P.S. and Y.C.; writing—original draft preparation, P.S., Y.C. and S.S.N.M.; writing—review and editing, S.S.N.M., A.M. and J.-K.L.; visualization, P.S. and A.M.; supervision, S.S.N.M.; funding acquisition, S.S.N.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Talent Introduction and Cultivation Project, University of Electronic Science and Technology of China, grant number A1098531023601245.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Acknowledgments
We thank the University of Electronic Science and Technology of China for funding this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gabel, A.W. Host-parasite relations of Phyllachora species on native grasses. Mycologia 1989, 81, 702–708. [Google Scholar] [CrossRef]
- Sutton, B.C.; Hodges, C.S. Hawaiian forest fungi. III. A new species, Gloeocoryneum hawaiiense, on Acacia koa. Mycologia 1983, 75, 280–284. [Google Scholar] [CrossRef]
- Parbery, D.G. Studies on graminicolous species of Phyllachora Nke. in Fckl. V. A taxonomic monograph. Aust. J. Bot. 1967, 15, 271–375. [Google Scholar] [CrossRef]
- Dayarathne, M.; Maharachchikumbura, S.; Jones, E.; Goonasekara, I.; Bulgakov, T.; Al-Sadi, A. Neophyllachora gen nov. (Phyllachorales), three new species of Phyllachora from Poaceae and resurrection of Polystigmataceae (Xylariales). Mycosphere 2017, 8, 1598–1625. [Google Scholar] [CrossRef]
- MacCready, J.S.; Roggenkamp, E.M.; Gdanetz, K.; Chilvers, M.I. Elucidating the obligate nature and biological capacity of an invasive fungal corn pathogen. Mol. Plant-Microbe Interact. 2023, 36, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Telenko, D.E.P.; Ross, T.J.; Shim, S.; Wang, Q.; Singh, R. Draft genome sequence resource for Phyllachora maydis—An obligate pathogen that causes tar spot of corn with recent economic impacts in the United States. Mol. Plant-Microbe Interact. 2020, 33, 884–887. [Google Scholar] [CrossRef]
- Bissonnette, S. Corn Disease Alert: New Fungal Leaf Disease “Tar Spot” Phyllachora Maydist Identified in 3 Northern Illinois Counties. The Bulletin: Pest Management and Crop Development Information for Illinois. Available online: https://farmdoc.illinois.edu/field-crop-production/diseases/corn-disease-alert-new-fungal-leaf-disease-tar-spot-phyllachora-maydis-identified-in-3-northern-illinois-counties.html (accessed on 16 January 2025).
- Roggenkamp, E.M.; Check, J.C.; Biswal, A.K.; Floyd, C.M.; Miles, L.A.; Nicolli, C.P.; Shim, S.; Salgado-Salazar, C.; Alakonya, A.E.; Malvick, D.K.; et al. Development of a qPCR assay for species-specific detection of the tar spot pathogen Phyllachora maydis. PhytoFrontiers 2023, 4, 61–71. [Google Scholar] [CrossRef]
- Theissen, F.; Sydow, H. Die Dothideales. Ann. Mycol. 1915, 13, 147–746. [Google Scholar]
- Hyde, K.; Noordeloos, M.E.; Savchenko, K. The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 2024, 15, 5146–6239. [Google Scholar] [CrossRef]
- Dos Santos, M.D.M.; de Noronha Fonseca, M.E.; Silva Boiteux, L.; Câmara, P.E.A.; Dianese, J.C. ITS phylogeny and taxonomy of Phyllachora species on native Myrtaceae from the Brazilian Cerrado. Mycologia 2016, 108, 1141–1164. [Google Scholar]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi, 10th ed.; CABI Publishing: London, UK, 2008; p. 708. [Google Scholar]
- Li, J.C.; Wu, H.X.; Li, Y.; Li, X.H.; Song, J.Y.; Suwannarach, N.; Wijayawardene, N.N. Taxonomy, phylogenetic and ancestral area reconstruction in Phyllachora, with four novel species from Northwestern China. J. Fungi 2022, 8, 520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, T. Flora Fungorum Sinicorum. Phyllachora; Science Press: Beijing, China, 2014; Volume 46. [Google Scholar]
- Li, X.L.; Wu, S.R.; Wang, C.L.; Feng, Y.L.; Zhao, C.Y.; Chen, Z.Q.; Yu, J.F.; Luo, R.; Promputtha, I.; Sun, D.F. Two new species of Phyllachora (Phyllachoraceae, Phyllachorales) on bamboo from China. Phytotaxa 2019, 425, 78–86. [Google Scholar] [CrossRef]
- Sun, Q.R.; Xu, X.L.; Zeng, Z.; Deng, Y.; Liu, F.; Gao, L.P.; Wang, F.H.; Yan, Y.Q.; Liu, Y.G.; Yang, C.L. Six species of Phyllachora with three new taxa on grass from Sichuan Province, China. J. Fungi 2024, 10, 588. [Google Scholar] [CrossRef]
- Bundhun, D.; Jeewon, R.; Dayarathne, M.C.; Bulgakov, T.S.; Khramtsov, A.K.; Aluthmuhandiram, J.; Pem, D.; To-Anun, C.; Hyde, K.D. A morpho-molecular re-appraisal of Polystigma fulvum and P. rubrum (Polystigma, Polystigmataceae). Phytotaxa 2019, 422, 209–224. [Google Scholar] [CrossRef]
- Jordi, L.P.; Zhang, F.M.; Ge, S. Plant biodiversity in China: Richly varied, endangered, and in need of conservation. Biodivers. Conserv. 2006, 15, 3983–4026. [Google Scholar]
- Tian, W.H.; Liu, J.W.; Jin, Y.; Chen, Y.P.; Zhou, Y.F.; Wu, K.; Su, P.W.; Guo, X.Y.; Wanasinghe, D.N.; Hyde, K.D.; et al. Morphological and phylogenetic studies of Ascomycota from gymnosperms in Sichuan Province, China. Mycosphere 2024, 15, 1794–1900. [Google Scholar] [CrossRef]
- Yang, C.L.; Xu, X.L.; Liu, Y.G.; Hyde, K.D.; Mckenzie, E.H. A new species of Phyllachora (Phyllachoraceae, Phyllachorales) on Phyllostachys heteroclada from Sichuan, China. Phytotaxa 2019, 392, 186–196. [Google Scholar] [CrossRef]
- Wang, J.; Seyler, B.C.; Ticktin, T.; Zeng, Y.; Ayu, K. An ethnobotanical survey of wild edible plants used by the Yi people of Liangshan Prefecture, Sichuan Province, China. J. Ethnobiol. Ethnomed. 2020, 16, 10. [Google Scholar] [CrossRef]
- Zeng, Q.; Lv, Y.C.; Xu, X.L.; Deng, Y.; Wang, F.H.; Liu, S.Y.; Liu, L.J.; Yang, C.L.; Liu, Y.G. Morpho-molecular characterization of microfungi associated with Phyllostachys (Poaceae) in Sichuan, China. J. Fungi 2022, 8, 702. [Google Scholar] [CrossRef]
- Madhushan, A.; Weerasingha, D.B.; Ilyukhin, E.; Taylor, P.W.; Ratnayake, A.S.; Liu, J.K.; Maharachchikumbura, S.S.N. From Natural Hosts to Agricultural Threats: The Evolutionary Journey of Phytopathogenic Fungi. J. Fungi 2025, 11, 25. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-Coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed]
- Cubeta, M.; Echandi, E.; Abernethy, T.; Vilgalys, R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 1991, 81, 1395–1400. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.S. Phylogenetic evidence for horizontal transmission of group I introns in the nuclear ribosomal DNA of mushroom-forming fungi. Mol. Biol. Evol. 1996, 13, 903–917. [Google Scholar] [CrossRef]
- Simon, L.; Lalonde, M.; Bruns, T.D. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl. Environ. Microbiol. 1992, 58, 291–295. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and evolution in R language. Bioinform. 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Hennings, P. Einige neue Pilze aus Japan. II. Hedwigia 1904, 43, 140–146. [Google Scholar]
- Hosagoudar, V.B.; Manian, S.; Vasuki, S. Miscellaneous fungi from south india iv. J. Econ. Taxon. Bot. 1988, 12, 421–424. [Google Scholar]
- Fuckel, L. Symbolae mycologicae. Jahrbücher Des Nassau. Ver. Für Naturkunde 1870, 23, 216–219. [Google Scholar]
- Li, J.C.; Wu, H.X.; Song, J.Y. Two new Phyllachora species in Southwest China. Phytotaxa 2023, 578, 275–285. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Niskanen, T.; Suwannarach, N.; Wannathes, N.; Chen, Y.J.; McKenzie, E.H.C.; Maharachchikumbura, S.S.N.; Buyck, B.; Zhao, C.L.; Fan, Y.G.; et al. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022, 114, 387–462. [Google Scholar] [CrossRef]
- Tamakaew, N. Tar spot fungi from Thailand. Mycosphere 2017, 8, 1054–1058. [Google Scholar] [CrossRef]
- Li, X.L.; Zhang, T.; Zhang, Z.Y.; Yang, H.Q.; Teng, X.Q. Four new records of Phyllachora from China. Mycosystema 2010, 29, 918–919. [Google Scholar]
- Li, X.; Yang, Z.; Wang, X.; Wang, Y.; Li, C. Two new species of graminicolous Phyllachora (Phyllachoraceae, Ascomycota). Mycosystema 2018, 37, 1127–1132. [Google Scholar]
- Liu, N.; Li, M. Phyllachora jianfengensis sp. nov. from China. Mycotaxon 2016, 130, 1039–1043. [Google Scholar] [CrossRef]
- Liu, N.; Wang, L.; Huang, G.; Guo, L. Phyllachora hainanensis sp. nov. from China. Mycotaxon 2015, 130, 237–239. [Google Scholar] [CrossRef]
- Mardones, M.; Trampe-Jaschik, T.; Oster, S.; Elliott, M.; Urbina, H.; Schmitt, I.; Piepenbring, M. Phylogeny of the order Phyllachorales (Ascomycota, Sordariomycetes): Among and within order relationships based on five molecular loci. Persoonia 2017, 39, 74–90. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hawksworth, D.L.; Hyde, K.D.; Jones, E.G.; Maharachchikumbura, S.S.N.; Manamgoda, D.S.; Thambugala, K.M.; Udayanga, D.; Camporesi, E.; Daranagama, A.; et al. Epitypification and neotypification: Guidelines with appropriate and inappropriate examples. Fungal Divers. 2014, 69, 57–91. [Google Scholar] [CrossRef]
- Cannon, P.F. A Revision of Phyllachora and Some Similar Genera on the Host Family Leguminosae; Mycol. Papers No.163; CAB International: Wallingford, UK, 1991; p. 302. [Google Scholar]
- Chen, Y.P.; Su, P.W.; Hyde, K.D.; Maharachchikumbura, S.S.N. Phylogenomics and diversification of Sordariomycetes. Mycosphere 2023, 14, 414–451. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).