Abstract
The unique biology of flies and their omnipresence in the environment of people and animals makes them ideal candidates to be important vectors of antimicrobial resistance genes. Consequently, there has been increasing research on the bacteria and antimicrobial resistance genes that are carried by flies and their role in the spread of resistance. In this review, we describe the current knowledge on the transmission of bacterial pathogens and antimicrobial resistance genes by flies, and the roles flies might play in the maintenance, transmission, and surveillance of antimicrobial resistance.
1. Introduction
Flies are insects in the order Diptera that have one pair of wings for flight and a residual second pair of wings, known as knobs, which are used for balance [1]. Over 125,000 species of Dipterans have been identified, including gnats, midges, mosquitoes, leaf miners, horse flies, houseflies, blowflies, and fruit flies. Houseflies, Musca domestica Linnaeus (Diptera: Muscidae), are of particular importance as they are notorious “pests” that can transmit a variety of bacterial pathogens [2]. They are thought to have originated in the savannahs of Central Asia and later spread worldwide [3], particularly in tropical and subtropical areas where they are mostly associated with people and domestic animals in both rural and urban areas [4].
Flies have four life stages—eggs, larvae/maggots, pupae, and adults [5]. Female houseflies lay their eggs in compost, trash, soiled bedding, or manure containing moist and microbial-rich decaying organic matter near people’s houses and farms [2]. Each female can oviposit four to six times in her lifetime, each time producing 100–150 eggs [2,6]. The eggs usually hatch within 8–12 h if the environment is moist and at an optimal temperature of 25 °C to 30 °C [2]. The first-instar larvae feed on bacteria in nutrient-rich environments and pass through a further two instars before becoming larvae/maggots that migrate into a dark, dry, and cool place where they pupate [2,6]. Adult flies emerge around 2–4 days later when ambient temperatures are 32 °C to 37 °C, meaning the entire life cycle of houseflies is very rapid, ranging from 10–21 days [7].
The behavior of houseflies promotes their ability to transmit bacterial pathogens [8,9]. They live in close proximity to people (synanthropic) or in their dwellings (endophilic cosmopolitan), and they often feed on animal and human feces (coprophagic) and decaying matter, such as garbage [7]. All life stages can thus be exposed to a variety of pathogens in unsanitary environments, and these can then be mechanically transmitted to people [10]. Adult flies can move over distances of up to 20 miles in their lifetime, which means they are ubiquitous in the environment and well capable of disseminating pathogens from unsanitary areas into people’s homes and places of work and leisure [11].
2. Flies as Vectors of Bacterial Pathogens
Flies can carry a surprising diversity and number of pathogens. One systemic review revealed more than 130 human pathogens have been identified in houseflies [3], including bacteria, fungi, viruses, and parasites [3,12] The predominant pathogens are bacteria, including Klebsiella spp. [13], Salmonella [14], Pseudomonas aeruginosa [15], Campylobacter jejuni [16], Edwardisella spp. [17], Clostridium spp. [18], Yersinia enterocolitica [19], and Burkholderia pseudomalliei [20]. Recently, Balaraman et al. reported that houseflies acquired and harbored infectious SARS-CoV-2 for up to 24 h post-exposure. They could mechanically transmit SARS-CoV-2 genomic RNA to the surrounding environment for up to 24 h post-exposure [12].
2.1. External Carriage of Bacterial Pathogens
Flies have unique body structures that enable them to effectively carry bacteria. For example, bacteria readily become attached to the sticky leg pads, hairs, electrostatically charged exoskeleton, and sponging mouthparts of flies [8,21,22]. A study quantifying the transfer of fluorescence-labeled E. coli from sugar, milk, steak, and potato salad to houseflies revealed a single housefly can carry up to 2 × 1012 E. coli and approximately 0.1 mg of food between landing sites [23]. Flies were also found to externally carry Enterococcus faecium in poultry farms [24], Klebsiella pneumoniae in kitchens and farms [25], Salmonella enterica in swine farms [26], and Staphylococcus aureus in urban areas [19]. Female flies carry more bacteria than males because they visit oviposition sites that are heavily contaminated with bacteria [27].
2.2. Internal Carriage of Bacterial Pathogens
Although most studies have focused on bacteria carried on the body surface, some researchers have investigated bacteria carried internally within the digestive tract [28,29,30,31] Ingested material containing bacteria is initially stored in the crop, from where it passes down the digestive tract through the proventriculus/foregut, midgut, hindgut, and rectum [32]. Whereas epithelial cells in the foregut and hindgut are covered with a protective cuticle [33], the midgut is lined with a unique structure, named the “peritorphic matrix”, peritrophic envelope, or peritrophic membrane (PM) [29,34] (Figure 1). This is a double-layered, noncellular structure composed of chitin, proteoglycans, and various proteins [33] that serves as a physical barrier to prevent microbes in the ingesta from invading epithelial cells and causing damage [34,35]. The PM has gaps, ranging from 2 to 10 nm, which enable digestive enzymes, acid, and secretions to enter the endoperitrophic space and digest food materials [34,36]. At the same time, as part of an innate immune response, antimicrobial peptides, reactive oxygen species, and other epithelial secretions can enter the lumen and kill and digest the trapped bacteria [34]. Not all bacteria are killed, however, with species, such as Pseudomonas aeruginosa [28], Salmonella enterica serovar Typhimurium [29], and Aeromonas caviae [30,31], being able to be ingested and proliferate in the midgut before being shed in the feces in high numbers. The survival rate of ingested bacteria is dose-dependent [37] and also dependent on competition with the commensal microbiota [38]. Studies have shown that the numbers of pathogens in the digestive tract are three times higher than on the body surface, probably due to the multiplication of the pathogens in the digestive tract [39,40,41].
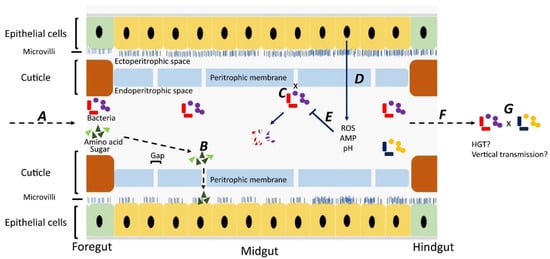
Figure 1.
Fate of bacteria in the digestive tract (midgut) of flies. A: Ingested food with bacteria is predigested with saliva in the crop. The epithelial cells in the foregut are covered by a cuticle, which prevents bacterial invasion. B: In the midgut, digestion products can pass through gaps in the peritrophic membrane and enter the ectoperitrophic space to be absorbed by epithelial cells. C: Bacteria cannot pass through the gaps on the peritrophic membrane and remain in the endoperitrophic space. D: Bacteria are trapped in the endoperitrophic space, triggering an innate immune response in epithelial cells to produce reactive oxygen species (ROS) and antimicrobial peptides (AMP). E: Trapped bacteria are killed by ROS, AMP, pH changes, and digestive enzymes. F: Some bacteria survive in the hostile environment, pass through to the hindgut, and are shed. G: There might be horizontal gene transfer between bacteria surviving in the digestive tract and bacteria may be transmitted vertically to offspring.
3. Transmission of Antimicrobial Resistance by Flies
3.1. Horizontal Transmission
It is now well known that bacterial pathogens with antimicrobial-resistant genes can be transmitted mechanically on the surfaces of flies, or in their feces, to new environments and animals/people [42]. Since 2009, however, there has been growing evidence that flies might be more than just mechanical vectors [43,44,45]. They might also act as vessels providing a suitable environment for ingested bacteria to transfer antimicrobial resistance genes to closely related bacterial species [43,44,45]. These can be shed in the feces and in this way spread antimicrobial resistance genes between different environments [46].
There is growing evidence the digestive tract of flies might serve as a suitable site for horizontal gene transfer [43,44,45]. Horizontal transfer of tetM on plasmid pCF10 among Enterococcus faecalis [44] and genes coding Shiga toxin and conferring antibiotic resistance in E. coli [45] have been demonstrated to occur in the gut of houseflies. In addition, an in vivo study showed that third-generation cephalosporin resistance genes, including blaCTX-M and blaCMY-2, were transferred successfully from E. coli to Achromobacter sp. and Pseudomonas fluroresens within the intestines of houseflies [43].
3.2. Vertical Transmission
There is also evidence that vertical transmission of antimicrobial resistance can occur in houseflies. When houseflies were fed with different concentrations of Salmonella enterica, Cronobacter sakazakii, Escherichia coli O157:H7, and Listeria monocytogenes, the organisms could be detected in the eggs of the next generation [47]. The same study demonstrated that Salmonella enterica and Cronobacter sakazakii fed to adult flies could be transmitted to the F1 [47]. A later study showed this can also occur with bacteria carrying antimicrobial resistance genes [48]. E. coli-containing plasmids with antimicrobial-resistant genes fed to houseflies were found in the subsequent immature and adult life stages [48]. In chickens, to which the immature stages were fed, the resistant E. coli persisted in the cecal contents for at least 16 days. There is thus growing evidence that houseflies are not only mechanical, but also biological vectors and, as such, they might facilitate the maintenance of antimicrobial-resistant genes [8,21,23,47,48]
4. Potential of Flies to Be Sentinels for Antimicrobial Resistance
Antimicrobial resistance remains one of the biggest threats to public health despite decades of efforts to lower the selection and transfer of resistance through more judicious use of antimicrobials [49]. There are more than two million illnesses and 23,000 deaths attributed to infections with antimicrobial-resistant bacteria every year in the USA [50]. In total, antibiotic resistance is estimated to add USD 20 billion annually to the direct healthcare costs in the USA, with additional costs to society resulting from lost productivity, which might be USD 35 billion a year [51].
Early detection of antimicrobial resistance in bacteria and ongoing surveillance are critical as they provide the information needed to monitor and develop therapy guidelines, infection control policies, and public health interventions [52]. Two surveillance systems have been commonly adopted: passive and active [53,54]. Although both can be used to monitor the prevalence of antimicrobial resistance in people and animals, they provide different ways to interpret surveillance and control strategies [54,55]. Passive surveillance involves the monitoring of resistance in routine samples collected from clinically ill patients [55,56]. It provides information on the current state of resistance in a naturally infected population [55,56]. In active surveillance, however, attempts are made to address a specific question by actively collecting data on resistance in defined infected and non-infected populations and locations [57]. Compared with passive surveillance, active surveillance is more labor intensive and costly and sample collection might be intrusive and involve ethical and personal issues [53,55]. Although the AMR surveillance approaches vary greatly in different countries, many unique phenotypes have been identified through the passive surveillance of pathogenic bacteria isolated from clinical specimens [53]. Furthermore, specific active surveillance programs for emerging resistant bacteria have been developed, for example for the ESBL-producing Enterobacteriaceae, methicillin-resistant Staphylococcus aureus (MRSA), and carbapenem-resistant Gram-negative bacteria [58].
Flies would appear to be useful surveillance vectors for tracking antimicrobial resistance. As mechanical vectors they can carry multiple bacterial pathogens and their resistance genes both externally and internally during all life stages [2]. Moreover, flies live in close association with people and their dwellings and can thus be easily and cheaply trapped for analysis. Methods for collecting flies include using aspirators [59], gauze traps over a bait [60,61], individual sweep/aerial nets [62,63], fly trap paper [64,65], jug traps [24,66], and the QuikStrike Abatement strip [67].
There are now many studies reporting on antimicrobial resistance genes in flies, reflecting the increasing awareness that they might play important roles in resistance transmission and maintenance [3,26,68,69]. Following an early study in 1983, in which nalidixic-resistant Campylobacter was reported in flies in Norway [70], there was little interest until 2005. Thereafter there have been numerous reports on a variety of bacteria and resistance genes in flies from Libya [19], North America [24,71,72,73,74], Morocco [75], Taiwan [26], Japan [76], the Netherlands [77], Spain [78], Zambia [79], Germany [80,81], Brazil [82,83], Bangladesh [84], Ethiopia [85], Thailand [68,86], Nigeria [69], and India [81] (Table 1). Flies were collected in multiple places, including hospitals, streets, abattoirs, poultry farms, cattle farms, pig farms, fish, and fast-food restaurants. Houseflies were the majority species during the collection and intestinal microbiota was frequently cultured in the collected flies against more than one antibiotic. Some bacteria harbor multiple antimicrobial-resistant genes, with the extended-spectrum β-lactamases and mobilized colistin resistance the most commonly observed.

Table 1.
Overview of multidrug antimicrobial resistance in flies.
5. Conclusions
Future studies, including epidemiological investigations and animal models, are warranted to explore the bacterial pathogens and resistance genes in the different fly species. Particularly, investigations need to be performed to accurately define if bacteria and resistance genes are effectively transmitted from flies to animals and people. The data from these studies are imperative to determine the definite roles flies might play in disseminating resistant bacteria and the threats flies pose to public health. As flies are ubiquitous, easy, and cheap to capture and process, the recent suggestion that they would seem to be ideal sentinels for antimicrobial resistance [74] warrants further investigation.
Over the past decades, there has been considerable research on the role flies might play in antimicrobial resistance. It is now known flies can carry a variety of bacteria and their antimicrobial resistance genes. Although commonly recognized as mechanical vectors, evidence is accumulating that flies are involved in the horizontal and vertical transmission of antimicrobial resistance genes. There is also some evidence that flies might be useful sentinels of antibiotic resistance in the environment and in animals and people. Further studies will more clearly define the roles played by flies in the transmission of antibiotic resistance between the environment, animals, and people and also on their usefulness as sentinels for resistance.
Author Contributions
Original draft preparation: J.-H.Y.; Review and editing: P.J.K. and C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the USDA Agricultural Research Service program (58-6040-9-017), and by the Alabama Agricultural Experiment Station and the USDA National Institute of Food and Agriculture, Hatch project (ALA052-1-17026).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented were obtained from all subjects involved in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yarger, A.M.; Fox, J.L. Dipteran halteres: Perspectives on function and integration for a unique sensory organ. Integr. Comp. Biol. 2016, 56, 865–876. [Google Scholar] [CrossRef]
- Nayduch, D.; Burrus, R.G. Flourishing in filth: House fly—Microbe interactions across life history. Ann. Entomol. Soc. Am. 2017, 110, 6–18. [Google Scholar] [CrossRef]
- Khamesipour, F.; Lankarani, K.B.; Honarvar, B.; Kwenti, T.E. A systematic review of human pathogens carried by the housefly (Musca domestica L.). BMC Public Health 2018, 18, 1049. [Google Scholar] [CrossRef]
- Ommi, D. Molecular detection and antimicrobial resistance of Aeromonas from houseflies (Musca domestica) in Iran. Rev. MVZ Córdoba 2015, 20, 4929–4936. [Google Scholar] [CrossRef][Green Version]
- de Jonge, N.; Michaelsen, T.Y.; Ejbye-Ernst, R.; Jensen, A.; Nielsen, M.E.; Bahrndorff, S.; Nielsen, J.L. Housefly (Musca domestica L.) associated microbiota across different life stages. Sci Rep. 2020, 10, 7842. [Google Scholar] [CrossRef]
- Cortinhas, L.B.; Martins Mendonça, P.; Braga, M.V.; Queiroz, M.M.C. Ultrastructure of the immature stages of Musca domestica (Diptera: Muscidae: Muscinae). J. Med. Entomol. 2020, 57, 1712–1721. [Google Scholar] [CrossRef]
- Stafford, K. Fly Management Handbook: A Guide to Biology, Dispersal, and Management of the House Fly and Related Flies for Farmers, Municipalities, and Public Health Officials; The Connecticut Agricultural Experiment Station: New Haven, CT, USA, 2008. [Google Scholar]
- Onwugamba, F.C.; Fitzgerald, J.R.; Rochon, K.; Guardabassi, L.; Alabi, A.; Kühne, S.; Grobusch, M.P.; Schaumburg, F. The role of filth ‘flies’ in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 2018, 22, 8–17. [Google Scholar] [CrossRef]
- Förster, M.; Klimpel, S.; Mehlhorn, H.; Sievert, K.; Messler, S.; Pfeffer, K. Pilot study on synanthropic flies (e.g. Musca, Sarcophaga, Calliphora, Fannia, Lucilia, Stomoxys) as vectors of pathogenic microorganisms. Parasitol. Res. 2007, 101, 243–246. [Google Scholar] [CrossRef]
- Chaiwong, T.; Srivoramas, T.; Sukontason, K.; Sanford, M.R.; Moophayak, K.; Sukontason, K.L. Survey of the synanthropic flies associated with human habitations in ubon ratchathani province of northeast Thailand. J. Parasitol. Res. 2012, 2012, 613132. [Google Scholar] [CrossRef]
- Murvosh, C.M.; Thaggard, C.W. Ecological studies of the house fly. Ann. Entomol. Soc. Am. 1966, 59, 533–547. [Google Scholar] [CrossRef]
- Balaraman, V.; Drolet, B.S.; Mitzel, D.N.; Wilson, W.C.; Owens, J.; Gaudreault, N.N.; Meekins, D.A.; Bold, D.; Trujillo, J.D.; Noronha, L.E.; et al. Mechanical transmission of SARS-CoV-2 by house flies. Parasites Vectors 2021, 14, 214. [Google Scholar] [CrossRef]
- Fotedar, R.; Banerjee, U.; Samantray, S.J.C. Vector potential of hospital houseflies with special reference to Klebsiella species. Epidemiol. Infect. 1992, 109, 143–147. [Google Scholar]
- Mawak, J.D.; Olukose, O.J. Vector potential of houseflies (Musca domestica) for pathogenic organisms in Jos, Nigeria. J. Pest Dis. Vector Manag. 2006, 7, 418–423. [Google Scholar]
- Hemmatinezhad, B.; Ommi, D.; Hafshejani, T.T.; Khamesipour, F. Molecular detection and antimicrobial resistance of Pseudomonas aeruginosa from houseflies (Musca domestica) in Iran. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 18. [Google Scholar] [CrossRef][Green Version]
- Gill, C.; Bahrndorff, S.; Lowenberger, C. Campylobacter jejuni in Musca domestica: An examination of survival and transmission potential in light of the innate immune responses of the house flies. Insect Sci. 2017, 24, 584–598. [Google Scholar] [CrossRef]
- Satish, S.; Saksham, C.; Ther, S.V.; Rakesh, S.; Ravi, S.; Central Poultry Diagnostic Laboratory (Phoenix Group). Isolation and identification of enterobacterial species from Musca domestica in broiler farms of Madhya Pradesh. Vet. Pract. 2013, 14, 239–241. [Google Scholar]
- Bahrndorff, S.; de Jonge, N.; Skovgård, H.; Nielsen, J.L. Bacterial communities associated with houseflies (Musca domestica L.) sampled within and between farms. PLoS ONE 2017, 12, e0169753. [Google Scholar] [CrossRef]
- Rahuma, N.; Ghenghesh, K.S.; Ben Aissa, R.; Elamaari, A. Carriage by the housefly (Musca domestica) of multiple-antibiotic-resistant bacteria that are potentially pathogenic to humans, in hospital and other urban environments in Misurata, Libya. Ann. Trop. Med. Parasitol. 2005, 99, 795–802. [Google Scholar] [CrossRef]
- Sulaiman, S.; Othman, M.Z.; Aziz, A.H. Isolations of enteric pathogens from synanthropic flies trapped in downtown Kuala Lumpur. J. Vector Ecol. 2000, 25, 90–93. [Google Scholar]
- Graczyk, T.K.; Knight, R.; Gilman, R.H.; Cranfield, M.R. The role of non-biting flies in the epidemiology of human infectious diseases. Microbes Infect. 2001, 3, 231–235. [Google Scholar] [CrossRef]
- Wiktorczyk-Kapischke, N.; Skowron, K.; Kwiecińska-Piróg, J.; Białucha, A.; Wałecka-Zacharska, E.; Grudlewska-Buda, K.; Kraszewska, Z.; Gospodarek-Komkowska, E. Flies as a potential vector of selected alert pathogens in a hospital environment. Int. J. Environ. Health Res. 2021, 1–20. [Google Scholar] [CrossRef]
- De Jesús, A.J.; Olsen, A.R.; Bryce, J.R.; Whiting, R.C. Quantitative contamination and transfer of Escherichia coli from foods by houseflies, Musca domestica L. (Diptera: Muscidae). Int. J. Food Microbiol. 2004, 93, 259–262. [Google Scholar] [CrossRef]
- Graham, J.P.; Price, L.B.; Evans, S.L.; Graczyk, T.K.; Silbergeld, E.K. Antibiotic resistant enterococci and staphylococci isolated from flies collected near confined poultry feeding operations. Sci. Total Environ. 2009, 407, 2701–2710. [Google Scholar] [CrossRef]
- Ranjbar, R.; Izadi, M.; Hafshejani, T.T.; Khamesipour, F. Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. J. Infect. Public Health 2016, 9, 499–505. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Chang, Y.-C.; Chuang, H.-L.; Chiu, C.-C.; Yeh, K.-S.; Chang, C.-C.; Hsuan, S.-L.; Lin, W.-H.; Chen, T.-H. Transmission of Salmonella between swine farms by the housefly (Musca domestica). J. Food Prot. 2011, 74, 1012–1016. [Google Scholar] [CrossRef]
- Neupane, S.; White, K.; Thomson, J.L.; Zurek, L.; Nayduch, D. Environmental and sex effects on bacterial carriage by adult house flies (Musca domestica L.). Insects 2020, 11, 401. [Google Scholar] [CrossRef]
- Joyner, C.; Mills, M.K.; Nayduch, D. Pseudomonas aeruginosa in Musca domestica L.: Temporospatial examination of bacteria population dynamics and house fly antimicrobial responses. PLoS ONE 2013, 8, e79224. [Google Scholar] [CrossRef]
- Chifanzwa, R.; Nayduch, D. Dose-dependent effects on replication and persistence of Salmonella enterica serovar Typhimurium in house flies (Diptera: Muscidae). J. Med. Entomol. 2018, 55, 225–229. [Google Scholar] [CrossRef]
- Nayduch, D.; Honko, A.; Noblet, G.P.; Stutzenberger, F. Detection of Aeromonas caviae in the common housefly Musca domestica by culture and polymerase chain reaction. Epidemiol. Infect. 2001, 127, 561–566. [Google Scholar] [CrossRef]
- Nayduch, D.; Noblet, G.P.; Stutzenberger, F.J. Vector potential of houseflies for the bacterium Aeromonas caviae. Med. Vet. Entomol. 2002, 16, 193–198. [Google Scholar] [CrossRef]
- Doud, C.W.; Zurek, L. Enterococcus faecalis OG1RF:pMV158 survives and proliferates in the house fly digestive tract. J. Med. Entomol. 2012, 49, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Kelkenberg, M.; Odman-Naresh, J.; Muthukrishnan, S.; Merzendorfer, H. Chitin is a necessary component to maintain the barrier function of the peritrophic matrix in the insect midgut. Insect Biochem. Mol. Biol. 2015, 56, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Miguel-Aliaga, I. The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 2013, 47, 377–404. [Google Scholar] [CrossRef] [PubMed]
- Lehane, M.J.; Msangi, A.R. Lectin and peritrophic membrane development in the gut of Glossina m.morsitans and a discussion of their role in protecting the fly against trypanosome infection. Med. Vet. Entomol. 1991, 5, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Lehane, M.J. Peritrophic matrix structure and function. Annu. Rev. Entomol. 1997, 42, 525–550. [Google Scholar] [CrossRef]
- Kumar, N.H.; Nayduch, D. Dose-dependent fate of GFP-expressing Escherichia coli in the alimentary canal of adult house flies. Med. Vet. Entomol. 2016, 30, 218–228. [Google Scholar] [CrossRef]
- Greenberg, B.; Kowalski, J.A.; Klowden, M.J. Factors affecting the transmission of salmonella by flies: Natural resistance to colonization and bacterial interference. Infect. Immun. 1970, 2, 800–809. [Google Scholar] [CrossRef]
- Rochon, K.; Lysyk, T.J.; Selinger, L.B. Retention of Escherichia coli by house fly and stable fly (Diptera: Muscidae) during pupal metamorphosis and eclosion. J. Med. Entomol. 2005, 42, 397–403. [Google Scholar] [CrossRef][Green Version]
- Mramba, F.; Broce, A.B.; Zurek, L. Vector competence of stable flies, Stomoxys calcitrans L. (Diptera: Muscidae), for Enterobacter sakazakii. J. Vector Ecol. 2007, 32, 134–139. [Google Scholar] [CrossRef]
- Pava-Ripoll, M.; Pearson, R.E.; Miller, A.K.; Ziobro, G.C. Prevalence and relative risk of Cronobacter spp., Salmonella spp., and Listeria monocytogenes associated with the body surfaces and guts of individual filth flies. Appl. Environ. Microbiol. 2012, 78, 7891–7902. [Google Scholar] [CrossRef]
- Gwenzi, W.; Chaukura, N.; Muisa-Zikali, N.; Teta, C.; Musvuugwa, T.; Rzymski, P.; Abia, A.L.K. Insects, rodents, and pets as reservoirs, vectors, and sentinels of antimicrobial resistance. Antibiotics 2021, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Usui, M.; Okubo, T.; Tamura, Y. Horizontal transfer of plasmid-mediated cephalosporin resistance genes in the intestine of houseflies (Musca domestica). Microb. Drug Resist. 2016, 22, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Hirt, H.; Zurek, L. Horizontal transfer of the tetracycline resistance gene tetM mediated by pCF10 among Enterococcus faecalis in the house fly (Musca domestica L.) alimentary canal. Microb. Ecol. 2009, 58, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Petridis, M.; Bagdasarian, M.; Waldor, M.K.; Walker, E. Horizontal transfer of Shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J. Med. Entomol. 2006, 43, 288–295. [Google Scholar] [CrossRef]
- Zurek, L.; Ghosh, A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microbiol. 2014, 80, 3562–3567. [Google Scholar] [CrossRef]
- Pava-Ripoll, M.; Pearson, R.E.; Miller, A.K.; Tall, B.D.; Keys, C.E.; Ziobro, G.C. Ingested Salmonella enterica, Cronobacter sakazakii, Escherichia coli O157:H7, and Listeria monocytogenes: Transmission dynamics from adult house flies to their eggs and first filial (F1) generation adults. BMC Microbiol. 2015, 15, 150. [Google Scholar] [CrossRef]
- Fukuda, A.; Usui, M.; Okamura, M.; Dong-Liang, H.; Tamura, Y. Role of flies in the maintenance of antimicrobial resistance in farm environments. Microb. Drug Resist. 2019, 25, 127–132. [Google Scholar] [CrossRef]
- Hoelzer, K.; Wong, N.; Thomas, J.; Talkington, K.; Jungman, E.; Coukell, A. Antimicrobial drug use in food-producing animals and associated human health risks: What, and how strong, is the evidence? BMC Vet. Res. 2017, 13, 211. [Google Scholar] [CrossRef]
- Dallal, M.M.S.; Motalebi, S.; Asl, H.M.; Yazdi, M.K.S.; Rahimi Forushani, A. Antimicrobial investigation on the multi-state outbreak of salmonellosis and shigellosis in Iran. Med. J. Islam. Repub. Iran 2020, 34, 49. [Google Scholar] [CrossRef]
- Spellberg, B.; Blaser, M.; Guidos, R.J.; Boucher, H.W.; Bradley, J.S.; Eisenstein, B.I.; Gerding, D.; Lynfield, R.; Reller, L.B.; Rex, J.; et al. Combating antimicrobial resistance: Policy recommendations to save lives. Clin. Infect. Dis. 2011, 52 (Suppl. 5), S397–S428. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Mather, A.E.; Reeve, R.; Mellor, D.J.; Matthews, L.; Reid-Smith, R.J.; Dutil, L.; Haydon, D.T.; Reid, S.W. Detection of rare antimicrobial resistance profiles by active and passive surveillance approaches. PLoS ONE 2016, 11, e0158515. [Google Scholar] [CrossRef] [PubMed]
- Cheah, A.L.Y.; Cheng, A.C.; Spelman, D.; Nation, R.L.; Kong, D.C.M.; McBryde, E.S. Mathematical modelling of vancomycin-resistant enterococci transmission during passive surveillance and active surveillance with contact isolation highlights the need to identify and address the source of acquisition. BMC Infect. Dis. 2018, 18, 511. [Google Scholar] [CrossRef] [PubMed]
- Kadykalo, S.V.; Anderson, M.E.C.; Alsop, J.E. Passive surveillance of antimicrobial resistance in Salmonella and Escherichia coli isolates from Ontario livestock, 2007–2015. Can. Vet. J. 2018, 59, 617–622. [Google Scholar]
- Rempel, O.R.; Laupland, K.B. Surveillance for antimicrobial resistant organisms: Potential sources and magnitude of bias. Epidemiol. Infect. 2009, 137, 1665–1673. [Google Scholar] [CrossRef]
- Agarwal, R.; Mohapatra, S.; Rath, G.P.; Kapil, A. Active surveillance of health care associated infections in neurosurgical patients. J. Clin. Diagn. Res. 2017, 11, DC01–DC04. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Blaak, H.; de Jong, M.C.; Graat, E.A.; Vandenbroucke-Grauls, C.M.; de Roda Husman, A.M. Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Barro, N.; Aly, S.; Tidiane, O.C.; Sababénédjo, T.A. Carriage of bacteria by proboscises, legs, and feces of two species of flies in street food vending sites in Ouagadougou, Burkina Faso. J. Food Prot. 2006, 69, 2007–2010. [Google Scholar] [CrossRef]
- Schaumburg, F.; Onwugamba, F.C.; Akulenko, R.; Peters, G.; Mellmann, A.; Köck, R.; Becker, K. A geospatial analysis of flies and the spread of antimicrobial resistant bacteria. Int. J. Med. Microbiol. 2016, 306, 566–571. [Google Scholar] [CrossRef]
- Gupta, A.K.; Nayduch, D.; Verma, P.; Shah, B.; Ghate, H.V.; Patole, M.S.; Shouche, Y.S. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS Microbiol. Ecol. 2012, 79, 581–593. [Google Scholar] [CrossRef]
- Sproston, E.L.; Ogden, I.D.; MacRae, M.; Forbes, K.J.; Dallas, J.F.; Sheppard, S.K.; Cody, A.; Colles, F.; Wilson, M.J.; Strachan, N.J. Multi-locus sequence types of Campylobacter carried by flies and slugs acquired from local ruminant faeces. J. Appl. Microbiol. 2010, 109, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Pava-Ripoll, M.; Pearson, R.E.; Miller, A.K.; Ziobro, G.C. Detection of foodborne bacterial pathogens from individual filth flies. J. Vis. Exp. 2015, 96, e52372. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.; Ito, Y.; Saito, K.; Tsubokura, M.; Otsuki, K. Role of the fly in the transport of Yersinia enterocolitica. Appl. Environ. Microbiol. 1979, 38, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yao, Y.; Wei, P.; Song, C.; Wan, S.; Yang, S.; Zhu, G.M.; Liu, H.M. Housefly Phormicin inhibits Staphylococcus aureus and MRSA by disrupting biofilm formation and altering gene expression in vitro and in vivo. Int. J. Biol. Macromol. 2021, 167, 1424–1434. [Google Scholar] [CrossRef] [PubMed]
- Keen, J.E.; Wittum, T.E.; Dunn, J.R.; Bono, J.L.; Durso, L.M. Shiga-toxigenic Escherichia coli O157 in agricultural fair livestock, United States. Emerg. Infect. Dis. 2006, 12, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Holt, P.S.; Geden, C.J.; Moore, R.W.; Gast, R.K. Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Appl. Environ. Microbiol. 2007, 73, 6030–6035. [Google Scholar] [CrossRef]
- Punyadi, P.; Thongngen, P.; Kiddee, A.; Assawatheptawee, K.; Tansawai, U.; Bunchu, N.; Niumsup, P.R. Prevalence of bla(CTX-M) and emergence of bla(CTX-M-5)-carrying Escherichia coli in Chrysomya megacephala (Diptera: Calliphoridae), Northern Thailand. Microb. Drug Resist. 2021, 27, 698–705. [Google Scholar] [CrossRef]
- Ngbede, E.O.; Poudel, A.; Kalalah, A.; Yang, Y.; Adekanmbi, F.; Adikwu, A.A.; Adamu, A.M.; Mamfe, L.M.; Daniel, S.T.; Useh, N.M.; et al. Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents 2020, 56, 106108. [Google Scholar] [CrossRef]
- Rosef, O.; Kapperud, G. House flies (Musca domestica) as possible vectors of Campylobacter fetus subsp. jejuni. Appl. Environ. Microbiol. 1983, 45, 381–383. [Google Scholar] [CrossRef]
- Macovei, L.; Zurek, L. Influx of enterococci and associated antibiotic resistance and virulence genes from ready-to-eat food to the human digestive tract. Appl. Environ. Microbiol. 2007, 73, 6740–6747. [Google Scholar] [CrossRef]
- Macovei, L.; Miles, B.; Zurek, L. Potential of houseflies to contaminate ready-to-eat food with antibiotic-resistant enterococci. J. Food Prot. 2008, 71, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.; Hathcock, T.; Butaye, P.; Kang, Y.; Price, S.; Macklin, K.; Walz, P.; Cattley, R.; Kalalah, A.; Adekanmbi, F.; et al. Multidrug-resistant Escherichia coli, Klebsiella pneumoniae and Staphylococcus spp. in houseflies and blowflies from farms and their environmental settings. Int. J. Environ. Res. Public Health 2019, 16, 3583. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.; Kang, Y.; Mandal, R.K.; Kalalah, A.; Butaye, P.; Hathcock, T.; Kelly, P.; Walz, P.; Macklin, K.; Cattley, R.; et al. Comparison of microbiota, antimicrobial resistance genes and mobile genetic elements in flies and the feces of sympatric animals. FEMS Microbiol. Ecol. 2020, 96, fiaa027. [Google Scholar] [CrossRef]
- Bouamamaa, L.; Sorlozano, A.; Laglaoui, A.; Lebbadi, M.; Aarab, A.; Gutierrez, J. Antibiotic resistance patterns of bacterial strains isolated from Periplaneta americana and Musca domestica in Tangier, Morocco. J. Infect. Dev. Ctries. 2010, 4, 194–201. [Google Scholar] [CrossRef][Green Version]
- Usui, M.; Iwasa, T.; Fukuda, A.; Sato, T.; Okubo, T.; Tamura, Y. The role of flies in spreading the extended-spectrum β-lactamase gene from cattle. Microb. Drug Resist. 2013, 19, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Blaak, H.; Hamidjaja, R.A.; van Hoek, A.H.; de Heer, L.; de Roda Husman, A.M.; Schets, F.M. Detection of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli on flies at poultry farms. Appl. Environ. Microbiol. 2014, 80, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Solà-Ginés, M.; González-López, J.J.; Cameron-Veas, K.; Piedra-Carrasco, N.; Cerdà-Cuéllar, M.; Migura-Garcia, L. Houseflies (Musca domestica) as Vectors for Extended-Spectrum β-Lactamase-Producing Escherichia coli on Spanish Broiler Farms. Appl. Environ. Microbiol. 2015, 81, 3604–3611. [Google Scholar] [CrossRef] [PubMed]
- Songe, M.M.; Hang’ombe, B.M.; Knight-Jones, T.J.; Grace, D. Antimicrobial resistant enteropathogenic Escherichia coli and Salmonella spp. in houseflies infesting fish in food markets in Zambia. Int. J. Environ. Res. Public Health 2016, 14, 21. [Google Scholar] [CrossRef]
- Guenther, S.; Falgenhauer, L.; Semmler, T.; Imirzalioglu, C.; Chakraborty, T.; Roesler, U.; Roschanski, N. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017, 72, 1289–1292. [Google Scholar] [CrossRef]
- Wadaskar, B.; Kolhe, R.; Waskar, V.; Pawade, M.; Kundu, K. Isolation of Escherichia coli and Salmonella species in flies trapped at animal farm premises. J. Entomol. Zool. Stud. 2019, 7, 198–201. [Google Scholar]
- Carramaschi, I.N.; Lopes, J.C.O.; Leite, J.A.; Carneiro, M.T.; Barbosa, R.R.; Boas, M.H.V.; Rangel, K.; Chagas, T.P.G.; Queiroz, M.M.; Zahner, V. Surveillance of antimicrobial resistant bacteria in flies (Diptera) in Rio de Janeiro city. Acta Trop. 2021, 220, 105962. [Google Scholar] [CrossRef] [PubMed]
- Alves, T.D.S.; Lara, G.H.B.; Maluta, R.P.; Ribeiro, M.G.; Leite, D.D.S. Carrier flies of multidrug-resistant Escherichia coli as potential dissemination agent in dairy farm environment. Sci. Total Environ. 2018, 633, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Sobur, A.; Haque, Z.F.; Sabuj, A.A.; Ievy, S.; Rahman, A.T.; El Zowalaty, M.E.; Rahman, T. Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Future Microbiol. 2019, 14, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Tufa, T.B.; Fuchs, A.; Wienemann, T.; Eggers, Y.; Abdissa, S.; Schneider, M.; Jensen, B.O.; Bode, J.G.; Pfeffer, K.; Häussinger, D.; et al. Carriage of ESBL-producing Gram-negative bacteria by flies captured in a hospital and its suburban surroundings in Ethiopia. Antimicrob. Resist. Infect. Control 2020, 9, 175. [Google Scholar] [CrossRef]
- Yang, Q.E.; Tansawai, U.; Andrey, D.O.; Wang, S.; Wang, Y.; Sands, K.; Kiddee, A.; Assawatheptawee, K.; Bunchu, N.; Hassan, B.; et al. Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): An increasing public health risk. Environ. Int. 2019, 122, 281–290. [Google Scholar] [CrossRef]
- Heiden, S.E.; Kurz, M.S.E.; Bohnert, J.; Bayingana, C.; Ndoli, J.M.; Sendegeya, A.; Gahutu, J.B.; Eger, E.; Mockenhaupt, F.P.; Schaufler, K. Flies from a tertiary hospital in Rwanda carry multidrug-resistant Gram-negative pathogens including extended-spectrum beta-lactamase-producing E. coli sequence type 131. Antimicrob. Resist. Infect. Control 2020, 9, 34. [Google Scholar] [CrossRef]
- Wetzker, W.; Pfeifer, Y.; Wolke, S.; Haselbeck, A.; Leistner, R.; Kola, A.; Gastmeier, P.; Salm, F. Extended-spectrum Beta-Lactamase (ESBL)-producing Escherichia coli isolated from flies in the urban center of Berlin, Germany. Int. J. Environ. Res. Public Health 2019, 16, 1530. [Google Scholar] [CrossRef]
- Odetoyin, B.; Adeola, B.; Olaniran, O. Frequency and antimicrobial resistance patterns of bacterial species isolated from the body surface of the housefly (Musca domestica) in Akure, Ondo state, Nigeria. J. Arthropod Borne Dis. 2020, 14, 88–96. [Google Scholar] [CrossRef]
- Pileggi, M.T.; Chase, J.R.; Shu, R.; Teng, L.; Jeong, K.C.; Kaufman, P.E.; Wong, A.C.N. Prevalence of field-collected house flies and stable flies with bacteria displaying cefotaxime and multidrug resistance. J. Med. Entomol. 2021, 58, 921–928. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).