Simple Summary
Sea turtles are currently endangered due to several factors, including infectious and parasitic pathogens. Opportunistic and pathogenic bacteria are largely present in the marine environment and may infect and cause diseases in sea turtles and other animals. Even though infected sea turtles are not a relevant source of infections for humans if compared to other animals, they may be involved in the epidemiology of zoonotic bacterial agents, including those that are antimicrobial resistant; therefore, the direct and indirect contact of humans with sea turtles, their products, and the environment where they live may represent a One Health threat.
Abstract
Sea turtles are important for the maintenance of marine and beach ecosystems, but they are seriously endangered due to factors mainly related to human activities and climate change such as pollution, temperature increase, and predation. Infectious and parasitic diseases may contribute to reducing the number of sea turtles. Bacteria are widespread in marine environments and, depending on the species, may act as primary or opportunistic pathogens. Most of them are able to infect other animal species, including humans, in which they can cause mild or severe diseases. Therefore, direct or indirect contact of humans with sea turtles, their products, and environment where they live represent a One Health threat. Chlamydiae, Mycobacteria, and Salmonellae are known zoonotic agents able to cause mild or severe diseases in sea turtles, other animals, and humans. However, other bacteria that are potentially zoonotic, including those that are antimicrobially resistant, are involved in different pathologies of marine turtles.
1. Introduction
Sea turtles are currently classified into seven living species, belonging to six genera and grouped into the family Cheloniidae and Dermochelyidae. The family Cheloniidae comprises the green turtle Chelonia mydas (Linnaeus, 1758), loggerhead Caretta caretta (Linnaeus, 1758), hawksbill Eretmochelys imbricata (Linnaeus, 1766), Kemp’s ridley Lepidochelys kempii (Garman, 1880), olive ridley Lepidochelys olivacea (Eschscholtz, 1829), and flatback Natator depressus (Garman, 1880). The family Dermochelyidae comprises only the leatherback Dermochelys coriacea (Vandelli, 1761) [1].
Sea turtles are considered as part of two distinct ecosystems: the marine system and the beach and lower dune system. Their presence in both environments is fundamental: nesting sea turtles help beaches by depositing their eggs in the sand; and eggshells and unhatched eggs left behind provide important nutrients that nourish dune vegetation such as beach grasses, which stabilize dunes and help to prevent coastal erosion. Hatchlings are an important source of food for birds, fish, and mammals. Green turtles grazing on seagrass is an important way to keep seagrass beds healthy; leatherbacks help manage the number of jellyfish in the ocean, and hawksbills help reefs by eating sponges that compete with them for space [2].
These environmental balances are currently compromised because marine turtles are seriously endangered due to factors mainly related to human activities and climate change such as pollution, temperature increase, and predation [3]. Caretta caretta, D. coriacea, and L. olivacea are classed as ‘Vulnerable’; C. mydas as ‘Endangered’; L. kempii and E. imbricata as ‘Critically Endangered’; and N. depressus is ‘data deficient’ according to the IUCN (International Union for Conservation of Nature and Natural Resources) [3].
Infectious and parasitic diseases have an important impact on the health status of sea turtles and, consequently, may contribute to the reduction in their number. Bacteria are widespread in marine environments and, depending on the species, may act as primary or opportunistic pathogens. Most of them are able to infect other animals, including humans, which can cause mild or severe diseases. Therefore, direct or indirect contact of humans with sea turtles, their products, and environment where they live can represent a One Health threat [4].
The aim of the present narrative review article was to focus on cases of infections in sea turtles due to the main bacterial agents, taking in account the pathogenic role of the involved bacteria for the turtles’ health and highlighting the role of these animals as source of infections for humans.
2. Chlamydia spp.
Members of the order Chlamydiales are obligate intracellular Gram-negative bacteria that have been under constant change in nomenclature in recent years. To date, this order includes 9 families: Chlamydiaceae, Clavichlamydiaceae, Cribchlamydiaceae, Parachlamydiaceae, Parilichlamydiaceae, Piscichlamydiaceae, Rhabdochlamydiaceae, Simkaniaceae, and Waddliaceae. The family Chlamydiaceae contains the genus Chlamydia, including 14 species (C. abortus, C. avium, C. buteonis, C. caviae, C. felis, C. gallinacea, C. muridarum, C. pecorum, C. pneumoniae, C. poikilothermis, C. psittaci, C. serpentis, C. suis, and C. trachomatis) and 4 Candidatus species. Moreover, recently the new genus Chlamydiifrater gen. nov. with two new species, Chlamydiifrater phoenicopteri sp. nov. and Chlamydiifrater volucris sp. nov. has been added. Several Chlamydia-like, also called Chlamydia-related, organisms, frequently found in different animal species, have been included in the eight other families [5,6].
Chlamydia spp. have been recognized as agents capable of infecting mammals, birds, as well as reptiles [5]. Some reports described pathological forms in chelonians due to Chlamydiae [7,8,9] as well. In particular, terrapins and tortoises usually are asymptomatic carriers of these bacteria, but, in some cases, ocular and respiratory signs and lethal systematic infection have been observed [10,11,12]. Otherwise, data about Chlamydial infections in sea turtles are very scanty.
An outbreak of severe chlamydiosis was observed and investigated in maricultured-reared green sea turtles (C. mydas) in a farm located in the British West Indies [13]. Affected turtles had general symptoms such as weakness, lethargy, and inability to dive. Several animals died, and post-mortem examinations showed lesions reportable to systemic infection. Liver, spleen, and heart were the organs with the most severe alterations, similar to the lesions usually observed in mammals and birds affected by chlamydiosis. An agent defined as Chlamydia sp. was observed in the affected organs through Macchiavello’s stain, electron microscopy, and Chlamydial antigen detection by anti-Chlamydia monoclonal antibodies, but the Chlamydial species was not identified because of the insufficient methods available in the years of the study [13]. Successively, Bodetti et al. [14] carried out further analyses on stored blocks of paraffin embedded heart from one of those green turtles: PCR and sequencing analyses on DNA extracted from the samples allowed the identification of C. abortus, C. pneumoniae, and Neochlamydia sp. Chlamydia pneumoniae resulted as 100% similar to the human genotype A. These findings highlighted that pathogens able to infect other animals may be harbored by sea turtles. In particular, C. abortus is an abortive agent frequently encountered in small ruminants, which is able to cause disease also in other mammals, including humans [15,16]; C. pneumoniae is a human pathogen, causing respiratory disease [14].
Recently, molecular techniques have allowed for the detection of Chlamydial DNA in samples collected from loggerhead sea turtles, temporarily housed for rehabilitation at the Marine Turtle Research Centre in South Italy (Portici, Italy) [17]. During this investigation, ocular–conjunctival, oropharyngeal, and nasal swabs were collected from turtles without clinical signs and submitted for molecular analyses; samples from all investigated animals resulted PCR-positive for Chlamydiaceae, but the identification of the Chlamydial species was not possible. However, the findings highlighted the spreading of Chlamydial agents in marine turtle populations. In this study, DNA of Chlamydiae was found on the mucosa of the first tract of the respiratory system and on conjunctiva similarly to the Chlamydial localization in mammals and birds [5].
Further surveys to investigate the presence of Chlamydiae in both asymptomatic and symptomatic sea turtles and the identification of Chlamydial species involved are pivotal to understand the possible role of these marine animals as source of infection for other animals and humans, as well as to identify new agents able to infect and cause diseases in sea turtles. In fact, in recent years, several strains identified as Candidatus Chlamydia sp. or Chlamydia-like agents, as observed in the complex taxonomy above summarized, have been found in other animal species; therefore, it is reasonable that also sea turtles could harbor new Chlamydial bacteria.
3. Mycobacterium spp.
Bacteria of the genus Mycobacterium are acid fast, aerobic, non-motile, and no-spore-forming bacilli. The genus includes numerous species differentiated for pathogenic properties, environmental distribution, and host affinity. Mycobacterium spp. are classified into two groups: the Mycobacterium tuberculosis complex (MTC) and the non-tuberculous mycobacteria (NTM) [18].
NTM are largely distributed in different environments; in fact, they can be isolated from water, soil, dust, and plants [18]. NTM, including known and novel species, are also present in the marine environment, as demonstrated by the infections often diagnosed in fishes and bivalves [19,20,21,22].
Reptiles can be infected by different NTM in relation to the habitat where they live. Some reports of mycobacterial infections in tortoises and terrapins have been described [23,24,25,26,27]. Sea turtles have been found to be susceptible to some NTM species as well.
Mycobacterium chelonae is the most frequently encountered species in these animals. It was first isolated by Friedman in 1903 from the lung of a turtle identified at that time as Chelonia corticata (now Caretta caretta) [28]. Mycobacterium chelonae grows optimally in marine and other aquatic environments at temperatures ranging from 22 °C to 40 °C; therefore, its finding is not surprising in sea turtles. The ability of this mycobacterium, which usually acts as opportunistic pathogen, to cause disease seems to be related to an immunodeficiency status of the turtles, as in the cases of warm-blooded animals [29]. Mycobacterium chelonae often penetrates through skin lesions and, on the basis of the animal immunocompetency, can induce generalized infections.
A case of mycobacteriosis by M. chelonae was well described by Greer et al. [29] in L. kempii. The animal, recovered in a rescue center in Baltimore (USA), showed epidermal appendicular and plastral lesions and a swollen left elbow joint. Mycobacterium chelonae was isolated from the joint and a skin nodule; in addition, after the necropsy carried out on the animals who was euthanized due to poor prognosis for recovery, the pathogen was also cultured from lungs, liver, spleen, pericardium, and kidneys.
More recently, a case of M. chelonae infection in a C. caretta has been described. The animal was found stranded alive along the Italian Adriatic coast, displaying obtundation, tachypnea, and increased respiratory effort, but it died a few hours after. The postmortem examination revealed miliary nodules in the liver, heart, stomach, gut wall, and lungs. Histological observations detected, in all nodular lesions, a small central area of necrosis with acid-fast bacilli surrounded by epithelioid cells, macrophages, and lymphocytes. Bacteriological examinations on tissue samples isolated microorganisms that, submitted to molecular analyses, showed a restriction pattern identical to M. chelonae [30].
Mycobacterium marinum was identified in a C. caretta in 1977. In particular, a captive-born hatchling loggerhead sea turtle with stunted growth, emaciation, and weakness died in at the National Marine Fisheries Service in Galveston, Texas (USA); on postmortem examination, the turtle presented with a granulomatous pneumonia, from which M. marinum was isolated [31].
Mycobacterium haemophilum has been detected by PCR in the spinal cord of a juvenile leatherback turtle (D. coriacea) that was found stranded on the Atlantic coast of Florida (USA). A disseminated granulomatous inflammation involving the lungs, liver, spleen, kidneys, small intestine, pancreas, thymus, bone marrow, and nervous system was observed in the animal [32]. Even though available data about this bacterium included its presence in fresh or chlorinated water and water supplies, M. haemophylum is present in the marine environment as also demonstrated by an infection case in a man who developed chronic dermal granulomata in his right arm after receiving a coral injury in Thailand [33]. Mycobacterium haemophilum is known as a fish pathogen, mainly zebrafish [34]. Pathologies in sea turtles due to this mycobacterial species could be underestimated because of the difficulties to cultivate and type the microorganisms that have growth requirements different from other NTM: although the normal growth temperature for mycobacteria is 35–37 °C, M. haemophilum prefers a lower growth temperature of 30–32 °C and requires iron supplements such as hemin or ferric ammonium citrate; its growth is visible after about 8 weeks [35].
Surveys carried out in the past allowed researchers to detect bacteria identified as Mycobcaterium spp. in sea turtles, but the difficulty to culture and type these agents did not always allow for a correct identification of the agents. Keymer in 1978 [36] described a case of mycobacteriosis in a C. mydas, but the method of bacterial identification and anatomical location of infection were not identified. Glazebrook and Campbell in 1990 [37] identified two farmed C. mydas with mycobacteriosis; in both cases, the infections were diagnosed on the basis of the observation of acid-fast bacilli in lung tissues. Similarly, Brock et al. in 1976 [38] observed mycobacteria in the lungs collected from three reared hatchling Pacific green sea turtles, and, on the basis of bacteriological analysis, M. avium was presumed to be the etiologic agent.
4. Enterobacteriaceae
The family Enterobacteriaceae includes Gram-negative, facultatively anaerobic, and non-spore-forming rods grouped into numerous genera and species. These bacteria are ubiquitous in nature; many species exist as free living in diverse ecological niches, both terrestrial and aquatic environments, and some are associated with animals, plants, or insects only [39]. Members of this family are localized in the intestinal tract of mammals, birds, and reptiles that excrete the bacteria with their feces. Some species, such as Salmonella enterica, are well-known pathogens able to cause diseases in humans and other animals characterized by clinical forms of different severity. Other Enterobacteriaceae act as opportunistic pathogens, causing infections in different anatomical region [39].
Salmonella enterica and Salmonella bongori, with their numerous serotypes, are the most important pathogens of this family and frequently associated with infections in ophidians, saurians, and chelonians who may develop disease or serve as asymptomatic carriers [40,41,42,43].
Salmonellosis has been investigated in sea turtles that often resulted as involved in the epidemiology of these pathogens. Infected marine turtles may represent a serious threat for the health of other animals, mainly humans. In fact, they may contaminate the environment of rescue centers or natural parks where they are hosted as well as beach sands where free-ranging turtles arrive to deposit their eggs.
Conversely, Salmonellae are not always detected in samples collected from investigated sea turtles [44,45,46,47]. This could mean absence of infection, but it could also be related to latent infection; in fact, reptiles excrete Salmonella spp. in their feces intermittently, and the bacterial load they shed increases during periods of stress [48].
A relevant issue is related to the human consumption of sea turtles’ meat and eggs. In fact, despite some national regulations restricting the capture of sea turtles, these species remain an important resource for communities worldwide [49]. Reports of ongoing sea turtle consumption (legal and illegal) have emerged from many parts of the world [50], and human cases of salmonellosis due to the consumption of meat have been reported [51,52]. In particular, serotypes Chester [51] and Muenchen [52] caused salmonellosis in persons who ate green turtles’ meat in Australia. Salmonellae have been also isolated from leatherback turtle eggs in Grenada (West Indies), where the local population usually consumes these products [53], suggesting a real risk of infection related to this feeding habit.
Salmonella infection can be a threat for marine turtle health too, even though few cases have been reported in the literature. Work and collaborators [54] associated S. enterica serotype Typhimurium with frequent cases of granulomatous nephritis in both pelagic (7%, 9/127 analyzed animals) and stranded (47%, 21/44 analyzed animals) L. olivacea turtles. In severely affected turtles, the kidneys were bilaterally enlarged by chronic granulomas characterized as central areas of caseous material surrounded by fibrous connective tissue, sometimes with associated fluid-filled cysts; milder cases consisted of single or few isolated lesions, where much of the kidneys were unaffected. It was not clear if the renal localization was due to septicemia or an ascendent passage from bowel to kidneys, as ureters connect directly to the intestinal lumen in sea turtles. Granulomatous nephritis is a common pathology in marine turtles [55], but cases associated with Salmonella spp. have been documented only in L. olivacea species [54].
Isolated strains were submitted to molecular analyses and resulted in belonging to a S. Typhimurium variant likely adapted to olive ridley turtles [54]; however, the risk of transmission of this strain to other animals, including humans, cannot be excluded.
No chelonian-specific Salmonella serotype is known; therefore, salmonella infection in these animals, including sea turtles, seem related to non-specific serotypes. However, S. enterica Choleraesuis, a serotype known as swine specific, was cultured from the mouth of one river turtle Podocnemis expansa and one Podocnemis unifilis, as well as from eggs of P. unifilis, on the beaches in the National Park of Araguaia, Brazil [56]. These findings, even though they regard no-marine turtles, suggest paying attention to the role of all chelonians in the spreading of all Salmonellae.
All reptilian species are known as reservoirs of Salmonellae; they often harbor these pathogens, even though they are clinically healthy. This is probably true for sea turtles; in fact, some surveys detected Salmonellae in asymptomatic marine turtles [57].
Salmonella enterica was cultured from the cloaca of 3/21 (14.2%) leatherback sea turtles that used the island of St. Kitts, West Indies as a nesting ground during 2011 [58]. Successively, a survey carried out on cloacal swabs collected from sea turtles in the same island found Salmonellae through cultivation and molecular methods; 1/14 E. imbricata turtles resulted as PCR-positive, 3/9 leatherback turtles were PCR-positive, and, from two of them, S. enterica Newport was also cultured; on the other hand, the serotype Montevideo was isolated from a dry sand sample [59]. Both serotypes have been also reported in terrestrial and aquatic wild animals in the Caribbean islands; therefore, an epidemiological cycle between turtles and other animals could be assumed [60,61,62]. In addition, more recently, Edwards and collaborators [63], testing sea turtles on the same island, isolated S. typhimurium, S. montevideo, S. newport, and the serotypes S. I:6,7:-:- and S. I:4,5,12:-:- from cloacal swabs collected from 15/57 (26.3%) nesting leatherback turtles. The finding during the same survey of Salmonella-negative E. imbricata and C. mydas is interesting; in these cases, the cloacal samples were collected from foraging turtles. The obtained results suggested that egg laying could represent a stressful event that negatively interferes with the immune system with the consequent bacteria shedding. However, the results could be also related to the environment where turtles lived. Nesting leatherback turtles had contact with the sand while foraging hard-shell turtles did not, especially male and juvenile turtles, as only mature females return to land once hatched. Therefore, the bacterial contamination of the sand could result in the transmission of S. enterica to nesting turtles who, subsequently, come ashore to nest [63].
Salmonella enterica was isolated from the oviductal fluid from 3/20 nesting C. mydas in the Ras Al-Hadd Reserve (Oman) located between the Gulf of Oman and the Arabian Sea [64].
Recently, sea turtle fecal samples were collected from 20 loggerhead and 3 green sea turtles found stranded but still alive or caught by fisherman along the Gulf of Taranto (Ionian Sea coast, Italy); one loggerhead and two green sea turtles resulted as PCR-positive for S. enterica [65].
Other members of the family Enterobacteriaceae have been isolated from sea turtles on the occasion of sporadic investigation on asymptomatic animals, as well as from turtles with different pathologies [66]. Among these bacteria, Escherichia coli was the most frequently encountered agent. It is a bacterium usually present in the intestinal tract of all animals and, in some circumstances, may induce infections with enteric diseases when enteropathogens/enteroinvasive/enterotoxic strains are involved or infections of other anatomical sites, mainly in the genito-urinary tract of mammals [67].
A recent study investigated the presence of E. coli O157:H7, a zoonotic enterohaemorrhagic serotype, in C. caretta and C. mydas living in the Gulf of Taranto, but no strains were isolated [65]. However, other E. coli strains have been associated with some lesions observed in sea turtles, such as necrotizing and granulomatous splenitis and hepatitis, fibrinous perihepatitis, fibrinous and necrotizing enteritis, fibrinous and ulcerative esophagitis, and perinephric abscesses [55].
Furthermore, E. coli, as well as Enterobacter cloacae, was frequently found in unhatched eggs and in nesting sand samples [68].
Citrobacter spp. are other Enterobacteriaceae members frequently isolated from sea turtles with and without lesions. Citrobacter freundii, together with Enterobacter hormaechei, was related to septic arthritis, which progressed to generalized coelomitis and death in C. mydas [69]; Citrobacter spp. were cultured from cases of cystitis, esophagitis, ulcerative stomatitis, and the granulomatous inflammation of the lungs, spleen, liver, and kidneys [55,70].
Morganella morganii, Serratia marcescens, and Proteus spp. were isolated from different tissues of dead D. coriacea turtles, which, during post-mortem examination, showed ulcerative/purulent dermatitis, granulomatous hepatitis, splenitis and pneumonia, diverticulitis, and brain hemorrhage [71].
A fatal case of necrotizing tracheitis and bronchopneumonia caused by Serratia proteamaculans has been recently reported in a C. mydas in Australia [72]. This Enterobacteriaceae is an opportunistic agent frequently isolated from soil, plants, water, and insects [73].
Moreover, several Enterobacteriaceae, such as E. coli and species of the genera Klebsiella, Citrobacter, Enterobacter, Morganella, Proteus, Providencia, and Serratia, have been cultured during surveys about bacterial microbiota in the gastrointestinal tract and also on the nasal and oral mucosal surfaces, skin, and shell [74,75,76,77,78,79].
The findings of Enterobacteriaceae are of increasing interest not only for the health status of marine turtles but mainly for the threat related to the antibiotic-resistance frequently encountered in these isolates [57,75,76,80,81,82]. In recent years, sea turtles have been used as a sentinel species for monitoring antibiotic resistance in marine environments [82]. In fact, they are considered good sentinels of this issue because of their long lifespans and tendencies to bioaccumulate pathogens and contaminants, including antimicrobials [83,84,85].
5. Vibrio spp.
Members of the genus Vibrio, of the family Vibrionaceae, are Gram-negative and facultatively anaerobic bacteria; they are motile by polar flagella and naturally present in marine ecosystems and estuarine waters [86]. In areas with temperate climate, the abundance of Vibrio is strictly related to the seasons: during the Summer, Vibrios can easily be isolated from water, suspended particulate matter, plankton, algae, sediment, fish, shellfish, and benthic marine environments; during the Winter, the number of Vibrios noticeably decreases, and they can be found overwintering in sediments [86]. The genus includes several species, most of which are pathogenic for humans. Different types of relationships between Vibrio spp. and host are known: a mutualistic association between Vibrio fischeri and fish has been identified; a symbiotic association of V. fischeri with sepiolid squids, chitinous shellfish, has been observed; and parasitic relationships of some Vibrios typically affecting fish, frogs, and eels are known [86].
Sea turtles have been frequently identified as carriers of Vibrio spp., becoming potential sources of infections for humans. Vibrio mimicus is a human pathogen responsible for cholera-like diarrhea, due to the fact that some strains produce the choleric toxin [87]. It has been isolated from the sand of hatching nests, as well as the cloaca and eggs of L. olivacea in Costa Rica [88]. The detection of this bacterial species in eggs was of particular interest because the population in Costa Rica largely consumed eggs of olive ridley turtles. However, other Vibrio species were found during these surveys, as well as in other investigations regarding the other sea turtle species. Vibrio parahaemolyticus, V. vulnificus, V. fluvialis, V. metschnikovii, and V. cholerae were frequently isolated [46,57,88,89,90].
Furthermore, Vibrio spp. have been isolated from lesions of sea turtles of different species. Vibrio alginolyticus has been frequently encountered, although it could have been overestimated if it was not identified through multiple biochemical tests and/or molecular analysis [91]. Oros and collaborators [55] cultured V. alginolyticus from cases of ulcerative dermatitis, broncopneumonia, ulcerative and fibrinous esophagitis, myositis, necrotizing splenitis, necrotizing enteritis, granulomatous nephritis, and granulomatous and necrotizing hepatitis. V. alginolyticus, largely present in marine water, usually acts as an opportunistic pathogen after its entry through traumatic injuries of the skin, carapace, oral cavity, and esophagus; lesions observed in different tissues are related to septicemia or anatomical location [55]. Integumental ulcerative disease, characterized by many lesions, mainly hemorrhagic ulcers and necrosis, in the skin of the neck, tail, axillary and inguinal regions, and also in the shell plates, was described in a C. caretta at the Bermuda Aquarium (Bermuda), and it was associated with V. alginolyticus, even though other bacteria were isolated from the lesions [92]. This species was also related to ulcerative stomatitis, obstructive rhinitis, and pneumonia in hatchling and juvenile C. caretta and C. mydas [93].
Photobacterium damselae (formerly Vibrio damsela) was considered as the agent responsible for a fatal infection with valvular endocarditis and septicemia in a D. coriacea found dead along the coast of Tasmania [94].
Table 1 reports the documented cases of human infections directly associated with the consumption of meat or eggs of sea turtles.

Table 1.
Documented cases of human infections associated with sea turtles.
6. Aeromonas spp.
Species of the genus Aeromonas are Gram-negative, facultative anaerobic, and non-spore-forming bacteria. The genus, previously included in the family Vibrionaceae, has been classified as belonging to the Aeromonadaceae family [95]. Aeromonas bacteria are largely present in freshwater, estuarine, and marine environments. Their range of growth temperature is between 0 °C and 42 °C; therefore, these bacteria are distributed worldwide, although a mild climate usually favors their presence. Several Aeromonas species, such as A. hydrophila, A. veronii, and A. caviae, are responsible for human diseases, mainly gastrointestinal forms [96]; however, members of this genus have been traditionally also related to infections in reptiles, including chelonians [97], in which they usually act as opportunistic pathogens [55,98,99].
The investigations carried out in sea turtles usually detected Aeromonas without arriving at the species’ identification; however, when the isolates had been submitted to typing, A. hydrophila was the most frequently encountered species of this genus.
Aeromonas spp. have been associated with cases of several infections forms in different sea turtle species such as dermatitis, stomatitis, esophagitis, rhinitis, pneumonia, osteomyelitis, splenitis, hepatitis, and myositis [55,57,100,101,102]. These forms may be the beginning of septicemic forms, as demonstrated by a recent investigation on stranded C. caretta in South Italy that found these bacteria as the prevalent agents cultured from several organs (lung, heart, intestine, liver, spleen, kidney) and cloacal and oral swabs [81].
A relevant finding was the detection of A. hydrophila in severe purulent adenitis of salt glands that represented a cause of mortality in C. caretta. Oros and collaborators [103] carried out bacteriological examinations on stranded loggerhead turtles with evident salt gland adenitis and cultured some bacterial species; A. hydrophila was the most frequently isolated agent, both in pure and mixed cultures.
Bacteria of the genus Aeromonas have been also isolated from unhatched eggs of sea turtles; this finding could be related to passage of the bacteria from female turtles to the eggs, but aeromonas, as well as other bacteria, could also infect eggs by penetrating shell pores where they exploit interior substrates, allowing the bacteria to proliferate [104]. In fact, Aeromonas bacteria are often isolated from seawater and sand samples [81,105,106]; therefore, the risk of contamination of the laid eggs is very high.
7. Pseudomonas spp.
Pseudomonas spp. are ubiquitous Gram-negative bacteria belonging to the family Pseudomonadaceae; they inhabit soil and water as well as animal-, human-, and plant-host-associated environments [107]. Pseudomonas aeruginosa is the most frequently encountered species related to animal and human infections, probably because it is the most frequently researched, but other species are known. Reptiles often are asymptomatic carriers of these opportunistic pathogens [96], and different pathologies have been associated with them [108].
Different Pseudomonas species have been cultured from tissues and swabs collected from both asymptomatic and sick sea turtles [55,109].
Pseudomonas aeruginosa and P. putrefaciens have been cultured from bronchoalveolar lavage samples from C. caretta with pulmonary disease [110].
Pseudomonas fluorescens, P. putrefaciens, P. maltophila, P. putida, and P.stutzeri were isolated from C. mydas affected by fibropapillomatosis [109]. Pseudomonas spp., with Burkholderia cepacia and Staphylococcus spp., were found in eyes affected by keratoconjunctivitis and ulcerative keratitis [55]. In all cases, pseudomonas agents act as opportunistic pathogens, often after their entrance through traumatic injuries [55].
The role of Pseudomonas spp. as opportunistic pathogens also emerged in cases of mortality of hatchling turtles; these animals can be affected by several pathologies, including yolk sacculitis, dermatitis, rhinitis, stomatitis, and pneumonia, caused or complicated by bacteria present in the environment [111].
8. Enterococcus spp.
The genus Enterococcus includes several species of Gram-positive bacteria usually present in the intestinal tract of all animals, but they can be also found on skin and oral cavities [112]. Usually, they bring benefits to their host related to probiotic activity and bacteriocins production, but, occasionally, they act as pathogens causing infections in humans and animals, with clinical forms ranging from mild to severe symptomatology [113]. In addition, they are largely distributed in different environments; in fact, they are found in soil, water, food, sewage, and plants [112].
Enterococcal infections in turtles have not been largely investigated, but it can be supposed that the bacteria act as opportunistic pathogens also in these animals. Tsai et al. [114] described a case of an olive ridley turtle (L. olivacea) admitted to a sea turtle rehabilitation facility at the National Museum of Marine Biology and Aquarium in Taiwan, showing lack of appetite and bilateral elbow joint swelling; Enterococcus faecalis was isolated from the affected joints, and an antibiotic-sensitivity test detected resistance to several antimicrobials. Resulting from the antibiotic resistance, the turtle died after antibiotic treatment for more than one month. This case report confirmed enterococci as multi-drug resistant bacteria, as it had been largely demonstrated in other animal species [115].
Enterococcus fecalis seems to be the species most frequently involved in enterococcal infections of marine turtles. It was found in different organs (bladder, brain, intestines, kidneys, liver, lungs, and muscles), suggesting a septicemic form of a C. caretta found dead along the coast in northern–central Italy [46].
Similarly, E. faecalis was isolated in cases of septicemia and osteomyelitis in cold-stunned Kemp’s ridley turtles (L. kempii) during their rehabilitation [116] and from a deep tissue infection in a loggerhead sea turtle caused by spotted eagle ray spines [117]. Infection cases observed in marine turtles seemed related to the introduction of enterococci via gastrointestinal perforation or through wounds from environmental contamination [117]. The presence of E. fecalis in the seawater environment has been directly demonstrated by Meena et al. [118], and its detection in fecal samples of wild sea turtles, such as hawksbill and green turtles from the southern coast of Brazil [119], corroborated these findings.
Other enterococcal species have been found in sea turtles. Enterococcus cloacae was cultured from one Hawaiian green turtle (C. mydas) afflicted with fibropapillomas, as well as one without this pathology [109]. Furthermore, Al-Bahry and collaborators found E. cloacae in oviductal fluid collected from green turtles, during the egg-laying process, in Ras Al-Hadd Reserve, located between the Gulf of Oman and the Arabian Sea [76].
Enterococcus faecalis, E. faecium, and E. hirae were the most common species identified in the fecal samples of wild sea turtles (C. mydas and E. imbricata) [119]; the same enterococcal species were also cultured from seabirds (S. magellanicus, S. trudeaui, and H. melanurus), marine mammals (B. acutorostrata, M. novaeangliae, and G. griseus), sea lions (Zalophus californianus), harbor seals (Phoca vitulina), and northern elephant seals (Mirounga angustirostris), suggesting the common circulation of these bacteria in marine environments [119,120,121].
9. Staphylococcus spp. and Streptococcus spp.
Members of the Staphylococcus genus are Gram-positive coccal bacteria; they are ubiquitous microorganisms, and most of them are animal commensals that colonize niches such as skin, nares, and diverse mucosal membranes. They act as opportunistic pathogens and have been largely associated with infections in humans and other mammalians [122].
Bacteria of the genus Streptococcus are Gram-positive cocci characterized by different virulence factors; they are categorized on the basis of their hemolytic pattern on blood agar as α-hemolytic, β-hemolytic, or γ-hemolytic (nonhemolytic); β-hemolytic streptococci, such as S. pyogenes, are typically pathogenic. Streptococcus spp. are common opportunistic pathogens, mainly from mammals, and are associated with a variety of diseases affecting multiple organ systems [123].
Staphylococcus spp. can be found as normal members of the bacterial flora of sea turtles, similarly to what has been observed in other reptiles, birds, and mammals [124,125]. Staphylococcus spp. were cultured from nasal and cloacal swabs collected from green turtles [126] and olive ridley turtles [127]. Different staphylococcal species, including S. aureus and S. epidermidis, have been found as components of the normal conjunctival flora of marine turtles [128], but they may be also involved in cases of keratoconjunctivitis [55].
Staphylococcus spp. were isolated also from cases of dermatitis, pneumonia, pleuritis, pericarditis, splenitis, nephritis, and myositis [55], and S. lentus has been associated with ulcerative stomatitis in green turtles [70]. Recently, S. haemolyticus was isolated from eggs of C. mydas that failed to hatch and relative nesting sites [129].
Staphylococcus spp. and Streptococcus spp. may be simultaneously involved in infections of sea turtles; in fact, they have been isolated from lesions of the digestive tract of these animals. Esophagitis, gastritis, enteritis, and hepatitis were associated with these bacteria, even though other Gram-positive and Gram-negative bacteria were cultured [44]. Streptococcus spp. were cultured from shoulders affected by chronic inflammation of a stranded C. mydas [130] and from wounds of olive ridley sea turtles [131].
Recently, Streptococcus iniae was isolated from tissues (kidney, liver, heart, spleen) of three dead N. depressus. Turtles were in an autolysis status when they were found; therefore, no gross or microscopic pathological abnormalities were observed [132]. Streptococcus iniae is an important aquatic pathogen responsible for disease outbreaks in freshwater and marine fishes, although disease is primarily reported in aquaculture; it is considered a zoonotic agent and has also been isolated from other aquatic animals, such as dolphins (Inia geoffrensis and Tursiops truncates), bullfrogs (Lithobates catesbeianus), and olive sea snakes (Aipysurus laevis) [132,133].
10. Bacillus spp.
The genus Bacillus includes several species of Gram-positive, rod-shaped, aerobic or facultatively anaerobic, spore-forming bacteria. Bacillus spp. are largely distributed in different environments being present in the soil, water, and other ecological niches [134].
Bacillus spp. have been often isolated from different samples collected from sea turtles. These bacteria were dominant in feces and the nasal cavities of investigated turtle species, such as C. mydas, L. olivacea, and D. coriacea [126,127,135]. Bacillus spp. may act as opportunistic pathogens in marine turtles, as suggested by their findings in cases of enteritis, splenitis, and pneumonia [44,55].
Different species of the genus Bacillus, including B. anthracis, B. subtilis, and B. cereus, which are well-known human pathogens, were also frequently found in unhatched eggs and sand from nesting sites [136,137].
11. Other Bacterial Agents
Occasionally, infections due to other bacterial agents, unusually for sea turtles in some cases, have been reported in the literature. The first case of listeriosis in a sea turtle has been recently reported. An adult female C. caretta stranded alive along the coast of the Abruzzo region (Italy) in Summer 2021 died after some days due to respiratory failure. The necropsy showed widespread organ lesions, mainly necrosis in many organs, gastrointestinal erosions, pericarditis, and granulomatous pneumonia, and Listeria monocytogenes, an agent responsible for severe disease in humans and other mammals, was isolated from the involved organs [138].
Carapacial ulcers have been associated with Psychrobacter sp. and Coriobacteriaceae spp., which seem to act as opportunistic pathogens; in fact, they can be isolated also from healthy green sea turtles and seawater samples [139]. However, these bacteria pose a potential risk of human infection too; Psychrobacter spp. may infect humans after surgery, blood transfusion, or exposure to the marine environment [140,141,142]; members of Coriobacteriaceae family, even though they are normal dwellers of mammalian body habitats such as the oral cavity and gastrointestinal and genital tracts, may cause bacteremia, periodontitis, and vaginosis [143].
Cardiobacteriaceae agents were found in carapacial ulcers of sea turtles, although they seem to have a simple role as contaminant [139]. However, bacteria of this family, also found in healthy penguins, dolphins, and whales, may cause endocarditis and wound infections in humans [144].
Lactococcus garvieae is a fish pathogen, also isolated from kidney and liver of a C. caretta [46]. Chryseobacterium meningosepticum has been cultured from C. caretta with myositis [55] and organs (lung, liver, kidney, heart, skin) of dead captive posthatchling leatherback turtles [145]. Both agents are potentially zoonotic, even if cases of infection in humans are rare [146,147].
A case of floppy flipper in a C. mydas has been associated with Clostridium botulinum [148], a zoonotic agent responsible for botulism [149]. The floppy flipper is an important clinical change associated with progressive paresis and paralysis of the flippers in sea turtles and causes death as paresis and paralysis extend to the neck, and animals are unable to lift their heads to breathe [148].
Brucella spp., including B. anthropi (formerly Ochrobacterium anthropi), have been isolated from nest and eggs of sea turtles [136,150,151]. The role of these pathogens in the failure of hatchings has been assumed because they were found in the eggs but not in the sand external to the nest; in addition, this finding suggests that brucellae derived from the mothers [151]. The zoonotic risk related to brucellae found in marine environments has not been clarified, but not excluded [152].
Table 2 summarizes the bacterial agents detected in unhatched eggs, hatchling turtles, and sand sampled from sea turtle nesting beaches.

Table 2.
Bacterial pathogens isolated from sea turtle eggs, hatchling turtles, and nest sand.
12. Conclusions
Sea turtle populations are threatened by several negative factors. Entanglement/fishing interaction, boat strike, plastic ingestion, coastal development affecting critical turtle habitat, marine pollution by inorganic elements, and organic contaminants are known as the main anthropogenic causes [71]. Moreover, there is a market for sea turtle products; the demand for sea turtle leather and shell products continues to exist despite the species’ protection under the Endangered Species Act and IUCN Red Listing. The hawksbill sea turtle (currently listed as critically endangered) is especially threatened by the illegal wildlife trade. Poachers kill the turtles for their shells and sell them as jewelry or souvenirs [153] (Figure 1).
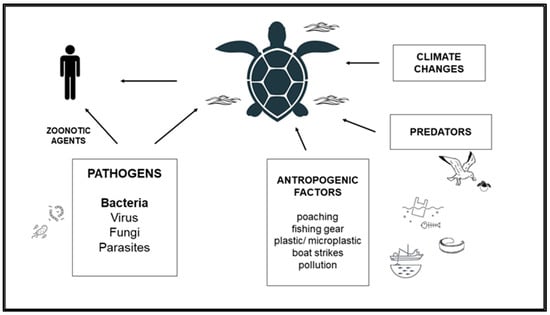
Figure 1.
Main threats to sea turtles.
Several pathogens may affect sea turtles causing pathologies that may be lethal in some cases. In fact, pathogens can directly cause fatal diseases or weaken turtles, which more easily may be victims of predators or injuries. Furthermore, pathogens may cause the death of embryos and hatching turtles. Among pathogens, bacteria often represent a severe threat not only for marine turtles’ health, but also for humans that come into contact with them, their products, and the environments where they live [4,154]. Sand contaminated by bacteria excreted with feces, eggs, and oviductal exudate represents a source of infection for people, mainly ecotourists, during their activity on the beach. Personnel working in rescue centers and sea turtle farms may be at risk of infection, as well.
The consumption of meat, eggs, and other products from infected sea turtles represents the major threat from a One Health perspective. Some coastal communities, especially in Central America and Asia, consume sea turtles, utilizing the entire animal. Turtle meat is eaten directly, whereas internal organs such as the kidneys and liver are used for soup [49], and the blood is drunk raw as a remedy for anemia. In addition, fat is used for extraction of oil employed as a cure for respiratory problems, especially in children [155,156]. Additionally, eggs, also valued as an aphrodisiac, are largely consumed [50]. Salmonella spp. and Vibrio spp. are considered the major bacterial hazards related to the consumption of meat and eggs, but other bacteria harbored by sea turtles may cause infection in humans through direct or indirect contact.
Bacteria harbored by sea turtles are frequently characterized by resistance to several antimicrobials; their presence in marine animals acts as an indicator of pollution levels in the surrounding area, but they also show that these animals are involved in their spreading [82]. Furthermore, antibiotic-resistant bacteria, of both human and animal origin, harbor transmissible antibiotic resistance genes. In the natural environments, including the marine one, the transfer of genetic determinants can occur [157]; therefore, new bacterial strains, pathogenetic and opportunistic, may become resistant to one or more antimicrobials.
Antibiotic-resistant bacteria should be taken into particular account as a major health concern of sea turtles because antibiotic treatment of hospitalized turtles may be unsuccessful [102,110,114,116,130,158]. However, evolution and severity of the infections, also those caused by opportunistic pathogens, are strongly related to sea turtles’ immune system status; the immune function is sensitive to fluctuations in temperature and to pollutants in the environment; and rapid changes in the marine environment are considered responsible for higher vulnerability to infections and diseases [159].
In conclusion, different pathogenic and opportunistic bacteria are able to cause infections and diseases in marine turtles. Even though infected sea turtles are not a relevant source of infections for humans if compared to other animals, they may be involved in the epidemiology of zoonotic bacterial agents, including those that are antimicrobially resistant.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflict of interest.
References
- Sea Turtle Conservancy. 2023. Available online: https://conserveturtles.org/information-sea-turtles-species-world (accessed on 6 March 2023).
- The IUCN Red List of Threatened Species. Available online: https://www.iucnredlist.org/en (accessed on 14 March 2023).
- SEE Turtles. 2023. Available online: https://www.seeturtles.org/why-are-sea-turtles-important (accessed on 6 March 2023).
- Mashkour, N.; Jones, K.; Kophamel, S.; Hipolito, T.; Ahasan, S.; Walker, G.; Jakob-Hoff, R.; Whittaker, M.; Hamann, M.; Bell, I.; et al. Disease risk analysis in sea turtles: A baseline study to inform conservation efforts. PLoS ONE 2020, 15, e0230760. [Google Scholar] [CrossRef] [PubMed]
- Borel, N.; Polkinghorne, A.; Pospischil, A. A Review on Chlamydial Diseases in Animals: Still a Challenge for Pathologists? Vet. Pathol. 2018, 55, 374–390. [Google Scholar] [CrossRef] [PubMed]
- Vorimore, F.; Hölzer, M.; Liebler-Tenorio, E.M.; Barf, L.M.; Delannoy, S.; Vittecoq, M.; Wedlarski, R.; Lécu, A.; Scharf, S.; Blanchard, Y.; et al. Evidence for the existence of a new genus Chlamydiifrater gen. nov. inside the family Chlamydiaceae with two new species isolated from flamingo (Phoenicopterus roseus): Chlamydiifrater phoenicopteri sp. nov. and Chlamydiifrater volucris sp. nov. Syst. Appl. Microbiol. 2021, 44, 126200. [Google Scholar] [CrossRef]
- Vanrompay, D.; De Meurichy, W.; Ducatelle, R.; Haesebrouck, F. Pneumonia in Moorish tortoises (Testudo graeca) associated with avian serovar A Chlamydia psittaci. Vet. Rec. 1994, 135, 284–285. [Google Scholar] [CrossRef]
- Frutos, M.C.; Monetti, M.S.; Re, V.E.; Cuffini, C.G. Molecular evidence of Chlamydophila pneumoniae infection in reptiles in Argentina. Rev. Argent. Microbiol. 2014, 46, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, H.; Sato, S.; Maruyama, S. Prevalence and characterization of Chlamydia DNA in zoo animals in Japan. Microbiol. Immunol. 2015, 59, 507–515. [Google Scholar] [CrossRef]
- Hotzel, H.; Blahak, S.; Diller, R.; Sachse, K. Evidence of infection in tortoises by Chlamydia-like organisms that are genetically distinct from known Chlamydiaceae species. Vet. Res. Commun. 2005, 29 (Suppl. S1), 71–80. [Google Scholar] [CrossRef]
- Mitura, A.; Niemczuk, K.; Zaręba, K.; Zając, M.; Laroucau, K.; Szymańska-Czerwińska, M. Free-living and captive turtles and tortoises as carriers of new Chlamydia spp. PLoS ONE 2017, 12, e0185407. [Google Scholar] [CrossRef]
- Laroucau, K.; Ortega, N.; Vorimore, F.; Aaziz, R.; Mitura, A.; Szymanska-Czerwinska, M.; Cicerol, M.; Salinas, J.; Sachse, K.; Caro, M.R. Detection of a novel Chlamydia species in captive spur-thighed tortoises (Testudo graeca) in southeastern Spain and proposal of Candidatus Chlamydia testudinis. Syst. Appl. Microbiol. 2020, 43, 126071. [Google Scholar] [CrossRef]
- Homer, B.L.; Jacobson, E.R.; Schumacher, J.; Scherba, G. Chlamydiosis in mariculture-reared green sea turtles (Chelonia mydas). Vet. Pathol. 1994, 31, 1–7. [Google Scholar] [CrossRef]
- Bodetti, T.J.; Jacobson, E.; Wan, C.; Hafner, L.; Pospischil, A.; Rose, K.; Timms, P. Molecular evidence to support the expansion of the host range of Chlamydophila pneumoniae to include reptiles as well as humans, horses, koalas and amphibians. Syst. Appl. Microbiol. 2002, 25, 146–152. [Google Scholar] [CrossRef]
- Aitken, I.D.; Longbottom, D. Chlamydial abortion. In Diseases of Sheep; Aitken, I.D., Ed.; Blackwelll Publishing: Oxford, UK, 2007; pp. 105–112. [Google Scholar]
- Pichon, N.; Guindre, L.; Laroucau, K.; Cantaloube, M.; Nallatamby, A.; Parreau, S. Chlamydia abortus in Pregnant Woman with Acute Respiratory Distress Syndrome. Emerg. Infect. Dis. 2020, 26, 628–629. [Google Scholar] [CrossRef]
- Pace, A.; Vicari, N.; Rigamonti, S.; Magnino, S.; Borrelli, L.; Dipineto, L.; Fioretti, A.; Hochscheid, S.; Tavares, L.; Duarte, A. Detection of Chlamydial DNA from Mediterranean Loggerhead Sea Turtles in Southern Italy. Animals 2022, 12, 715. [Google Scholar] [CrossRef]
- Sharma, S.K.; Upadhyay, V. Epidemiology, diagnosis & treatment of non-tuberculous mycobacterial diseases. Indian J. Med. Res. 2020, 152, 185–226. [Google Scholar] [CrossRef] [PubMed]
- Levi, M.H.; Bartell, J.; Gandolfo, L.; Smole, S.C.; Costa, S.F.; Weiss, L.M.; Johnson, L.K.; Osterhout, G.; Herbst, L. Characterization of Mycobacterium montefiorense sp. nov., a novel pathogenic mycobacterium from moray eels that is related to Mycobacterium triplex. J. Clin. Microbiol. 2003, 41, 2147–2152. [Google Scholar] [CrossRef]
- Rhodes, M.W.; Kator, H.; Kotob, S.; van Berkum, P.; Kaattari, I.; Vogelbein, W.; Quinn, F.; Floyd, M.M.; Butler, W.R.; Ottinger, C.A. Mycobacterium shottsii sp. nov., a slowly growing species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 2003, 53, 421–424. [Google Scholar] [CrossRef]
- Rhodes, M.W.; Kator, H.; McNabb, A.; Deshayes, C.; Reyrat, J.M.; Brown-Elliott, B.A.; Wallace, R.; Trott, K.A.; Parker, J.M.; Lifland, B.; et al. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 2005, 55, 1139–1147. [Google Scholar] [CrossRef]
- Carella, F.; Aceto, S.; Pollaro, F.; Miccio, A.; Iaria, C.; Carrasco, N.; Prado, P.; De Vico, G. A mycobacterial disease is associated with the silent mass mortality of the pen shell Pinna nobilis along the Tyrrhenian coastline of Italy. Sci. Rep. 2019, 9, 2725. [Google Scholar] [CrossRef] [PubMed]
- Oros, J.; Acosta, B.; Gaskin, J.M.; Deniz, S.; Jensen, H.E. Mycobacterium kansasii infection in a Chinese soft shell turtle (Pelodiscus sinensis). Vet. Rec. 2003, 152, 474–476. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.; Waliszewski, N.T.; Garner, M.M.; Tseng, F.S. Sepsis and disseminated intravascular coagulation in an eastern spiny softshell turtle (Apalone spinifera spinifera) with acute mycobacteriosis. J. Zoo Wildl. Med. 2009, 40, 572–575. [Google Scholar] [CrossRef]
- Ebani, V.V.; Fratini, F.; Bertelloni, F.; Cerri, D.; Tortoli, E. Isolation and identification of mycobacteria from captive reptiles. Res. Vet. Sci. 2012, 93, 1136–1138. [Google Scholar] [CrossRef] [PubMed]
- Muro, J.; Canturri, A.; Velarde, R.; Martínez-Silvestre, A.; Pastor, J.; Lavín, S.; Marco, I. Atypical systemic mycobacteriosis in Hermann’s tortoises (Testudo hermanni). J. Exot. Pet. Med. 2020, 32, 8–12. [Google Scholar] [CrossRef]
- Ollé, R.D.; Sevilla, I.A.; Juan-Sallés, C.; Garrido, J.M.; Santamaria, J.J. Mycobacterial Cloacitis with Systemic Dissemination in a Hermann’s Tortoise (Testudo hermanni). J. Herpetol. Med. Surg. 2022, 32, 162–170. [Google Scholar] [CrossRef]
- Friedmann, F. Spontane lungentuberkulose Schildkröten und die Stellung des tuberkelbazil system. ZTuberk 1903, 4, 439–457. [Google Scholar]
- Greer, L.L.; Strandberg, J.D.; Whitaker, B.R. Mycobacterium chelonae osteoarthritis in a Kemp’s ridley sea turtle (Lepidochelys kempii). J. Wildl. Dis. 2003, 39, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Nardini, G.; Florio, D.; Di Girolamo, N.; Gustinelli, A.; Quaglio, F.; Fiorentini, L.; Leopardi, S.; Fioravanti, M.L. Disseminated mycobacteriosis in a stranded loggerhead sea turtle (Caretta caretta). J. Zoo Wildl. Med. 2014, 45, 357–360. [Google Scholar] [CrossRef]
- Leong, J.K.; Smith, D.L.; Révéra, D.B.; Clary, J.C.; Lewis, D.H.; Scott, J.L.; DiNuzzo, A.R. Health care and diseases of captive-reared loggerhead and Kemp’s ridley sea turtles. In Proceedings of the First International Symposium on Kemp’s Ridley Sea Turtle Biology, Conservation and Management, Galveston, TX, USA, 1–4 October 1985; pp. 178–201. [Google Scholar]
- Donnelly, K.; Waltzek, T.B.; Wellehan, J.F., Jr.; Stacy, N.I.; Chadam, M.; Stacy, B.A. Mycobacterium haemophilum infection in a juvenile leatherback sea turtle (Dermochelys coriacea). J. Vet. Diagn. Investig. 2016, 28, 718–721. [Google Scholar] [CrossRef]
- Smith, S.; Taylor, G.D.; Fanning, E.A. Chronic cutaneous Mycobacterium haemophilum infection acquired from coral injury. Clin. Infect. Dis. 2003, 37, e100–e101. [Google Scholar] [CrossRef]
- Whipps, C.M.; Dougan, S.T.; Kent, M.L. Mycobacterium haemophilum infections of zebrafish (Danio rerio) in research facilities. FEMS Microbiol. Lett. 2007, 270, 21–26. [Google Scholar] [CrossRef]
- Dawson, D.J.; Jennis, F. Mycobacteria with a growth requirement for ferric ammonium citrate, identified as Mycobacterium haemophilum. J. Clin. Microbiol. 1980, 11, 190–192. [Google Scholar] [CrossRef]
- Keymer, I.F. Diseases of chelonians: (2) Necropsy survey of terrapins and turtles. Vet. Rec. 1978, 103, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.S.; Campbell, R.S.F. A survey of the diseases of marine turtles in northern Australia. I. Farmed turtles. Dis. Aquat. Organ. 1990, 9, 83–95. [Google Scholar] [CrossRef]
- Brock, J.A.; Nakamura, R.M.; Miyahara, A.Y.; Chang, E.M.L. Tuberculosis in Pacific Green Sea Turtles, Chelonia mydas. Trans. Am. Fish. Soc. 1976, 105, 564–566. [Google Scholar] [CrossRef]
- Octavia, S.; Lan, R. The Family Enterobacteriaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Habib, I.; Shahabi, M.P.; Dybing, N.A.; Wang, P.; Bruce, M. A Review of the Public Health Challenges of Salmonella and Turtles. Vet. Sci. 2020, 7, 56. [Google Scholar] [CrossRef]
- Ebani, V.V.; Cerri, D.; Fratini, F.; Meille, N.; Valentini, P.; Andreani, E. Salmonella enterica isolates from faeces of domestic reptiles and a study of their antimicrobial in vitro sensitivity. Res. Vet. Sci. 2005, 78, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Bertelloni, F.; Chemaly, M.; Cerri, D.; Gall, F.L.; Ebani, V.V. Salmonella infection in healthy pet reptiles: Bacteriological isolation and study of some pathogenic characters. Acta Microbiol. Immunol. Hung. 2016, 63, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Zając, M.; Puchalski, A.; Szczepaniak, K.; Urban-Chmiel, R. Pet Reptiles in Poland as a Potential Source of Transmission of Salmonella. Pathogens 2022, 11, 1125. [Google Scholar] [CrossRef]
- Oros, J.; Deniz, S.; Calabuig, P. Digestive pathology of sea turtles stranded in the Canary Islands between 1993 and 2001. Vet. Rec. 2004, 155, 169–174. [Google Scholar] [CrossRef]
- Foti, M.; Bottari, T.; Coci, G.; Daidone, A.; Pennisi, M.G. Enterobacteriaceae isolates in cloacal swabs from live-stranded internally-hooked Loggerhead Sea turtles, Caretta caretta, in the Central Mediterranean Sea. J. Herpetol. Med. Surg. 2008, 17, 125–128. [Google Scholar] [CrossRef]
- Fichi, G.; Cardeti, G.; Cersini, A.; Mancusi, C.; Guarducci, M.; Di Guardo, G.; Terracciano, G. Bacterial and viral pathogens detected in sea turtles stranded along the coast of Tuscany, Italy. Vet. Microbiol. 2016, 185, 56–61. [Google Scholar] [CrossRef]
- Pace, A.; Rinaldi, L.; Ianniello, D.; Borrelli, L.; Cringoli, G.; Fioretti, A.; Hochscheid, S.; Dipineto, L. Gastrointestinal investigation of parasites and Enterobacteriaceae in loggerhead sea turtles from Italian coasts. BMC Vet. Res. 2019, 15, 370. [Google Scholar] [CrossRef] [PubMed]
- Scheelings, T.F.; Lightfoot, D.; Holz, P. Prevalence of Salmonella in Australian reptiles. J. Wildl. Dis. 2011, 47, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Duplaix, N.; Wells, S. Sea turtles, animals of divisible parts: International trade in sea turtle products. In Biology and Conservation of Sea Turtles; Bjorndal, K., Ed.; Smithsonian Institution Press: Washington, DC, USA, 1982; pp. 545–565. [Google Scholar]
- Aguirre, A.A.; Gardner, S.C.; Marsh, J.C.; Delgado, S.G.; Limpus, C.J.; Nichols, W.J. Hazards Associated with the Consumption of Sea Turtle Meat and Eggs: A Review for Health Care Workers and the General Public. EcoHealth 2006, 3, 141–153. [Google Scholar] [CrossRef]
- O’Grady, K.A.; Krause, V. An outbreak of salmonellosis linked to a marine turtle. Southeast Asian J. Trop. Med. Public Health 1999, 30, 324–327. [Google Scholar] [PubMed]
- Draper, A.D.K.; James, C.L.; Pascall, J.E.; Shield, K.J.; Langrell, J.; Hogg, A. An outbreak of Salmonella Muenchen after consuming sea turtle, Northern Territory, Australia, 2017. Commun. Dis. Intell. Q. Rep. 2017, 41, E290–E294. [Google Scholar] [PubMed]
- Zieger, U.; Trelease, H.; Winkler, N.; Mathew, V.; Sharma, R.N. Bacterial contamination of leatherback turtle (Dermochelys coriacea) eggs and sand in nesting chambers at Levera Beach, Grenada, West Indies preliminary study. West Indian Vet. J. 2009, 9, 21–26. [Google Scholar]
- Work, T.M.; Dagenais, J.; Stacy, B.A.; Ladner, J.T.; Lorch, J.M.; Balazs, G.H.; Barquero-Calvo, E.; Berlowski-Zier, B.M.; Breeden, R.; Corrales-Gómez, N.; et al. A novel host-adapted strain of Salmonella Typhimurium causes renal disease in olive ridley turtles (Lepidochelys olivacea) in the Pacific. Sci. Rep. 2019, 9, 9313. [Google Scholar] [CrossRef]
- Oros, J.; Torrent, A.; Calabuig, P.; Deniz, S. Diseases and causes of mortality among sea turtles stranded in the Canary Islands, Spain (1998–2001). Dis. Aquat. Org. 2005, 63, 13–24. [Google Scholar] [CrossRef]
- de Morais, P.B.; de Souza, D.R.; de Sousa, F.M.; de Oliveira, K.W.; Pimenta, R.S. Enterobacteriaceae in mouth and cloaca of Podocnemis expansa and P. unifilis (testudines:chelonia) populations of national park of Araguaia plains. Brazil. Braz. J. Microbiol. 2011, 42, 526–530. [Google Scholar] [CrossRef]
- Trotta, A.; Marinaro, M.; Sposato, A.; Galgano, M.; Ciccarelli, S.; Paci, S.; Corrente, M. Antimicrobial Resistance in Loggerhead Sea Turtles (Caretta caretta): A Comparison between Clinical and Commensal Bacterial Isolates. Animals 2021, 11, 2435. [Google Scholar] [CrossRef]
- Dutton, C.S.; Revan, F.; Wang, C.; Xu, C.; Norton, T.M.; Stewart, K.M.; Kaltenboeck, B.; Soto, E. Salmonella enterica prevalence in leatherback sea turtles (Dermochelys coriacea) in St. Kitts, West Indies. J. Zoo Wildl. Med. 2013, 44, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Ives, A.K.; Antaki, E.; Stewart, K.; Francis, S.; Jay-Russell, M.T.; Sithole, F.; Kearney, M.T.; Griffin, M.J.; Soto, E. Detection of Salmonella enterica Serovar Montevideo and Newport in Free-ranging Sea Turtles and Beach Sand in the Caribbean and Persistence in Sand and Seawater Microcosms. Zoonoses Public Health 2017, 64, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.; Hariharan, H.; Matthew, V.; Chappell, S.; Davies, R.; Parker, R.; Sharma, A. Prevalence, serovars, and antimicrobial susceptibility of Salmonella isolated from blue land crabs (Cardisoma guanhumi) in Grenada, West Indies. J. Food Prot. 2013, 76, 1270–1273. [Google Scholar] [CrossRef] [PubMed]
- Drake, M.; Amadi, V.; Zieger, U.; Johnson, R.; Hariharan, H. Prevalence of Salmonella spp. in cane toads (Bufo marinus) from Grenada, West Indies, and their antimicrobial susceptibility. Zoonoses Public Health 2013, 60, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Amadi, V.; Stone, D.; Johnson, R.; Hariharan, H.; Zieger, U. Prevalence and antimicrobial susceptibility of Salmonella spp. in small Indian mongooses (Herpestes auropunctatus) in Grenada, West Indies. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 205–210. [Google Scholar] [CrossRef]
- Edwards, J.J.; Amadi, V.A.; Soto, E.; Jay-Russel, M.T.; Aminabadi, P.; Kenelty, K.; Charles, K.; Arya, G.; Mistry, K.; Nicholas, R.; et al. Prevalence and phenotypic characterization of Salmonella enterica isolates from three species of wild marine turtles in Grenada, West Indies. Vet. World 2021, 14, 222–229. [Google Scholar] [CrossRef]
- Al-Bahry, S.; Mahmoud, I.; Elshfie, A.; Al-Harthy, A.; Al-Ghafri, S.; Al-Amri, I.; Alkindi, A. Bacterial flora and antibiotic resistance from eggs of green turtles Chelonia mydas: An indication of polluted effluents. Mar. Pollut. Bull. 2009, 58, 720–725. [Google Scholar] [CrossRef]
- Marangi, M.; Carlucci, R.; Carlino, P.; Fanizza, C.; Cirelli, G.; Maglietta, R.; Beneduce, L. Dolphins and sea turtles may host zoonotic parasites and pathogenic bacteria as indicators of anthropic pressure in the Gulf of Taranto (Northern Ionian Sea, Central-Eastern Mediterranean Sea). Vet. Res. Commun. 2022, 46, 1157–1166. [Google Scholar] [CrossRef]
- Raidal, S.R.; Ohara, M.; Hobbs, R.P.; Prince, R.I. Gram-negative bacterial infections and cardiovascular parasitism in green sea turtles (Chelonia mydas). Aust. Vet. J. 1998, 76, 415–417. [Google Scholar] [CrossRef]
- Mueller, M.; Tainter, C.R. Escherichia coli. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK564298/ (accessed on 5 February 2023).
- Praja, R.N.; Yudhana, A.; Haditanojo, W.; Oktaviana, V. Short communication: Antimicrobial properties in cloacal fluid of olive ridley sea turtle (Lepidochelys olivacea). Biodiversitas 2021, 22, 3671–3676. [Google Scholar] [CrossRef]
- Goldberg, D.W.; Fernandes, M.R.; Sellera, F.P.; Costa, D.G.C.; Loureiro Bracarense, A.P.; Lincopan, N. Genetic background of CTX-M-15-producing Enterobacter hormaechei ST114 and Citrobacter freundii ST265 co-infecting a free-living green turtle (Chelonia mydas). Zoonoses Public Health 2019, 66, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Vega-Manriquez, D.X.; Dávila-Arrellano, R.P.; Eslava-Campos, C.A.; Salazar Jiménez, E.; Negrete-Philippe, A.C.; Raigoza-Figueras, R.; Muñoz-Tenería, F.A. Identification of bacteria present in ulcerative stomatitis lesions of captive sea turtles Chelonia mydas. Vet. Res. Commun. 2018, 42, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Orós, J.; Camacho, M.; Calabuig, P.; Rial-Berriel, C.; Montesdeoca, N.; Déniz, S.; Luzardo, O.P. Postmortem investigations on leatherback sea turtles (Dermochelys coriacea) stranded in the Canary Islands (Spain) (1998–2017): Evidence of anthropogenic impacts. Mar. Pollut. Bull. 2021, 167, 112340. [Google Scholar] [CrossRef]
- Hall, J.; Bender, H.; Miller, N.; Thompson, P. Fatal Bronchopneumonia and Tracheitis in a Green Turtle (Chelonia mydas) Caused by Serratia proteamaculans. Animals 2022, 12, 1891. [Google Scholar] [CrossRef] [PubMed]
- Mahlen, S.D. Serratia infections: From military experiments to current practice. Clin. Microbiol. Rev. 2011, 24, 755–791. [Google Scholar] [CrossRef]
- Santoro, M.; Hernández, G.; Caballero, M.; García, F. Aerobic bacterial flora of nesting green turtles (Chelonia mydas) from Tortuguero National Park, Costa Rica. J. Zoo Wildl. Med. 2006, 37, 549–552. [Google Scholar] [CrossRef]
- Foti, M.; Giacopello, C.; Bottari, T.; Fisichella, V.; Rinaldo, D.; Mammina, C. Antibiotic Resistance of Gram Negatives isolates from loggerhead sea turtles (Caretta caretta) in the central Mediterranean Sea. Mar. Pollut. Bull. 2009, 58, 1363–1366. [Google Scholar] [CrossRef]
- Al-Bahry, S.N.; Al-Zadjali, M.A.; Mahmoud, I.Y.; Elshafie, A.E. Biomonitoring marine habitats in reference to antibiotic resistant bacteria and ampicillin resistance determinants from oviductal fluid of the nesting green sea turtle, Chelonia mydas. Chemosphere 2012, 87, 1308–1315. [Google Scholar] [CrossRef]
- Pace, A.; Dipineto, L.; Fioretti, A.; Hochscheid, S. Loggerhead sea turtles as sentinels in the western Mediterranean: Antibiotic resistance and environment-related modifications of Gram-negative bacteria. Mar. Pollut. Bull. 2019, 149, 110575. [Google Scholar] [CrossRef]
- Tsai, M.A.; Chang, C.C.; Li, T.H. Antimicrobial-resistance profiles of gram-negative bacteria isolated from green turtles (Chelonia mydas) in Taiwan. Environ. Poll. 2021, 277, 116870. [Google Scholar] [CrossRef]
- Kuschke, S.G. What lives on and in the sea turtle? A literature review of sea turtle bacterial microbiota. Anim. Microbiome 2022, 4, 52. [Google Scholar] [CrossRef] [PubMed]
- Blasi, M.F.; Migliore, L.; Mattei, D.; Rotini, A.; Thaller, M.C.; Alduina, R. Antibiotic Resistance of Gram-Negative Bacteria from Wild Captured Loggerhead Sea Turtles. Antibiotics 2020, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Alduina, R.; Gambino, D.; Presentato, A.; Gentile, A.; Sucato, A.; Savoca, D.; Filippello, S.; Visconti, G.; Caracappa, G.; Vicari, D.; et al. Is Caretta caretta a Carrier of Antibiotic Resistance in the Mediterranean Sea? Antibiotics 2020, 9, 116. [Google Scholar] [CrossRef]
- Drane, K.; Huerlimann, R.; Power, M.; Whelan, A.; Ariel, E.; Sheehan, M.; Kinobe, R. Testudines as Sentinels for Monitoring the Dissemination of Antibiotic Resistance in Marine Environments: An Integrative Review. Antibiotics 2021, 10, 775. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.R.; Mahmoud, I.Y.; Al-Musharafi, S.K.; Al-Bahry, S.N. Antibiotic resistant bacteria in the environment as bio-indicators of pollution. Open Biotechnol. J. 2016, 10, 342–351. [Google Scholar] [CrossRef]
- Ahasan, M.S.; Waltzek, T.B.; Huerlimann, R.; Ariel, E. Comparative analysis of gut bacterial communities of green turtles (Chelonia mydas) pre-hospitalization and post-rehabilitation by high-throughput sequencing of bacterial 16S rRNA gene. Microbiol. Res. 2018, 207, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Palit, A.; Nair, G.B. Bacteria: Other Vibrios. In Encyclopedia of Food Safety; Motarjemi, Y., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 570–573. [Google Scholar] [CrossRef]
- Campos, E.; Bolaños, H.; Acuña, M.T.; Díaz, G.; Matamoros, M.C.; Raventós, H.; Sánchez, L.M.; Sánchez, O.; Barquero, C. Vibrio mimicus diarrhea following ingestion of raw turtle eggs. Appl. Environ. Microbiol. 1996, 62, 1141–1144. [Google Scholar] [CrossRef]
- Acuña, M.T.; Díaz, G.; Bolaños, H.; Barquero, C.; Sánchez, O.; Sánchez, L.M.; Mora, G.; Chaves, A.; Campos, E. Sources of Vibrio mimicus contamination of turtle eggs. Appl. Environ. Microbiol. 1999, 65, 336–338. [Google Scholar] [CrossRef]
- Vallini, C.; Rubini, S.; Tarricone, L.; Mazziotti, C.; Gaspari, S. Unusual Stranding of Live, Small, Debilitated Loggerhead Turtles along the Northwestern Adriatic Coast. Mar. Turt. Newsl. 2011, 131, 25–28. [Google Scholar]
- Zavala-Norzagaray, A.A.; Aguirre, A.A.; Velazquez-Roman, J.; Flores-Villaseñor, H.; León-Sicairos, N.; Ley-Quiñonez, C.P.; Hernández-Díaz, L.J.; Canizalez-Roman, A. Isolation, characterization, and antibiotic resistance of Vibrio spp. in sea turtles from Northwestern Mexico. Front. Microbiol. 2015, 6, 635. [Google Scholar] [CrossRef] [PubMed]
- Noguerola, I.; Blanch, A.R. Identification of Vibrio spp. with a set of dichotomous keys. J. Appl. Microbiol. 2008, 105, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Wiles, M.; Rand, T.G. Integumental ulcerative disease in a loggerhead turtle Caretta caretta at the Bermuda Aquarium: Microbiology and histopathology. Dis. Aquat. Org. 1987, 3, 85–90. [Google Scholar] [CrossRef]
- Glazebrook, J.S.; Campbell, R.S.F.; Thomas, A.D. Studies on an ulcerative stomatitis-obstructive rhinitis-pneumonia disease complex in hatchling and juvenile sea turtles Chelonia mydas and Caretta caretta. Dis. Aquat. Org. 1993, 16, 133–147. [Google Scholar] [CrossRef]
- Obendorf, D.L.; Carson, J.; McManus, T.J. Vibrio damsela Infection in a Stranded Leatherback Turtle (Dermochelys coriacea). J. Wildl. Dis. 1987, 23, 666–668. [Google Scholar] [CrossRef]
- Colwell, R.R.; MacDonell, M.R.; De Ley, J. Proposal to recognize the family Aeromonadaceae. Int. J. Syst. Bacteriol. 1986, 36, 473. [Google Scholar] [CrossRef]
- Pessoa, R.B.G.; de Oliveira, W.F.; Correia, M.T.D.S.; Fontes, A.; Coelho, L.C.B.B. Aeromonas and Human Health Disorders: Clinical Approaches. Front. Microbiol. 2022, 13, 868890. [Google Scholar] [CrossRef]
- Ebani, V.V.; Fratini, F.; Ampola, M.; Rizzo, E.; Cerri, D.; Andreani, E. Pseudomonas and Aeromonas isolates from domestic reptiles and study of their antimicrobial in vitro sensitivity. Vet. Res. Commun. 2008, 32 (Suppl. S1), S195–S198. [Google Scholar] [CrossRef]
- Nyman, S. Mass Mortality in Larval Rana sylvatica Attributable to the Bacterium, Aeromonas hydrophila. J. Herpetol. 1986, 20, 196–201. [Google Scholar] [CrossRef]
- Musgrave, K.E.; Diehl, K.; Mans, C. Aeromonas hydrophila Keratitis in Freshwater Turtles. J. Exotic. Pet. Med. 2016, 25, 26–29. [Google Scholar] [CrossRef]
- Clary, J.C.; Leong, J.K. Disease studies aid Kemp’s ridley sea turtle headstart research. Herp. Rev. 1984, 15, 69–70. [Google Scholar]
- Glazebrook, J.S.; Campbell, R.S.F. A survey of the diseases of marine turtles in northern Australia. II. Oceanarium—Reared and wild turtles. Dis. Aquat. Org. 1990, 9, 97–104. [Google Scholar] [CrossRef]
- Pace, A.; Meomartino, L.; Affuso, A.; Mennonna, G.; Hochscheid, S.; Dipineto, L. Aeromonas induced polyostotic osteomyelitis in a juvenile loggerhead sea turtle Caretta caretta. Dis. Aquat. Organ. 2018, 132, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Orós, J.; Camacho, M.; Calabuig, P.; Arencibia, A. Salt gland adenitis as only cause of stranding of loggerhead sea turtles Caretta caretta. Dis. Aquat. Organ. 2011, 95, 163–166. [Google Scholar] [CrossRef]
- Soslau, G.; Spotila, J.R.; Chun, A.; Yi, S.; Weber, K.T. Potentially lethal bacteria in leatherback turtle eggs in the wild threaten both turtles and conservationists. J. Exp. Mar. Biol. Ecol. 2011, 410, 101–106. [Google Scholar] [CrossRef]
- Dumontet, S.; Krovacek, K.; Svenson, S.B.; Pasquale, V.; Baloda, S.B.; Figliuolo, G. Prevalence and diversity of Aeromonas and Vibrio spp. in coastal waters of Southern Italy. Comp. Immunol. Microbiol. Infect. Dis. 2000, 23, 53–72. [Google Scholar] [CrossRef]
- Fiorentini, C.; Barbieri, E.; Falzano, L.; Matarrese, P.; Baffone, W.; Pianetti, A.; Katouli, M.; Kühn, I.; Möllby, R.; Bruscolini, F. Occurrence, diversity and pathogenicity of mesophilic Aeromonas in estuarine waters of the Italian coast of the Adriatic Sea. J. Appl. Microbiol. 1998, 85, 501–511. [Google Scholar] [CrossRef]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33, Erratum in Microbiology 2021, 167, 001073. [Google Scholar] [CrossRef]
- Xiong, Y.; Wu, Q.; Qin, X.; Yang, C.; Luo, S.; He, J.; Cheng, Q.; Wu, Z. Identification of Pseudomonas aeruginosa from the Skin Ulcer Disease of Crocodile Lizards (Shinisaurus crocodilurus) and Probiotics as the Control Measure. Front. Vet. Sci. 2022, 9, 850684. [Google Scholar] [CrossRef]
- Aguirre, A.A.; Balazs, G.H.; Zimmerman, B.; Spraker, T.R. Evaluation of Hawaiian green turtles (Chelonia mydas) for potential pathogens associated with fibropapillomas. J. Wildl. Dis. 1994, 30, 8–15. [Google Scholar] [CrossRef]
- Ciccarelli, S.; Valastro, C.; Di Bello, A.; Paci, S.; Caprio, F.; Corrente, M.L.; Trotta, A.; Franchini, D. Diagnosis and Treatment of Pulmonary Disease in Sea Turtles (Caretta caretta). Animals 2020, 10, 1355. [Google Scholar] [CrossRef]
- Rodríguez, P.S.; Stewart, K.; Picknell, A.; Pemberton, L.; Tepedino, A.; Capaldo, D.; Dennis, M.M. Pathology of hatchling hawksbill sea turtle (Eretmochelys imbricata) mortalities occurring while under rehabilitative care, 2015–2021. J. Wildl. Dis. 2023, 59, 109–120. [Google Scholar] [CrossRef]
- Raza, T.; Ullah, S.R.; Mehmood, K.; Andleeb, S. Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 2018, 68, 768–772. [Google Scholar] [PubMed]
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Igrejas, G.; Poeta, P. Enterococci, from harmless bacteria to a pathogen. Microorganisms 2020, 8, 1118. [Google Scholar] [CrossRef]
- Tsai, M.A.; Chang, C.C.; Li, T.H. Multiple-antibiotic resistance of Enterococcus faecalis in an endangered olive ridley sea turtle (Lepidochelys olivacea): A case report. Indian J. Anim. Res. 2019, 1064, 1–6. [Google Scholar] [CrossRef]
- Cagnoli, G.; Bertelloni, F.; Interrante, P.; Ceccherelli, R.; Marzoni, M.; Ebani, V.V. Antimicrobial-Resistant Enterococcus spp. in Wild Avifauna from Central Italy. Antibiotics 2022, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- Innis, C.J.; Braverman, H.; Cavin, J.M.; Ceresia, M.L.; Baden, L.R.; Kuhn, D.M.; Frasca, S., Jr.; McGowan, J.P.; Hirokawa, K.; Weber, E.S., 3rd; et al. Diagnosis and management of Enterococcus spp infections during rehabilitation of cold-stunned Kemp’s ridley turtles (Lepidochelys kempii): 50 cases (2006–2012). J. Am. Vet. Med. Assoc. 2014, 245, 315–323. [Google Scholar] [CrossRef]
- Bezjian, M.; Wellehan, J.F.X.; Walsh, M.T.; Anderson, E.; Jacobson, E. Management of wounds in a loggerhead sea turtle (Caretta caretta) caused by traumatic bycatch injury from the spines of a spotted eagle ray (Aetobatus narinari). J. Zoo Wildl. Med. 2014, 45, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Meena, B.; Anburajan, L.; Sathish, T.; Raghavan, R.V.; Jha, D.K.; Venkateshwaran, P.; Das, A.K.; Dheenan, P.S.; Vinithkumar, N.V.; Dharani, G.; et al. Enterococcus species diversity and molecular characterization of biomarker genes in Enterococcus faecalis in Port Blair Bay, Andaman and Nicobar Islands, India. Mar. Poll. Bull. 2015, 94, 217–227. [Google Scholar] [CrossRef]
- Prichula, J.; Pereira, R.I.; Wachholz, G.R.; Cardoso, L.A.; Tolfo, N.C.; Santestevan, N.A.; Medeiros, A.W.; Tavares, M.; Frazzon, J.; d’Azevedo, P.A.; et al. Resistance to antimicrobial agents among enterococci isolated from fecal samples of wild marine species in the southern coast of Brazil. Mar. Poll. Bull. 2016, 105, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.P.; Nolan, S.; Gulland, F.M. Antimicrobial susceptibility of bacteria isolated from pinnipeds stranded in central and northern California. J. Zoo Wildl. Med. 1998, 29, 288–294. [Google Scholar] [PubMed]
- Thornton, S.M.; Nolan, S.; Gulland, F.M. Bacterial isolates from California sea lions (Zalophus californianus), harbor seals (Phoca vitulina), and northern elephant seals (Mirounga angustirostris) admitted to a rehabilitation center along the central California coast, 1994-1995. J. Zoo Wildl. Med. 1998, 29, 171–176. [Google Scholar] [PubMed]
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in animals. Microbiol. Spectr. 2019, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lamm, C.G.; Ferguson, A.C.; Lehenbauer, T.W.; Love, B.C. Streptococcal Infection in Dogs: A Retrospective Study of 393 Cases. Vet. Pathol. 2010, 47, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Nagase, N.; Sasaki, A.; Yamashita, K.; Shimizu, A.; Wakita, Y.; Kitai, S.; Kawano, J. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 2002, 64, 245–250. [Google Scholar] [CrossRef]
- Jho, Y.S.; Park, D.H.; Lee, J.H.; Cha, S.Y.; Han, J.S. Identification of bacteria from the oral cavity and cloaca of snakes imported from Vietnam. Lab. Anim. Res. 2011, 27, 213–217. [Google Scholar] [CrossRef]
- Al-Bahry, S.N.; Mahmoud, I.Y.; Al-Zadjali, M.; Elshafie, A.; Al-Harthy, A.; Al-Alawi, W. Antibiotic resistant bacteria as bio-indicator of polluted effluent in the green turtles, Chelonia mydas in Oman. Mar. Environ. Res. 2011, 71, 139–144. [Google Scholar] [CrossRef]
- Santoro, M.; Orrego, C.M.; Hernández Gómez, G. Flora bacteriana cloacal y nasal de Lepidochelys olivacea (Testudines: Cheloniidae) en el pacíficonorte de Costa Rica. Biol. Trop. 2006, 54, 43–48. [Google Scholar] [CrossRef]
- Cardoso-Brito, V.; Raposo, A.C.S.; Pires, T.T.; Pinna, M.H.; Oria, A.P. Conjunctival bacterial flora and antimicrobial susceptibility of captive and free-living sea turtles in Brazil. Vet. Ophthalmol. 2019, 22, 246–255. [Google Scholar] [CrossRef]
- Candan, E.D.; Candan, O.; Çevik, Y.N. Bacterial diversity of loggerhead and green turtle eggs from two major nesting beaches from the Turkish coast of the Mediterranean. Arch. Microbiol. 2022, 204, 682. [Google Scholar] [CrossRef]
- Guthrie, A.; George, J.; de Maar, T.W. Bilateral Chronic Shoulder Infections in an Adult Green Sea Turtle (Chelonia mydas). J. Herpetol. Med. Surg. 2010, 20, 105–108. [Google Scholar] [CrossRef]
- Sagasiousman, R.; Khairani, S. Microbial Isolations from Olive Ridley Sea Turtle (Lepidochelys olivacea) in Alas Purwo National Park, Indonesia. In Proceedings of the 10th International Seminar and 12th Congress of Indonesian Society for Microbiology (ISISM 2019), Surakarta, Indonesia, 29–30 August 2019. Adv. Biol. Sci. Res. 2021, 12, 189–194. [Google Scholar] [CrossRef]
- Young, E.J. Health and Disease Status of Sea Turtles in Western Australia. Ph.D. Thesis, Murdoch University, Parkville, VIC, Australia, 2022. [Google Scholar]
- Young, E.; Bannister, J.; Buller, N.; Vaughan-Higgins, R.; Stephens, N.; Whiting, S.; Yeap, L.; Miller, T.; Warren, K. Streptococcus iniae associated mass marine fish kill off Western Australia. Dis. Aquat. Org. 2020, 142, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Skóra, M.; Zając, M.; Kwit, R.; Skarżyńska, M.; Pasim, P.; Mikos-Wojewoda, E.; Bomba, A.; Giza, A.; Chesneau, O.; Hendriksen, R.S.; et al. Draft Genome Sequences of Six Isolates of the Bacillus cereus Group Isolated from Pet Reptiles. Microbiol. Resour. Announc. 2022, 11, e0038522. [Google Scholar] [CrossRef]
- Santoro, H.; Hernandéz, G.; Caballero, M.; García, F. Potential bacterial pathogens carried by nesting leatherback turtles (Dermochelys coriacea) in Costa Rica. Chelonian Conserv. Biol. 2008, 7, 104–108. [Google Scholar] [CrossRef]
- Candan, O.; Candan, E.D. Bacterial diversity of the green turtle (Chelonia mydas) nest environment. Sci. Total Environ. 2020, 720, 137717. [Google Scholar] [CrossRef]
- Hoh, D.Z.; Lin, Y.F.; Liu, W.A.; Sidique, S.N.M.; Tsai, I.J. Nest microbiota and pathogen abundance in sea turtle hatcheries. Fungal Ecol. 2020, 47, 100964. [Google Scholar] [CrossRef]
- Di Renzo, L.; De Angelis, M.E.; Torresi, M.; Di Lollo, V.; Di Teodoro, G.; Averaimo, D.; Defourny, S.V.P.; Di Giacinto, F.; Profico, C.; Olivieri, V.; et al. First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing. Animals 2022, 12, 2364. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, H.; Liu, P.; Wang, F.; Li, L.; Ye, M.; Zhao, W.; Chen, J. Microbial composition of carapace, feces, and water column in captive juvenile green sea turtles with carapacial ulcers. Front. Vet. Sci. 2022, 9, 1039519. [Google Scholar] [CrossRef]
- Bonwitt, J.; Tran, M.; Droz, A.; Gonzalez, A.; Glover, W.A. Psychrobacter sanguinis wound infection associated with marine environment exposure, Washington, USA. Emerg. Infect. Dis. 2018, 24, 1942–1944. [Google Scholar] [CrossRef]
- Caspar, Y.; Recule, C.; Pouzol, P.; Lafeuillade, B.; Mallaret, M.-R.; Maurin, M.; Croize, J. Psychrobacter arenosus bacteremia after blood transfusion, France. Emerg. Infect. Dis. 2013, 19, 1118–1120. [Google Scholar] [CrossRef]
- Le Guern, R.; Wallet, F.; Vega, E.; Courcol, R.J.; Loiez, C. Psychrobacter sanguinis: An unusual bacterium for nosocomial meningitis. J. Clin. Microbiol. 2014, 52, 3475–3477. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Lepage, P.; Charrier, C. The Family Coriobacteriaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- McNally, K.L.; Mott, C.R.; Guertin, J.R.; Bowen, J.L. Microbial communities of wild-captured Kemp’s ridley (Lepidochelys kempii) and green sea turtles (Chelonia mydas). Endanger. Species Res. 2021, 45, 21–36. [Google Scholar] [CrossRef]
- Miller, D.L.; Wyneken, J.; Rajeev, S.; Perrault, J.; Mader, D.R.; Weege, J.; Baldwin, C.A. Pathologic findings in hatchling and posthatchling leatherback sea turtles (Dermochelys coriacea) from Florida. J. Wildl. Dis. 2009, 45, 962–971. [Google Scholar] [CrossRef]
- Wang, C.Y.; Shie, H.S.; Chen, S.C.; Huang, J.P.; Hsieh, I.C.; Wen, M.S.; Lin, F.C.; Wu, D. Lactococcus garvieae infections in humans: Possible association with aquaculture outbreaks. Int. J. Clin. Pract. 2007, 61, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ceyhan, M.; Celik, M. Elizabethkingia meningosepticum (Chryseobacterium meningosepticum) Infections in Children. Int. J. Pediatr. 2011, 2011, 215237. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Mustin, W.; McManamon, R.; Ritchie, B.W. Clinical Evidence for Association of Clostridium botulinum and Floppy Flipper in Green Sea Turtles (Chelonia mydas). In Proceedings of the 47th Annual IAAAM Meeting and Conference, Virginia Beach, VA, USA, 21–26 May 2016. [Google Scholar]
- Tiwari, A.; Nagalli, S. Clostridium botulinum. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Craven, K.S.; Awong-Taylor, J.; Griffiths, L.; Bass, C.; Muscarella, M. Identification of bacterial isolates from unhatched loggerhead (Caretta caretta) sea turtle eggs in Georgia, USA. Mar. Turt. Newsl. 2007, 115, 9–11. [Google Scholar]
- Capri, F.C.; Prazzi, E.; Casamento, G.; Gambino, D.; Cassata, G.; Alduina, R. Correlation Between Microbial Community and Hatching Failure in Loggerhead Sea Turtle Caretta caretta. Microb. Ecol. 2023, 2023, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Whatmore, A.M.; Dawson, C.E.; Groussaud, P.; Koylass, M.S.; King, A.C.; Shankster, S.J.; Sohn, A.H.; Probert, W.S.; McDonald, W.L. Marine mammal Brucella genotype associated with zoonotic infection. Emerg. Infect. Dis. 2008, 14, 517–518. [Google Scholar] [CrossRef]
- International Fund for Animal Welfare. Sea Turtles. Available online: https://www.ifaw.org/international/animals/sea-turtles (accessed on 10 March 2023).
- Warwick, C.; Arena, P.C.; Steedman, C. Health implications associated with exposure to farmed and wild sea turtles. J. R. Soc. Med. Sh. Rep. 2013, 4, 8. [Google Scholar] [CrossRef]
- Caldwell, D.K. The sea turtle fishery of Baja California, Mexico. Calif. Fish Game 1963, 49, 140–151. [Google Scholar]
- Felger, R.S.; Moser, M. Sea turtles in Seri Indian culture. Environ. Southwest 1987, 519, 18–21. [Google Scholar]
- Gambino, D.; Sciortino, S.; Migliore, S.; Galuppo, L.; Puleio, R.; Dara, S.; Vicari, D.; Seminara, S.; Gargano, V. Preliminary Results on the Prevalence of Salmonella spp. in Marine Animals Stranded in Sicilian Coasts: Antibiotic Susceptibility Profile and ARGs Detection in the Isolated Strains. Pathogens 2021, 10, 930. [Google Scholar] [CrossRef] [PubMed]
- Ahasan, M.S.; Picard, J.; Elliott, L.; Kinobe, R.; Owens, L.; Ariel, E. Evidence of antibiotic resistance in Enterobacteriales isolated from green sea turtles, Chelonia mydas on the Great Barrier Reef. Mar. Pollut. Bull. 2017, 120, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Nash, A.; Ryan, E.J. Immunity in Sea Turtles: Review of a Host-Pathogen Arms Race Millions of Years in the Running. Animals 2023, 13, 556. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).