Abstract
The SARS-CoV-2 coronavirus expresses two essential proteases: firstly, the 3Chymotrypsin-like protease (3CLpro) or main protease (Mpro), and secondly, the papain-like protease (PLpro), both of which are considered as viable drug targets for the inhibition of viral replication. In order to perform drug discovery assays for SARS-CoV-2, it is imperative that efficient methods are established for the production and purification of 3CLpro and PLpro of SARS-CoV-2, designated as 3CLpro-CoV2 and PLpro-CoV2, respectively. This article expands the data collected in the attempts to express SARS-CoV-2 proteases under different conditions and purify them under single-step chromatography. Data showed that the use of E. coli BL21(DE3) strain was sufficient to express 3CLpro-CoV2 in a fully soluble form. Nevertheless, the single affinity chromatography step was only applicable for 3CLpro-CoV2 expressed at 18 °C, with a yield and purification fold of 92% and 49, respectively. Meanwhile, PLpro-CoV2 was successfully expressed in a fully soluble form in either BL21(DE3) or BL21-CodonPlus(DE3) strains. In contrast, the single affinity chromatography step was only applicable for PLpro-CoV2 expressed using E. coli BL21-CodonPlus(DE3) at 18 or 37 °C, with a yield and purification fold of 86% (18 °C) or 83.36% (37 °C) and 112 (18 °C) or 71 (37 °C), respectively. The findings provide a guide for optimizing the production of SARS-CoV-2 proteases of E. coli host cells.
Dataset: 10.5281/zenodo.5503693
Dataset License: CC-BY
1. Summary
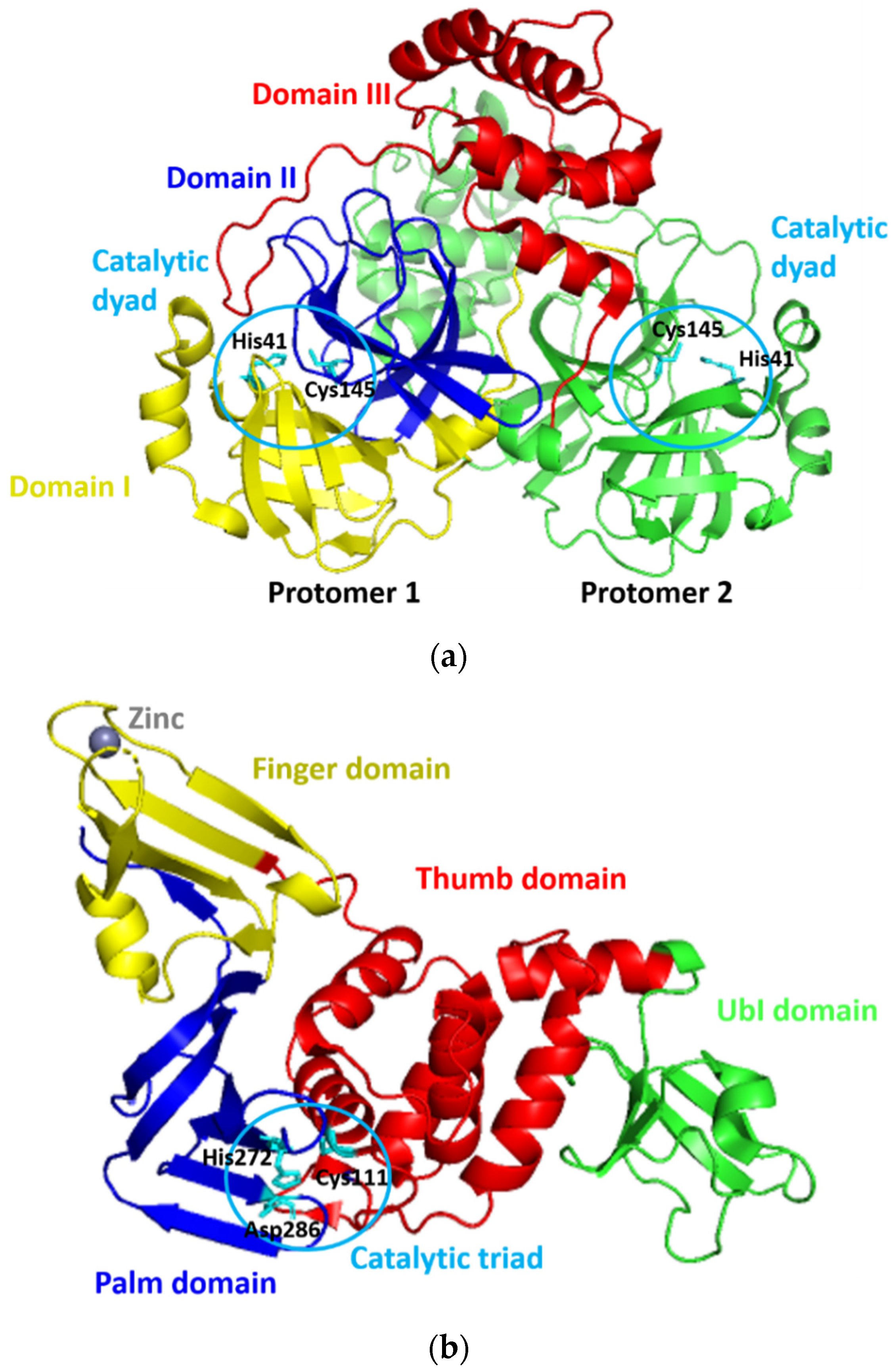
Since its first emergence in December 2019, a new coronavirus, namely, severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2), has become a global health issue [1,2,3]. The virus causes the coronavirus disease 2019 (COVID-19) [1,2], with the current total global death cases of more than 4 million people [3]. The fast and widespread spread of SARS-CoV-2 has prompted a rush to find promising targets for COVID-19 therapeutic development [4]. For this purpose, SARS-CoV-2 proteases have gotten a lot of attention for therapeutic development due to their critical functions during viral replication [5,6]. SARS-CoV-2 expresses two proteases of 3Chymotrypsin-like protease (3CLpro) or main protease (Mpro) and papain-like protease (PLpro), which are known to mediate the proteolytic processing during viral replication in the host cells. This processing occurs after the production of the 800 kDa polypeptide by the virus upon the translation of its material genetic inside the host cells. The 3CLpro cleaves the polypeptide at 11 positions to produce various essential structural and non-structural viral proteins [7]. Meanwhile, PLpro does similar things as 3CLpro, yet, with different cleavage sites. In addition to this function, PLpro deubiquitinates and deISGylates the proteins of the host cell by removing ISG15 and ubiquitin. This furthers assist the virus in dodging the host-innate immune response [8]. Because of their critical roles, the SARS-CoV-2 proteases are viable therapeutic targets for COVID-19 treatments [9,10,11,12]. The three-dimensional models of 3CLpro and PLpro of SARS-CoV-2 (designated as 3CLpro-CoV2 and PLpro-CoV2, respectively) were reported and available at the Protein Data Bank (Figure 1) which allows us to discover promising drug-able spots on these proteins. PLpro-CoV2 have a structure that resemble a right-hand fold, which comprises of four distinct domains. The thumb and palm (in which the catalytic triad is situated), the fingers (which include the Zn-binding sites), and an independent N-terminal domain which termed as Ubl domain [13]. Meanwhile, 3CLpro-CoV2 forms a dimer structure in which each protomer is comprised of three structural domains. The first two domains form a chymotrypsin-like fold that is responsible for catalytic reactions (where the catalytic dyad is situated), and the third domain is responsible for the enzyme dimerization [14].
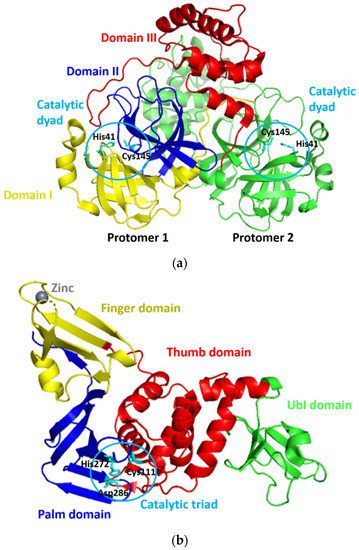
Figure 1.
The three-dimensional model structures of (a) 3CLpro-CoV2 (PDB ID: 6WTM) and (b) PLpro-CoV2 (PDB ID: 6W9C). The domain organization and catalytic residues of both proteases were also indicated for clarity.
Most drug discovery initiatives require the production of recombinant proteins, which is an indispensable step in the process. In particular, the screening step in the drug discovery pipeline requires the target protein with sufficient amount and quality. Similarly, this is also a prerequisite for many fundamental studies (structural and functional studies) of the target protein, which further serve as a platform for drug design and development. However, the production of target protein through recombinant technology is often challenged by the issues of the expression level and purification process [15]. Labor and costs should be maintained to a minimum level in the ideal production process. To note, the whole process to obtain high purity of target protein under recombinant technology involves multiple and lengthy steps. This includes, but is not limited to, gene cloning, transformation into the host cells, expression induction, host cells harvesting and lysis, removal of the cell debris, and end up with purification through one or more chromatography processes followed by the purity and yield determination [16]. Optimization at any step along this pipeline is therefore needed for obtaining the most efficient production of the target protein.
Escherichia coli is a widely accepted host organism to produce various recombinant proteins due to its technical and cost issues. Nevertheless, the expression of foreign proteins in the microbial system, including E. coli, is challenging since these proteins place a metabolic load on the host. Producing larger amounts of soluble heterologous protein is a substantial difficulty due to its foreignness. Because these foreign proteins have a proclivity for misfolding and form insoluble inclusions in the host’s cytoplasm [17,18]. The production and purification of recombinant 3CLpro-CoV2 and PLpro-CoV2 were widely reported; however, the majority of the reports used different techniques. As a result, extracting the conclusive demand for the most efficient strategy to be employed in future screening and studies is tough. So far, there has not been a study that details the technical data on the production and purifying processes of these proteins.
This paper, therefore, offers a comprehensive description of the data of protein produced obtained from multiple expression conditions of SARS-CoV-2 proteases (Table 1). The rest of the paper is arranged as follows: data description is done in Section 2 and a discussion of materials and methods used in this research is presented in Section 3.

Table 1.
Specifications table.
2. Data Description
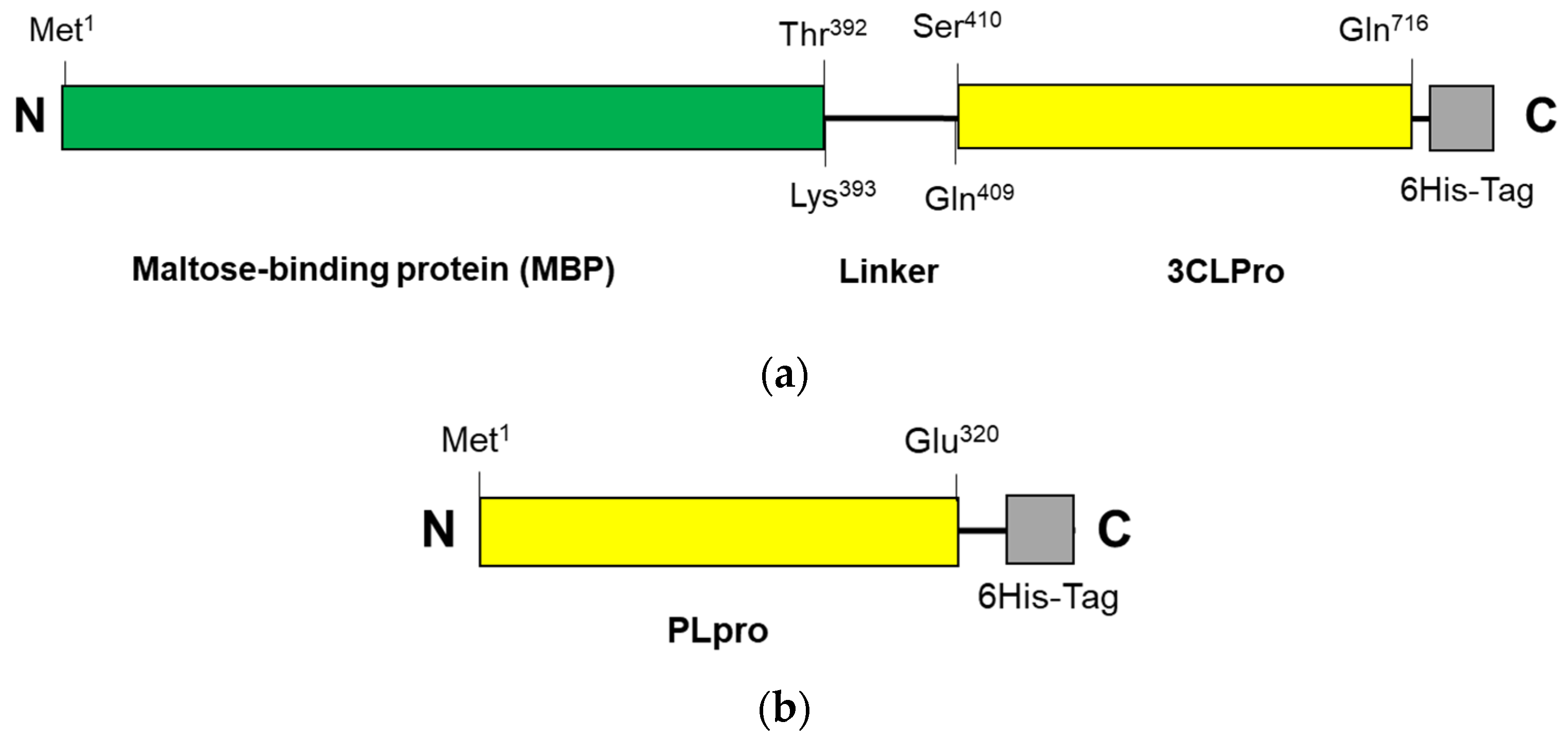
The construction systems for 3CLpro-CoV2 and PLpro-CoV2 were composed of the genes encoding 3CLpro (Ser1—Gln306) and PLpro (Met1—Glu320), respectively, which were then inserted into pD451-SR or pET21a expression plasmids, respectively. Under this system, 3CLpro-CoV2 was expressed in a fusion form to a maltose-binding protein (MBP), at the N-terminal, and a 6His-at the C-terminal, with a total theoretical size of 79 kDa. A linker sequence (LINGDGAGLEVLSAVLQ) was located between the Maltose-binding protein (MBP) and 3CLpro-CoV2, which also serves as the autocleavage site. The expressed 3CLpro-CoV2 was therefore expected to have a free N-terminus, with no MBP and linker fragment, due to the autocleavage event during the expression. Meanwhile, PLpro-CoV2 was expressed in a fusion form to a 6His-tag at its C-terminus, with the theoretical size of 37 kDa. The primary structures of 3CLpro-CoV2 and PLpro-CoV2 are shown in Figure 2. Table 2 shows the expression results of 3CLpro-CoV2 and PLpro-CoV2 under various expression conditions. Overall, all expression conditions lead to the successful expression of both proteins. For 3CLpro-CoV2, all expression conditions (1 and 2) in E. coli BL21(DE3) strain successfully led to the formation of 3CLpro-CoV2 in a fully soluble form. Nevertheless, PLpro-CoV2 was expressed in a fully soluble form only under four conditions (5, 6, 9 and 10), out of 8 tested conditions.
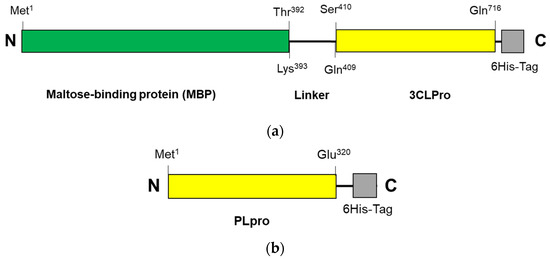
Figure 2.
The primary structure of SARS-CoV-2 proteases: (a) 3CLpro-CoV2 and (b) PLpro-CoV2. The linker sequence for connecting MBP and 3CLpro is LINGDGAGLEVLSAVLQ. The 6His-tag sequences for 3CLpro-CoV2 and PLpro-CoV2 are GPHHHHHH and HHHHHH, respectively. The figures are not drawn to scale.

Table 2.
Summary of expression conditions of SARS-CoV-2 proteases.
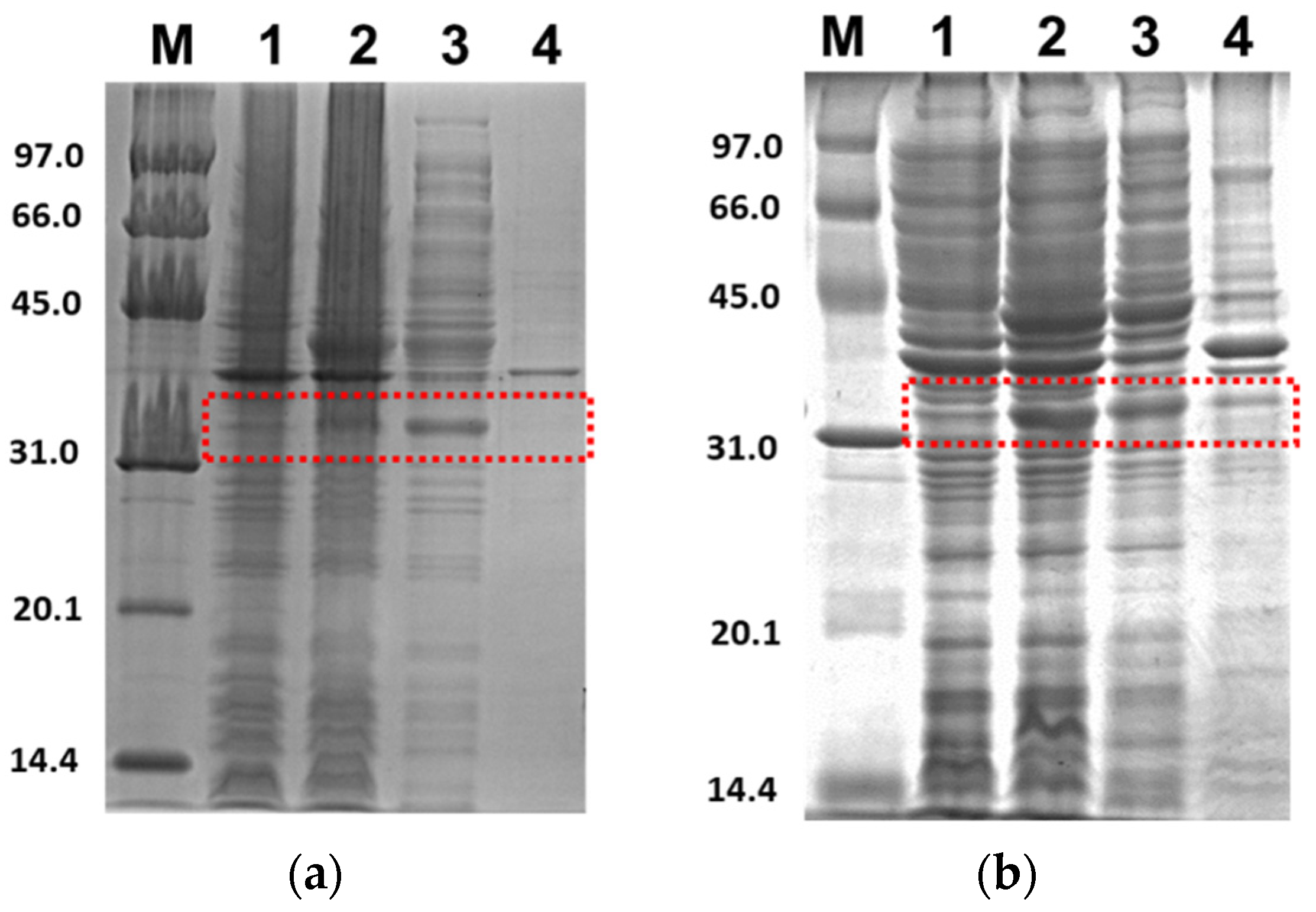
Figure 3 shows the SDS-PAGE for the expression profile of 3CLpro-CoV2 under E. coli BL21(DE3) cells with the given conditions 1 and 2. It clearly shows that the 3CLpro-CoV2 was indeed expressed after the induction of IPTG at condition 1 or 2, as indicated by the presence of the band corresponding to 3CLpro-CoV2 in the lane without IPTG. The apparent size of 3CLpro-CoV2 in the gel, as shown in Figure 3, was ~35 kDa, which was lower than its theoretical size (79 kDa). This is due to the autocleavage of MBP (~44 kDa) by 3CLpro-CoV2 during the expression.
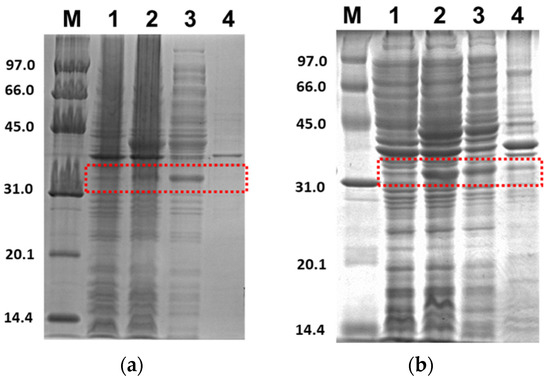
Figure 3.
Expression profile of 3CLpro-CoV2 in E. coli BL21 (DE3) under 15% SDS-PAGE. Lane 1: The cell before IPTG induction; Lane 2: The cell after IPTG induction; Lane 3: Soluble fraction of the cell obtained after the sonication; Lane 4: Insoluble fraction of the cell obtained after the sonication. The area that corresponds to the 3CLpro-CoV2 band is indicated by a red box: (a) The expression profile under condition 1; (b) The expression profile under condition 2. Details of the conditions are shown in Table 2.
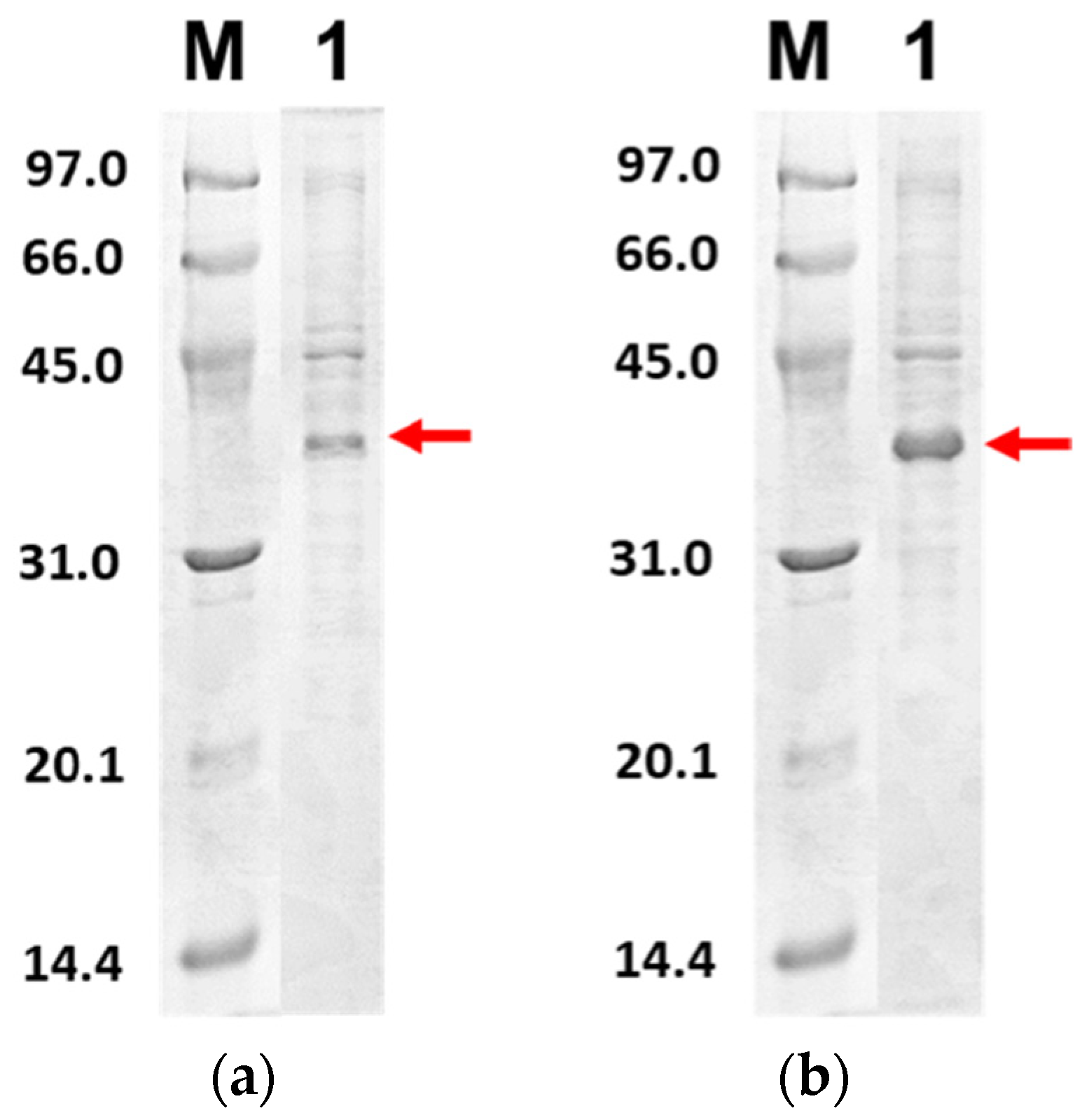
Figure 4 shows the SDS-PAGE of PLpro-CoV2 expression profile under E. coli BL21(DE3) or E. coli BL21-CodonPlus(DE3) strain with the expression conditions 5, 6, 9 and 10. The band corresponds to PLpro-CoV2, with the apparent size of ~37 kDa, which appeared only in lanes after IPTG induction and in soluble fraction. The apparent size of PLpro-CoV2 in the gel was comparable to its theoretical size which indicated that no cleavage occurred during the expression of this protein. To note, SDS-PAGE led to the insoluble form of PLpro-CoV2 was not shown in this report.
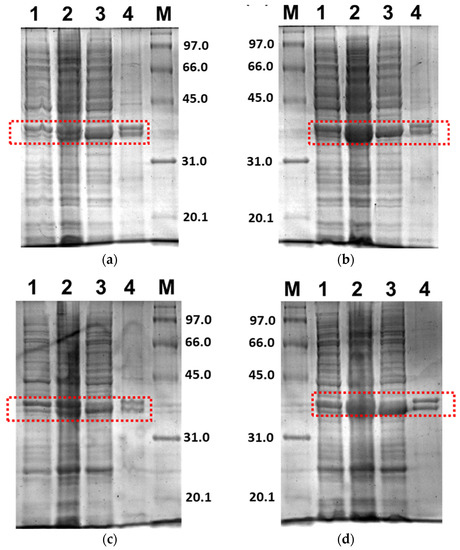
Figure 4.
Expression check of PLpro-CoV2 under 15% SDS-PAGE. Lane 1: The cell before IPTG induction; Lane 2: The cell after IPTG induction; Lane 3: Soluble fraction of the cell obtained after the sonication; Lane 4: Insoluble fraction of the cell obtained after the sonication. The area that corresponds to the PLpro-CoV2 band is indicated by a red box: (a) The expression profile under condition 5; (b) The expression profile under condition 6; (c) The expression profile under condition 9; (d) The expression profile under condition 10. Details of the conditions are shown in Table 2.
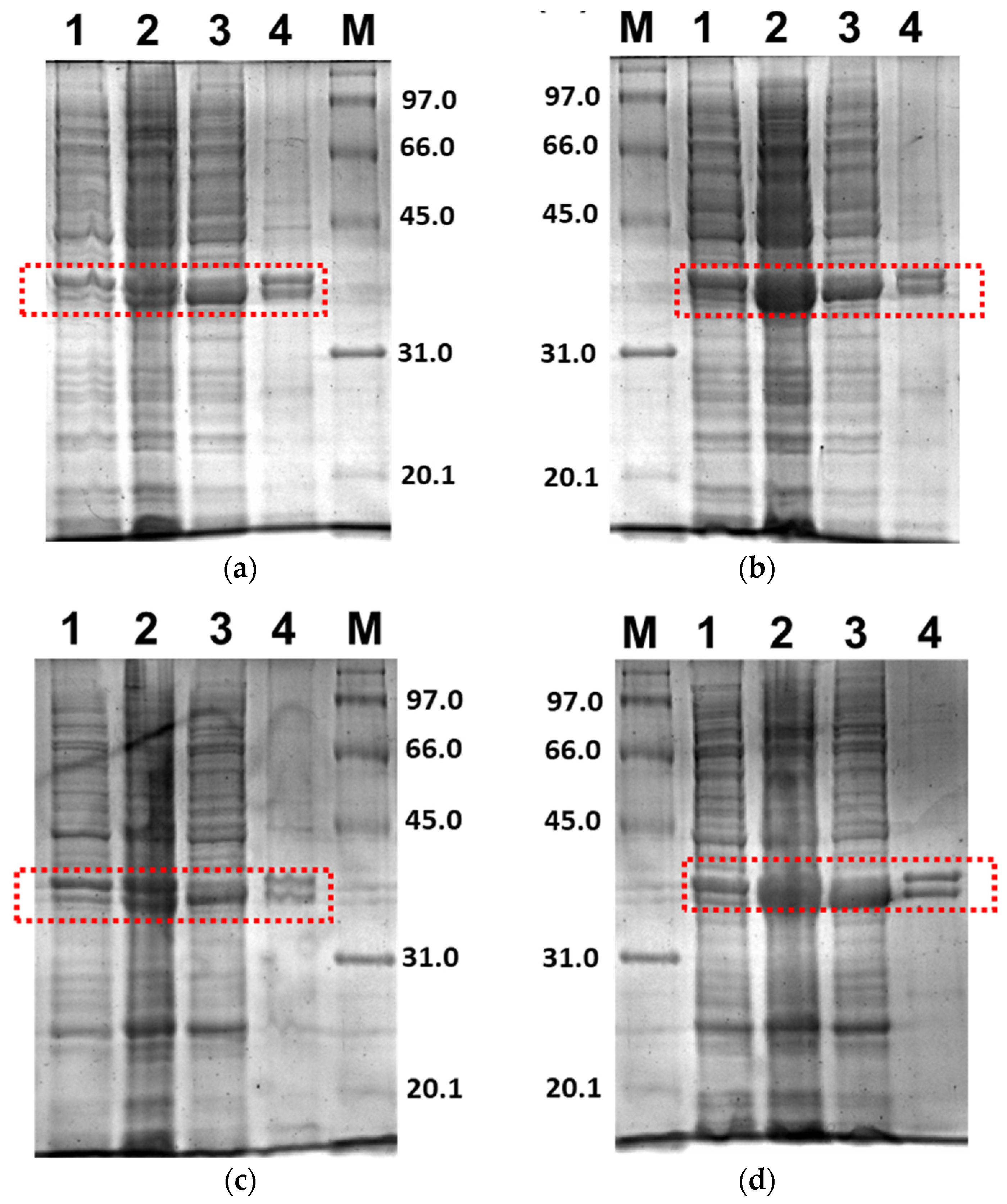
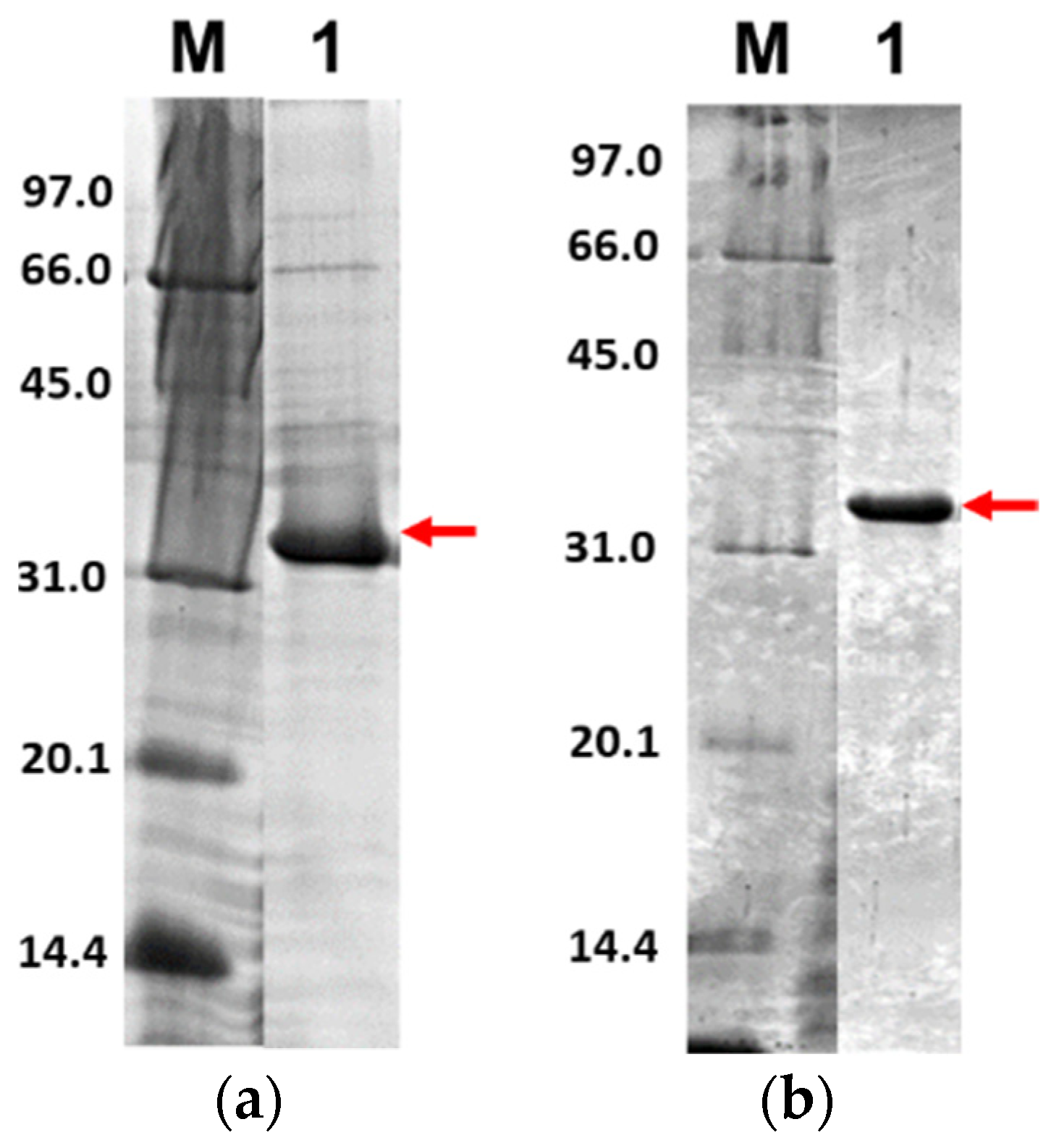
Further, only soluble fractions were proceeded to the purification process using Ni2+-NTA affinity chromatography to obtain pure proteins. Figure 5 shows the purified 3CLpro-CoV2 from the expression conditions 1 and 2 after the Ni2+-NTA chromatography. It shows that the presence of protein contaminants in Figure 5b was considerably undetectable under the gel. Meanwhile, the contaminants remain visible after the purification of 3CLpro-CoV2 expressed from condition 1 (Figure 5a). This may be due to that more indigenous proteins of E. coli host cell were expressed at 37 °C (condition 1) than at 18 °C (condition 2). The 3CLpro-CoV2 expressed from condition 1 was, therefore, considered to require further purification steps to be at a high purity level.
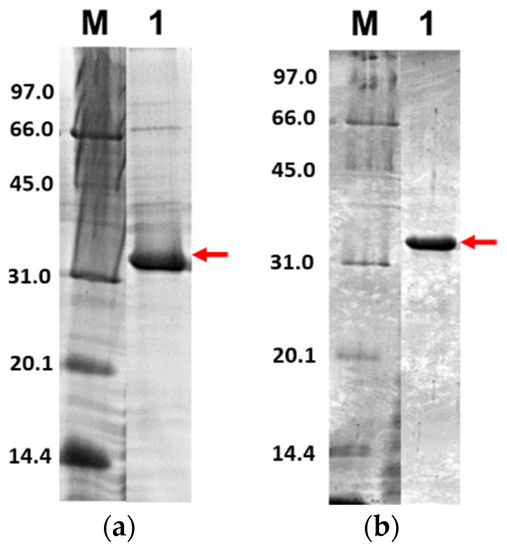
Figure 5.
The 15% SDS-PAGE analysis of purified 3CLpro-CoV2. Lane M: Protein marker; Lane 1: Purified protein after Ni2+-NTA chromatography: (a) Purified 3CLpro-CoV2 expressed under condition 1; (b) Purified 3CLpro-CoV2 expressed under condition 2. The band that corresponds to the 3CLpro-CoV2 is indicated by an arrow. Details of the conditions are shown in Table 2.
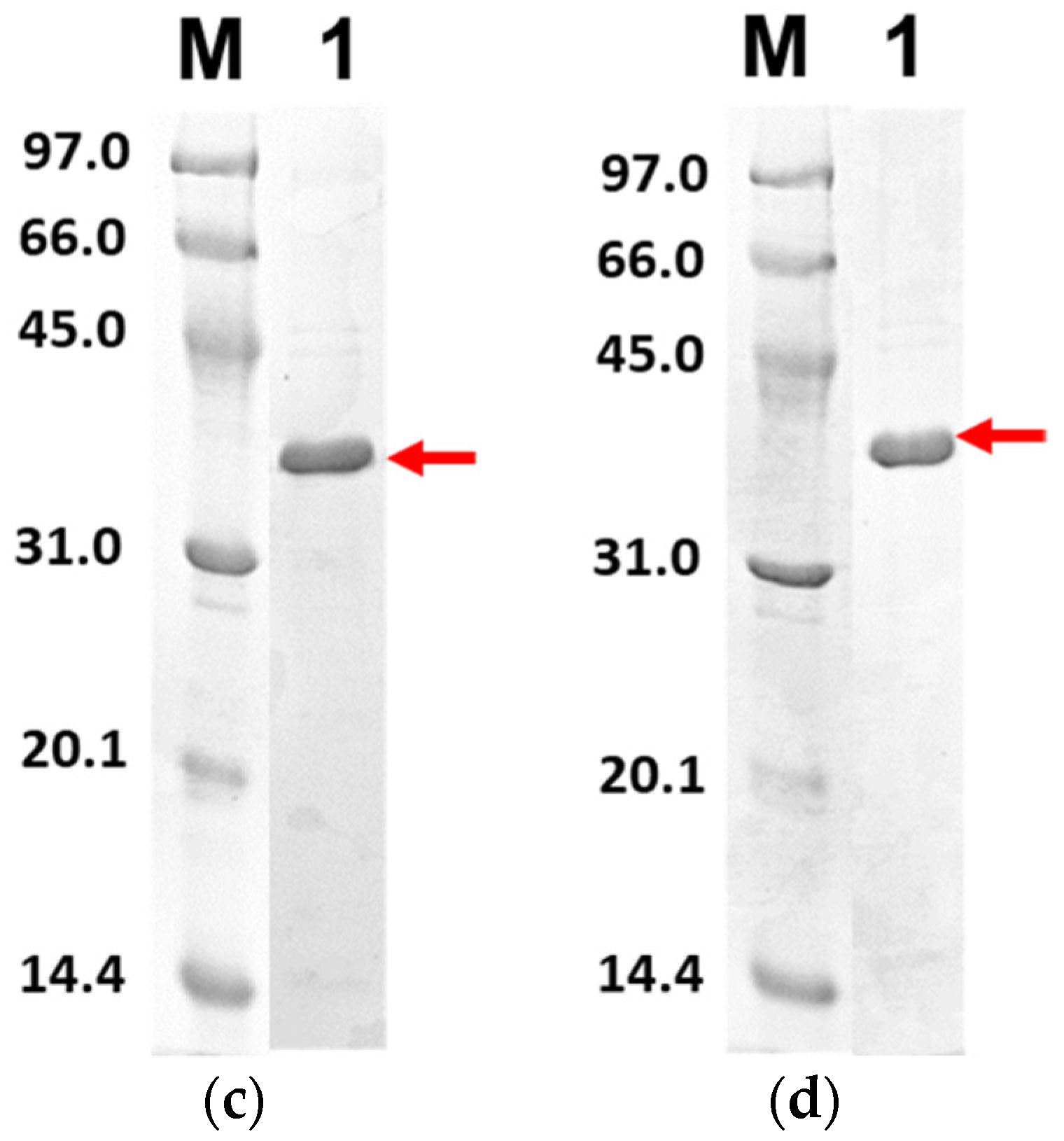
Figure 6 shows the purified PLpro-CoV2 from the expression conditions 5, 6, 9 and 10 after the Ni2+-NTA chromatography. It shows that only conditions 9 and 10 resulted in undetectable contaminants after the Ni2+-NTA chromatography (Figure 6c,d). Meanwhile, a remarkable presence of protein contaminants remained visible after the purification of PLpro-CoV2 expressed under conditions 5 and 6 (Figure 6a,b). This may be due to the E. coli BL21-CodonPlus(DE3) strain which expressed less protein at the given conditions. This result also suggests that the use of IPTG at low concentration (0.1 mM) with no ZnSO4 may be sufficient to produce PLpro-CoV2 in a fully soluble form and high purity under a single-step chromatography.
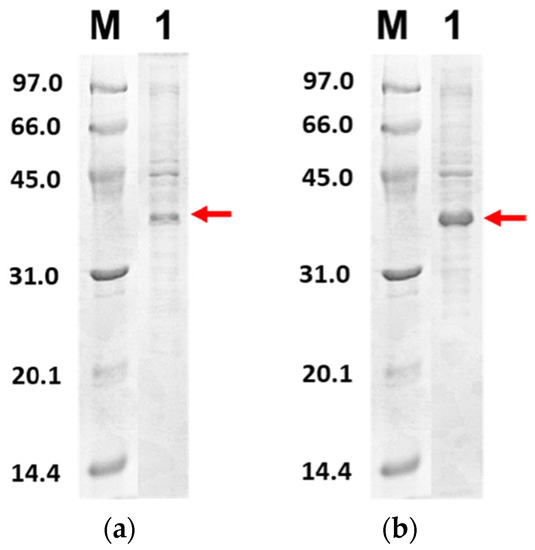
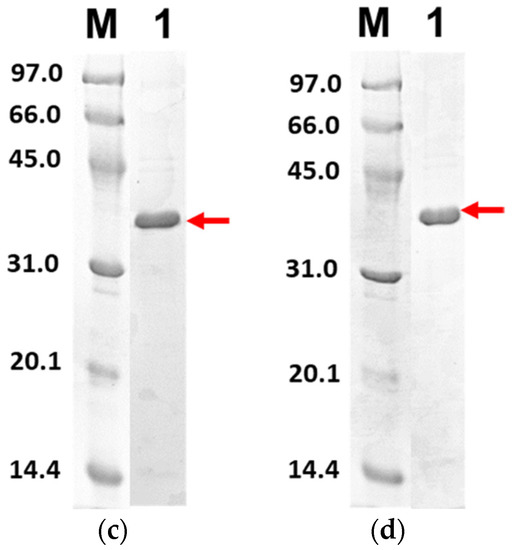
Figure 6.
The 15% SDS-PAGE analysis of purified PLpro-CoV2. Lane M: Protein marker; Lane 1: Purified protein after Ni2+-NTA chromatography: (a) Purified PLpro-CoV2 expressed under condition 5; (b) Purified PLpro-CoV2 expressed under condition 6; (c) Purified PLpro-CoV2 expressed under condition 9; (d) Purified PLpro-CoV2 expressed under condition 10. Details of the conditions are shown in Table 2.
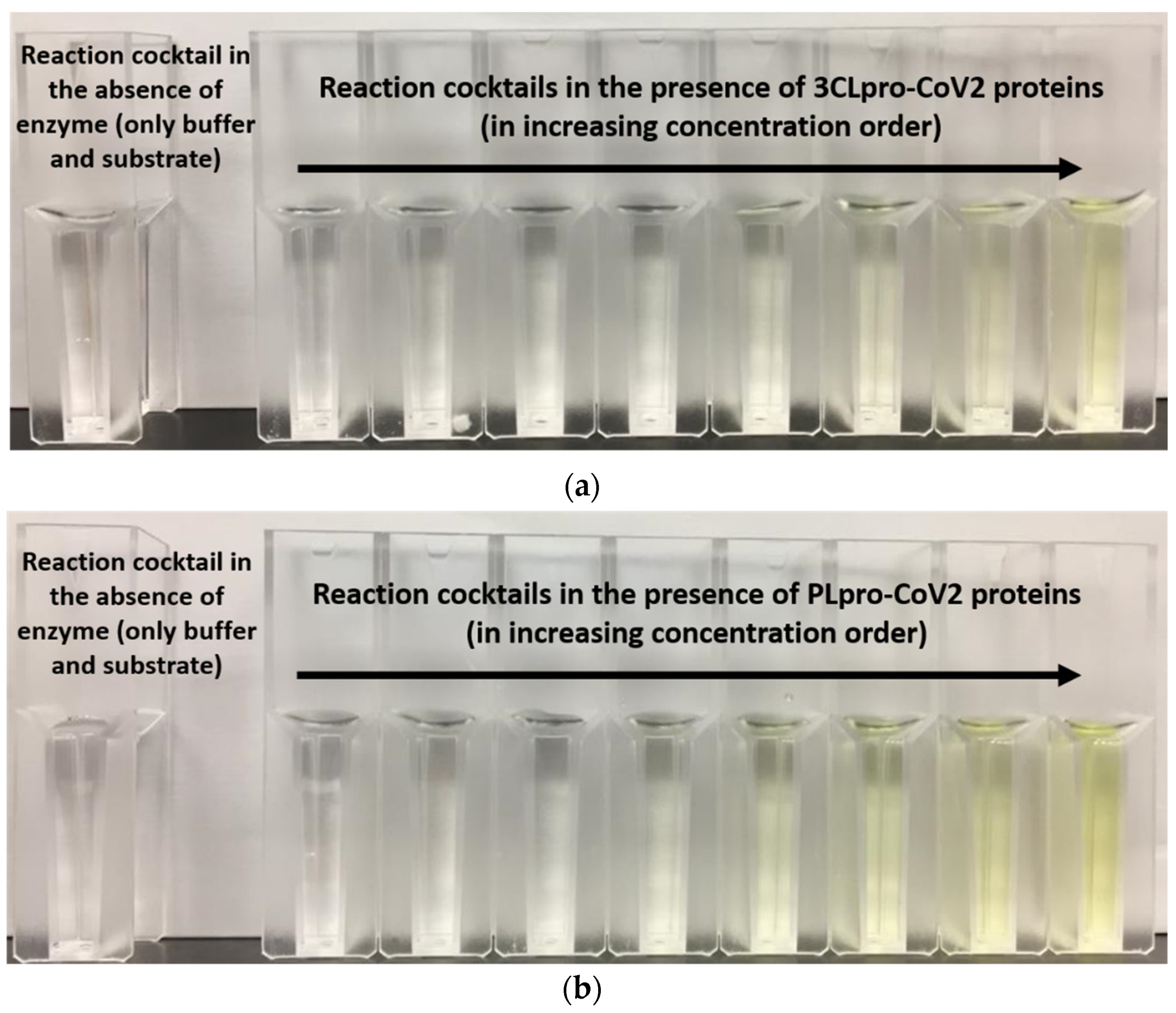
Purification profile for 3CLpro-CoV2 and PLpro–CoV2 were calculated (Table 3). This calculation was only done for the protein that was able to be purified using single-step Ni2+-NTA chromatography. Table 2 shows that under condition 2, with a single purification step, 3CLpro-CoV2 was able to be produced at the purification fold of close to 50, with a specific activity of 1.04 U/mg. Meanwhile, PLpro-CoV2 from condition 10 was able to be purified better than that of condition 9, with the purification fold of more than 100, with comparable specific activity to condition 9. Notably, the measurable specific activity of these proteases (Table 3) indicated that the purified 3CLpro-CoV2 and PLpro-CoV2 were enzymatically active. Qualitatively, the active forms of both proteases were also demonstrated by the changes on the assay cocktail, whereby the yellow color formation was observed in the presence of 3CLpro-CoV2 or PLpro-CoV2 (Figure 7). The yellow color is formed due to the release of pNA moiety of the substrate upon the cleavage of the substrate by the active protease.

Table 3.
Purification profile of SARS-CoV-2 proteases.
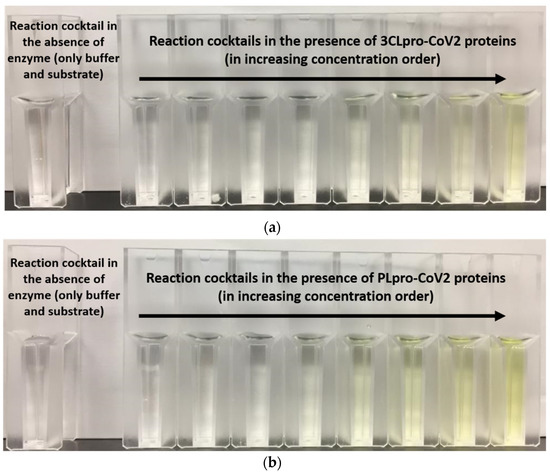
Figure 7.
The formation of yellow color in the reaction cocktails of (a) 3CLpro-CoV2 and (b) PLpro-CoV2.
3. Methods
3.1. Expression and Purification of 3CLpro-CoV2
The expression system of 3CLpro-CoV2 was obtained from Andrey Kovalevsky (Oak Ridge National Laboratory, Oak Ridge, TN, USA) as described in Kneller et al. [14]. In this system, the gene encoding of 3CLpro-CoV2 was inserted into the pD451-SR plasmid, resulting in an expression system of pD451-3CLpro. The pD451-3CLpro was transformed into E. coli strain BL21(DE3) [19]. The positive transformants were selected and cultured in Luria Bertani (LB) medium containing 35 µg/mL kanamycin at 37 °C, 180 rpm overnight. Approximately 2% of the bacterial suspensions were transferred into the larger culture volume of LB medium containing the antibiotic and incubated at 37 °C, 180 rpm. The protein expression was induced with 0.5 mM of isopropyl β-d-1-thiogalactosidase (IPTG) and incubated at two different conditions (refer to Table 4) once the OD600nm reached 0.8.

Table 4.
Expression conditions of SARS-CoV-2 proteases.
3.2. Expression and Purification of PLpro-CoV2
The expression system of PLpro-CoV2 was obtained from Prof. Shaun K. Olsen (University of South Carolina, USA) as described in Rut et al. [9]. Under this system, the gene encoding of PLpro-CoV2 was inserted into the pET21a plasmid, resulting in an expression system of pET21-PLpro. The pET21-PLpro was transformed into two E. coli strains of BL21(DE3) or BL21-CodonPlus(DE3). The positive transformants were selected and cultured in LB medium supplemented with respective antibiotics (100 µg/mL ampicillin for E. coli BL21(DE3); 100 µg/mL ampicillin with 25 µg/mL chloramphenicol for E. coli BL21-CodonPlus(DE3)) at 37 °C, 180 rpm for overnight. Similar to 3CLpro-CoV2, 2% of the bacterial suspensions were transferred into the larger culture volume of LB medium containing antibiotics and incubated at 37 °C, 180 rpm. The expressions of recombinant PLpro-CoV2 were conducted at eight different conditions (refer to Table 4).
3.3. Cell Harvesting
The cells were centrifuged at 8000× g at 4 °C for 10 min, followed by washing to completely remove the remaining medium. The washed cells were then suspended in lysis buffer and sonicated on ice. The cell debris from the sonication was removed through centrifugation at 35,000× g (Beckman Optima L-100K, Brea, CA, USA) for 30 min at 4 °C. The supernatant (soluble fraction) was then collected and used for purification steps [20].
3.4. Purification of Recombinant Proteins
The purifications of all recombinant proteins were conducted using an ÄKTA Pure liquid-chromatography system (GE Healthcare, Chicago, IL, USA) by Ni2+-NTA affinity chromatography. All purifications were run under the same flowrate. The 5 mL HisTrap HP column (GE Healthcare, USA) was firstly equilibrated with lysis buffer (20 mM Tris-HCl pH 8.0, 40 mM imidazole, 150 mM NaCl, 1 mM DTT). Before loading onto the column, the soluble fractions were firstly filtered using a 0.22 µm filter. The loading of the filtered sample onto the column was performed at the flow rate of 1 mL/min. The elution of bound proteins was conducted through a linear gradient of increasing concentration (0–500 mM) of imidazole in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT.
3.5. SDS-PAGE
Confirmation of expression, solubility and purity of the target proteins was done using 15% sodium SDS-PAGE [21]. The gel was stained with Coomassie staining and visualized using a Gel DocTM XR+ imager (Biorad, Hercules, CA, USA).
3.6. Purification Profiles
The concentration of proteins was calculated using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at 280 nm. For this purpose, the extinction coefficient (ε) at 0.1% (1 mg/mL) of 1.65 and 0.70 were used for 3CLpro-CoV2 and PLpro-CoV2, respectively, which were calculated using ε = 1576 M−1 cm−1 for Tyr and 5225 M−1 cm−1 for Trp at 280 nm [22]. The total activity protein was calculated based on the following formula (1):
To obtain unit activity, the activity of 3CLpro-CoV2 and PLpro-CoV2 was measured using Z-TSAVLQ-pNA and L-Pyroglutamyl-L-phenylalanyl-Leucine-pNA substrates, respectively, according to Cheng et al. [23] and Bala et al. [24]. One unit activity is defined as the amount of enzyme required to produce 1µmol of the product in 1 min reaction time. Further, the specific activity was then calculated based on the following formula (2):
The purification yield was calculated based on formula (3):
Meanwhile, the purification (fold) was calculated based on formula (4):
Author Contributions
Writing—original draft preparation, R.R.; Visualization, R.R.; Methodology, R.R.; Investigation, R.R.; Supervision, V.K.S. and C.B.; Writing—review and editing, V.K.S. and C.B.; Conceptualization, C.B.; Funding acquisition, C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research is under SDK0208-2020 Research Grant of Universiti Malaysia Sabah (V.K.S., C.B.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in https://doi.org/10.5281/zenodo.5503693.
Acknowledgments
We would like to acknowledge Universiti Malaysia Sabah for the funding support on this study (Skim Dana Khas/SDK 0208-2020). We thank Haslina Asis for the constructive comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorbalenya, A.E.; Haagmans, B.L.; Sola, I. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-Ncov and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Altay, O.; Mohammadi, E.; Lam, S.; Turkez, H.; Boren, J.; Nielsen, J.; Uhlen, M.; Mardinoglu, A. Current status of COVID-19 therapies and drug repositioning applications. iScience 2020, 23, 101303. [Google Scholar] [CrossRef] [PubMed]
- Ziebuhr, J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004, 7, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Ivanov, K.A.; Putics, Á.; Hertzig, T.; Schelle, S.; Bayer, S.; WeiBbrich, B.; Snijder, E.J.; Rabenau, H.; Doerr, H.W.; et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003, 84, 2305–2315. [Google Scholar] [CrossRef]
- Ul Qamar, M.H.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef]
- Alamri, M.A.; ul Qamar, M.T.; Mirza, M.U.; Alqahtani, S.M.; Froeyen, M.; Chen, L.L. Discovery of human coronaviruses pan-papain-like protease inhibitors using computational approaches. J. Pharm. Anal. 2020, 10, 546–559. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M.; et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef]
- Devaraj, S.G.; Wang, N.; Chen, Z.; Zhen, Z.; Tseng, M.; Barretto, N.; Lin, R.; Peters, C.J.; Tseng, C.K.; Baker, S.C.; et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007, 282, 32208–32221. [Google Scholar] [CrossRef] [Green Version]
- Clementz, M.A.; Chen, Z.; Banach, B.S.; Wang, Y.; Sun, L.; Ratia, K.; Baez-Santos, Y.M.; Wang, J.; Tukuyama, J.; Ghosh, A.K.; et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2019, 84, 4619–4629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frieman, M.; Ratia, K.; Johnston, R.E.; Mesecar, A.D.; Baric, R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappa B signaling. J. Virol. 2009, 83, 6689–6705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osipiuk, J.; Azizi, S.A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Kneller, D.W.; Phillips, G.; O’neil, H.M.; Jedrzejczak, R.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef]
- Forstner, M.; Leder, L.; Mayr, L.M. Optimization of protein expression systems for modern drug discovery. Expert Rev. Proteomic 2007, 4, 67–78. [Google Scholar] [CrossRef]
- Smith, C. Striving for purity: Advances in protein purification. Nat. Methods 2005, 2, 71–77. [Google Scholar] [CrossRef]
- De Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef] [Green Version]
- LaVallie, E.R.; DiBlasio, E.A.; Kovacic, S.; Grant, K.L.; Schendel, P.F.; McCoy, J.M. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology 1993, 11, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Froger, A.; Hall, J.E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 2007, 6, 253. [Google Scholar] [CrossRef] [PubMed]
- Razali, R.; Kumar, V.; Budiman, C. Structural insights into the enzymatic activity of protease bromelain of MD2 pineapple. Pak. J. Biol. Sci. 2020, 23, 829–838. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Goodwin, T.; Morton, R. The spectrophotometric determination of tyrosine and tryptophan in proteins. Biochem. J. 1946, 40, 628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, S.; Chang, G.; Chou, C. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 2010, 98, 1327–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bala, M.; Mel, M.; Jami, M.S.; Amid, A.; Salleh, H.M. Kinetic studies on recombinant stem bromelain. Adv. Enzym. Res. 2013, 1, 52–60. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).