GCSA-SegFormer: Transformer-Based Segmentation for Liver Tumor Pathological Images
Abstract
1. Introduction
2. Background and Related Work
3. Datasets and Method
3.1. Datasets
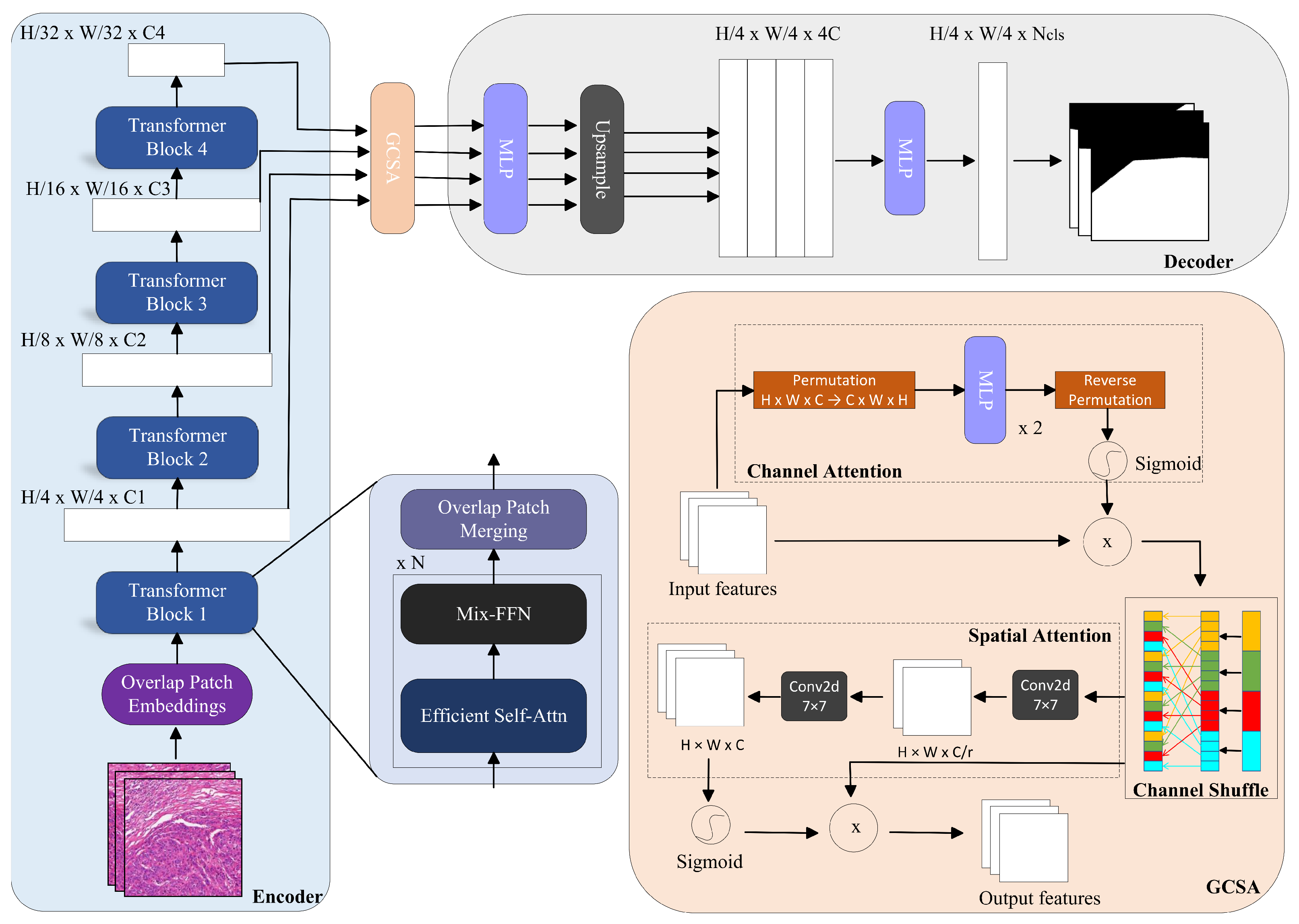
3.2. Method
- (1)
- Channel Attention
- (2)
- Channel Shuffling
- (3)
- Spatial Attention
4. Experiments and Results
4.1. Experimental Environment and Parameters
4.2. Evaluation Metrics
4.3. Results and Analysis
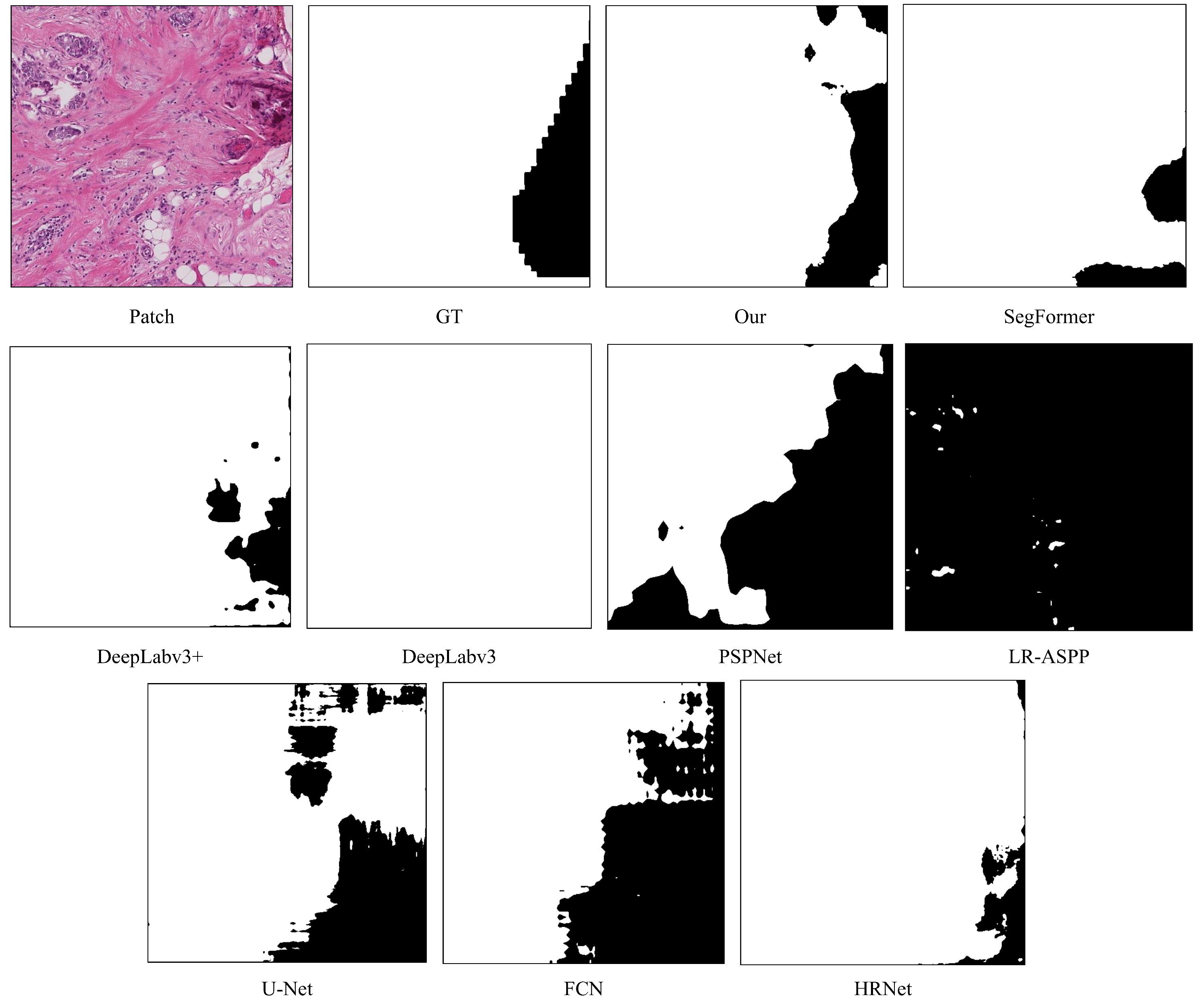
4.3.1. Liver Dataset
4.3.2. BACH Dataset
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, Y.; Wang, K.; Wang, Y.R.; Xiang, Y.J.; Liu, Z.H.; Feng, J.K.; Cheng, S.Q. Preoperative and Prognostic Prediction of Microvascular Invasion in Hepatocellular Carcinoma: A Review Based on Artificial Intelligence. Technol. Cancer Res. Treat. 2023, 22, 15330338231212726. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lauwers, G.Y.; Terris, B.; Balis, U.J.; Batts, K.P.; Regimbeau, J.M.; Chang, Y.; Graeme-Cook, F.; Yamabe, H.; Ikai, I.; Cleary, K.R.; et al. International Cooperative Study Group on Hepatocellular Carcinoma. Prognostic histologic indicators of curatively resected hepatocellular carcinomas: A multi-institutional analysis of 425 patients with definition of a histologic prognostic index. Am. J. Surg. Pathol. 2002, 26, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, V.; Llovet, J.M.; Miceli, R.; Bhoori, S.; Schiavo, M.; Mariani, L.; Camerini, T.; Roayaie, S.; Schwartz, M.E.; Grazi, G.L.; et al. Metroticket Investigator Study Group. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: A retrospective, exploratory analysis. Lancet Oncol. 2009, 10, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.C.; Chow, P.K.; Allen, J.C.; Chia, G.S.; Lim, M.; Cheow, P.C.; Chung, A.Y.; Ooi, L.L.; Tan, S.B. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg. 2011, 254, 108–113. [Google Scholar] [CrossRef]
- Nalisnik, M.; Gutman, D.A.; Kong, J.; Cooper, L.A. An Interactive Learning Framework for Scalable Classification of Pathology Images. Proc IEEE Int. Conf. Big Data 2015, 928–935. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khened, M.; Kori, A.; Rajkumar, H.; Krishnamurthi, G.; Srinivasan, B. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci. Rep. 2021, 11, 11579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tang, L.; Diao, S.; Li, C.; He, M.; Ru, K.; Qin, W. Global contextual representation via graph-transformer fusion for hepatocellular carcinoma prognosis in whole-slide images. Comput. Med. Imaging Graph. 2024, 115, 102378. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.C.; Picarsic, J.; McGenity, C.; Treanor, D.; Williams, B.; Sebire, N.J. Whole Slide Imaging, Artificial Intelligence, and Machine Learning in Pediatric and Perinatal Pathology: Current Status and Future Directions. Pediatr. Dev. Pathol. 2024, 18, 10935266241299073. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17), Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 6000–6010. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; An kumar, A.; Álvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. arXiv 2021, arXiv:2105.15203. [Google Scholar]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:abs/1505.04597. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar]
- Chen, L.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:abs/1706.05587. [Google Scholar]
- Chen, L.; Zhu, Y.; Papandreou, G.; Schroff, F.L.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. arXiv 2018, arXiv:1802.02611. [Google Scholar]
- Howard, A.G.; Sandler, M.; Chu, G.; Chen, L.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep High-Resolution Representation Learning for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 3349–3364. [Google Scholar] [CrossRef]
- Jiayin, Z.; Weimin, H.; Wei, X.; Chen, W.; Tian, Q. Segmentation of hepatic tumor from abdominal CT data using an improved support vector machine framework. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 3347–3350. [Google Scholar]
- Christ, P.F.; Elshaer, M.E.A.; Ettlinger, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; Rempfler, M.; Armbruster, M.; Hofmann, F.; D’Anastasi, M.; et al. Automatic liver and lesion segmentation in ct using cascaded fully convolutional neural networks and 3D conditional random fields. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Istanbul, Turkey, 17–21 October 2016; pp. 415–423. [Google Scholar]
- Chen, Y.; Zheng, C.; Hu, F.; Zhou, T.; Feng, L.; Xu, G.; Yi, Z.; Zhang, X. Efficient two-step liver and tumour segmentation on abdominal CT via deep learning and a conditional random field. Comput. Biol. Med. 2022, 150, 106076. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z. Computational Models for Automated Histopathological Assessment of Colorectal Liver Metastasis Progression. Ph.D. Thesis, Queen Mary University of London, London, UK, 2019. [Google Scholar]
- Zhai, Z.; Wang, C.; Sun, Z.; Cheng, S.; Wang, K. Deep Neural Network Guided by Attention Mechanism for Segmentation of Liver Pathology Image. In Proceedings of the 2021 Chinese Intelligent Systems Conference, Fuzhou, China, 16–17 October 2021; Springer: Singapore, 2022; pp. 425–435. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Han, X. Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv 2017, arXiv:1704.07239. [Google Scholar]
- Tian, J.; Li, C.; Shi, Z.; Xu, F. A diagnostic report generator from CT volumes on liver tumor with semi-supervised attention mechanism. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part II 11. Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 702–710. [Google Scholar]
- Abdalla, A.; Ahmed, N.; Dakua, S.; Balakrishnan, S.; Abinahed, J. A surgical-oriented liver segmentation approach using deep learning. In Proceedings of the 2020 IEEE International Conference on Informatics, IoT, and Enabling Technologies (ICIoT), Doha, Qatar, 2–5 February 2020. [Google Scholar]
- Zheng, S.; Fang, B.; Li, L.; Gao, M. Automatic liver lesion segmentation in CT combining fully convolutional networks and non-negative matrix factorization. In Proceedings of the Imaging for Patient-Customized Simulations and Systems for Point-of-Care Ultrasound: International Workshops, BIVPCS 2017 and POCUS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, 14 September 2017; Proceedings. Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 44–51. [Google Scholar]
- Wang, G.; Zhu, S.; Li, X. Comparison of values of CT and MRI imaging in the diagnosis of hepatocellular carcinoma and analysis of prognostic factors. Oncol. Lett. 2019, 17, 1184–1188. [Google Scholar] [CrossRef]
- Chlebus, G.; Meine, H.; Abolmaali, N.; Schenk, A. Automatic liver and tumor segmentation in late-phase MRI using fully convolutional neural networks. Proc. CURAC 2018, 195–200. [Google Scholar]
- Pan, F.; Huang, Q.; Li, X. Classification of liver tumors with CEUS based on 3D-CNN. In Proceedings of the 2019 IEEE 4th International Conference on Advanced Robotics and Mechatronics (ICARM), Toyonaka, Japan, 3–5 July 2019; pp. 845–849. [Google Scholar]
- Schmauch, B.; Herent, P.; Jehanno, P.; Dehaene, O.; Saillard, C.; Aubé, C.; Luciani, A.; Luciani, N.; Luciani, S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn. Interv. Imaging 2019, 100, 227–233. [Google Scholar] [CrossRef]
- Duanmu, H.; Bhattarai, S.; Li, H.; Shi, Z.; Wang, F.; Teodoro, G.; Gogineni, K.; Subhedar, P.; Kiraz, U.; Janssen, E.A.M.; et al. A spatial attention guided deep learning system for prediction of pathological complete response using breast cancer histopathology images. Bioinformatics 2022, 38, 4605–4612. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, T.; Liu, L.; Lu, H.; Niu, X. GIAE-Net: A gradient-intensity oriented model for multimodal lung tumor image fusion. Eng. Sci. Technol. Int. J. 2024, 54, 101727. [Google Scholar] [CrossRef]
- Liu, R.; Cheng, J.; Zhu, X.; Liang, H. Multi-modal brain tumor segmentation based on self-organizing active contour model. In Proceedings of the Pattern Recognition: 7th Chinese Conference, CCPR 2016, Chengdu, China, 5–7 November 2016; Proceedings, Part II 7. Springer: Singapore, 2016; pp. 486–498. [Google Scholar]


| Model | Backbone | MIoU (%) | MPA (%) | Accuracy (%) |
|---|---|---|---|---|
| DeepLabv3+ | MobileNetV2 | 86.25 | 92.80 | 95.60 |
| UNet | VGG-16 | 89.77 | 93.78 | 96.71 |
| PSPNet | MobileNetV2 | 87.58 | 94.52 | 96.14 |
| HRNet | HRNetV2 | 87.53 | 93.31 | 96.03 |
| FCN | ResNet-50 | 89.00 | 93.35 | 96.60 |
| DeepLabv3 | ResNet-50 | 83.40 | 88.22 | 94.80 |
| LR-ASPP | MobileNetV3 | 88.50 | 95.00 | 96.20 |
| SegFormer | MIT-B0 | 91.07 | 94.60 | 97.26 |
| Our Model | MIT-B0 | 92.19 | 95.75 | 97.58 |
| Model | Backbone | MIoU (%) | MPA (%) | Accuracy (%) |
|---|---|---|---|---|
| DeepLabv3+ | MobileNetV2 | 68.57 | 84.39 | 92.28 |
| UNet | VGG-16 | 68.74 | 86.99 | 92.63 |
| PSPNet | MobileNetV2 | 68.66 | 85.09 | 92.39 |
| HRNet | HRNetV2 | 70.68 | 87.02 | 93.02 |
| FCN | ResNet-50 | 62.50 | 66.95 | 91.90 |
| DeepLabv3 | ResNet-50 | 60.30 | 64.80 | 91.40 |
| LR-ASPP | MobileNetV3 | 68.30 | 72.50 | 93.10 |
| SegFormer | MIT-B0 | 70.45 | 89.78 | 93.24 |
| Our Model | MIT-B0 | 71.71 | 90.17 | 93.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, J.; Yang, S.; Li, W.; Cheng, S. GCSA-SegFormer: Transformer-Based Segmentation for Liver Tumor Pathological Images. Bioengineering 2025, 12, 611. https://doi.org/10.3390/bioengineering12060611
Wen J, Yang S, Li W, Cheng S. GCSA-SegFormer: Transformer-Based Segmentation for Liver Tumor Pathological Images. Bioengineering. 2025; 12(6):611. https://doi.org/10.3390/bioengineering12060611
Chicago/Turabian StyleWen, Jingbin, Sihua Yang, Weiqi Li, and Shuqun Cheng. 2025. "GCSA-SegFormer: Transformer-Based Segmentation for Liver Tumor Pathological Images" Bioengineering 12, no. 6: 611. https://doi.org/10.3390/bioengineering12060611
APA StyleWen, J., Yang, S., Li, W., & Cheng, S. (2025). GCSA-SegFormer: Transformer-Based Segmentation for Liver Tumor Pathological Images. Bioengineering, 12(6), 611. https://doi.org/10.3390/bioengineering12060611









