Feasibility Study of CLIP-Based Key Slice Selection in CT Images and Performance Enhancement via Lesion- and Organ-Aware Fine-Tuning
Abstract
1. Introduction
- CLIP-based slice selector for clinical CT.We present the first CLIP-based key slice selector that can be used to construct large-scale image–text pairs directly from routine CT studies. Feasibility is evaluated with both automatic metrics and radiologist review.
- Efficient dataset curation protocol.We describe and release a lightweight procedure that uses a small number of studies to generate balanced fine-tuning data containing both lesion-level and organ-level (negative) sentence–slice pairs.
- Radiologist acceptance gains.For abnormal findings, the expert “acceptance ratio” (i.e., the proportion of automatically selected slices judged correct) is summarized in Table 1.
2. Related Works
2.1. MedVQA Dataset Construction
2.2. Slice Selection
2.3. Multimodal Learning
3. Materials and Methods
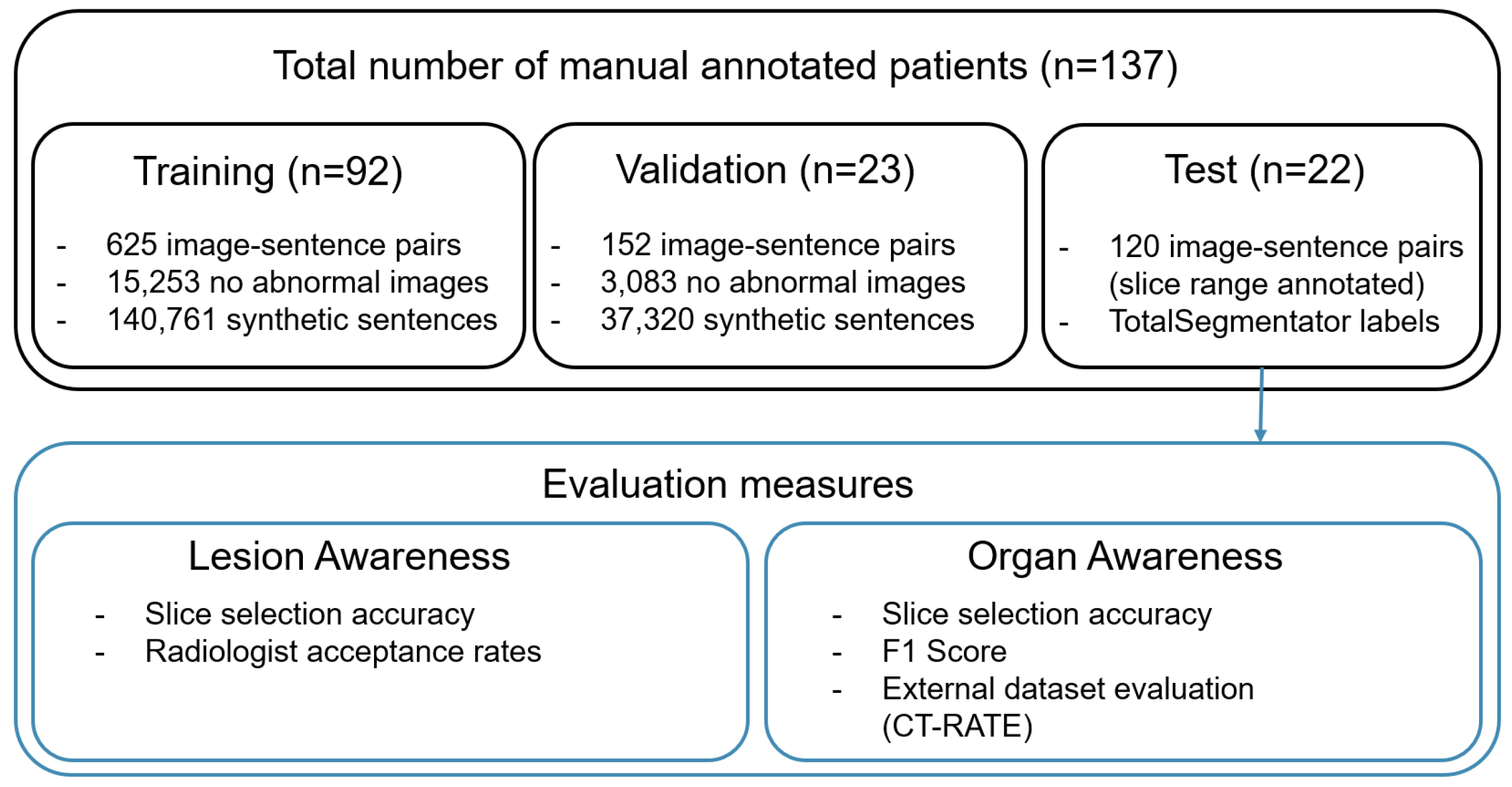
3.1. Dataset
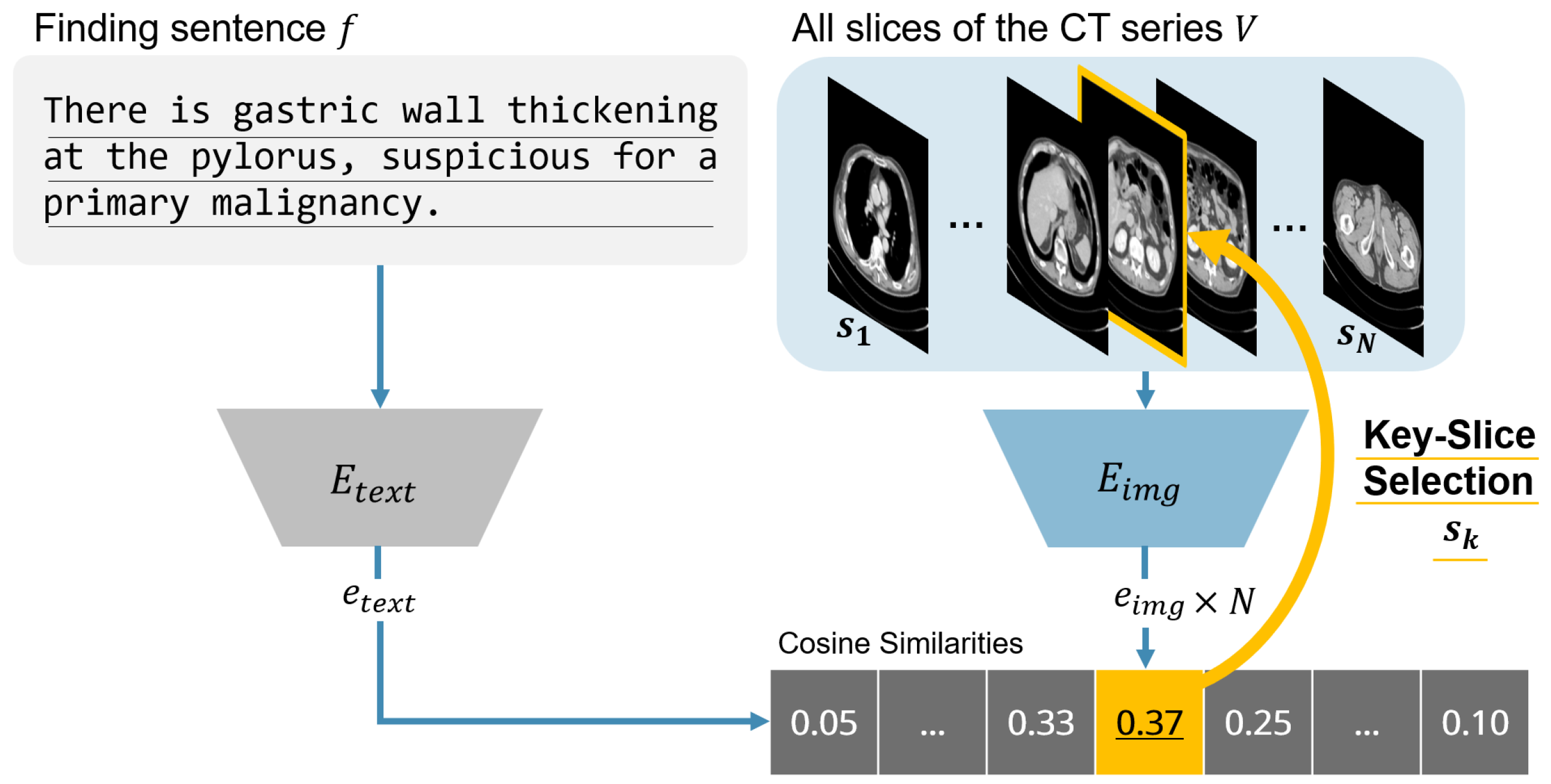
3.2. Slice Selection Algorithm
| Algorithm 1 CLIP-based key slice selection. |
|
3.3. Models and Training Details
4. Results
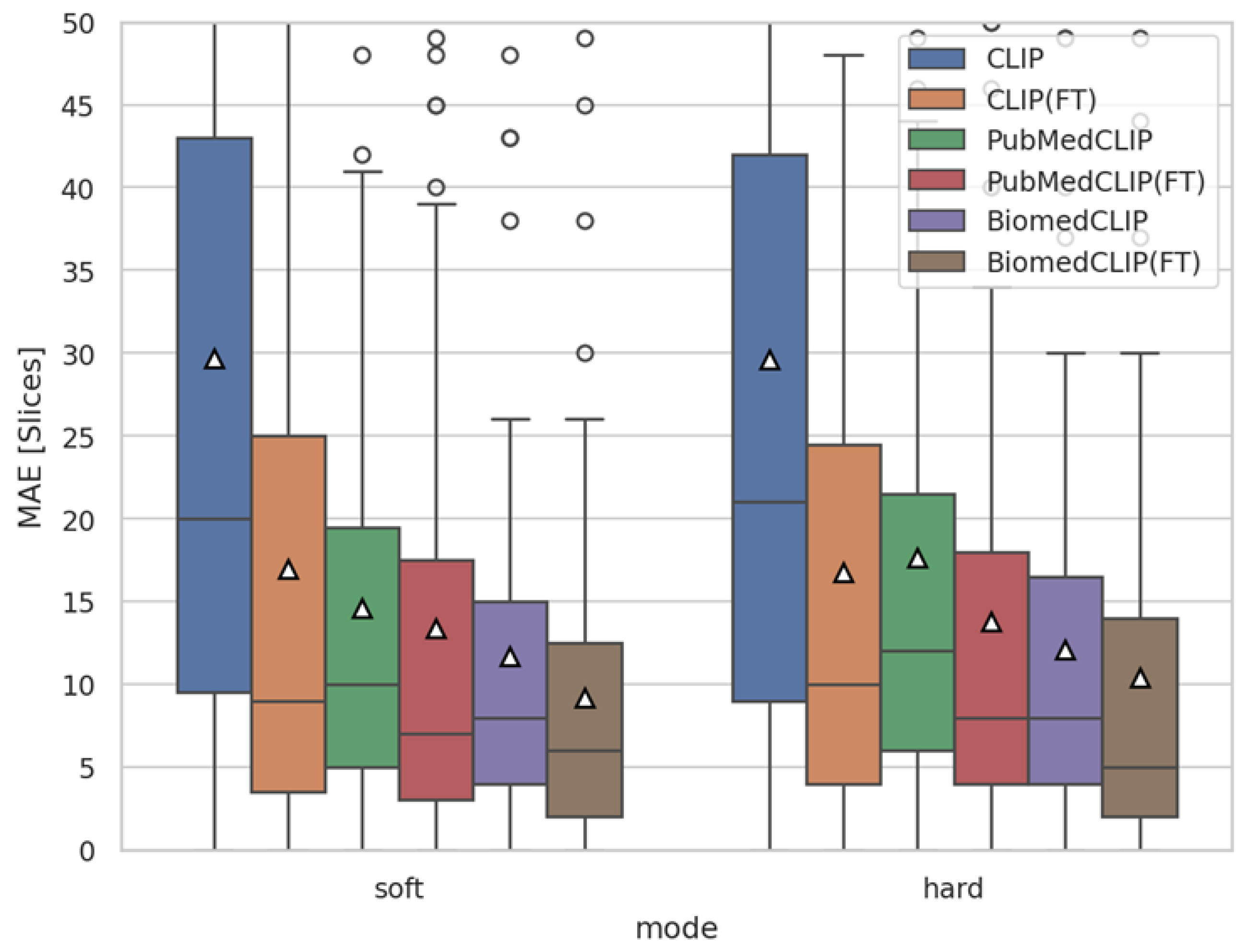
4.1. Training Verification via Slice-Level MAE
4.2. Lesion Awareness
4.3. Organ Awareness
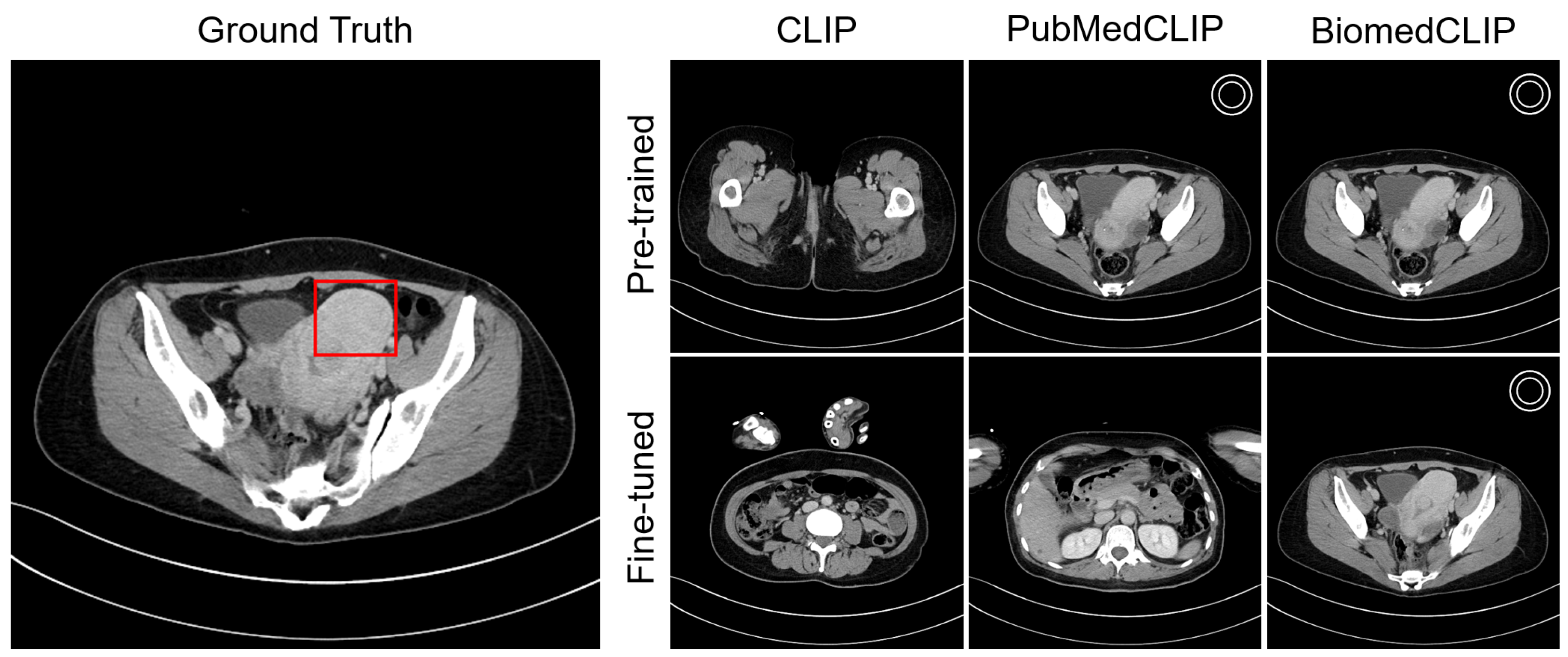
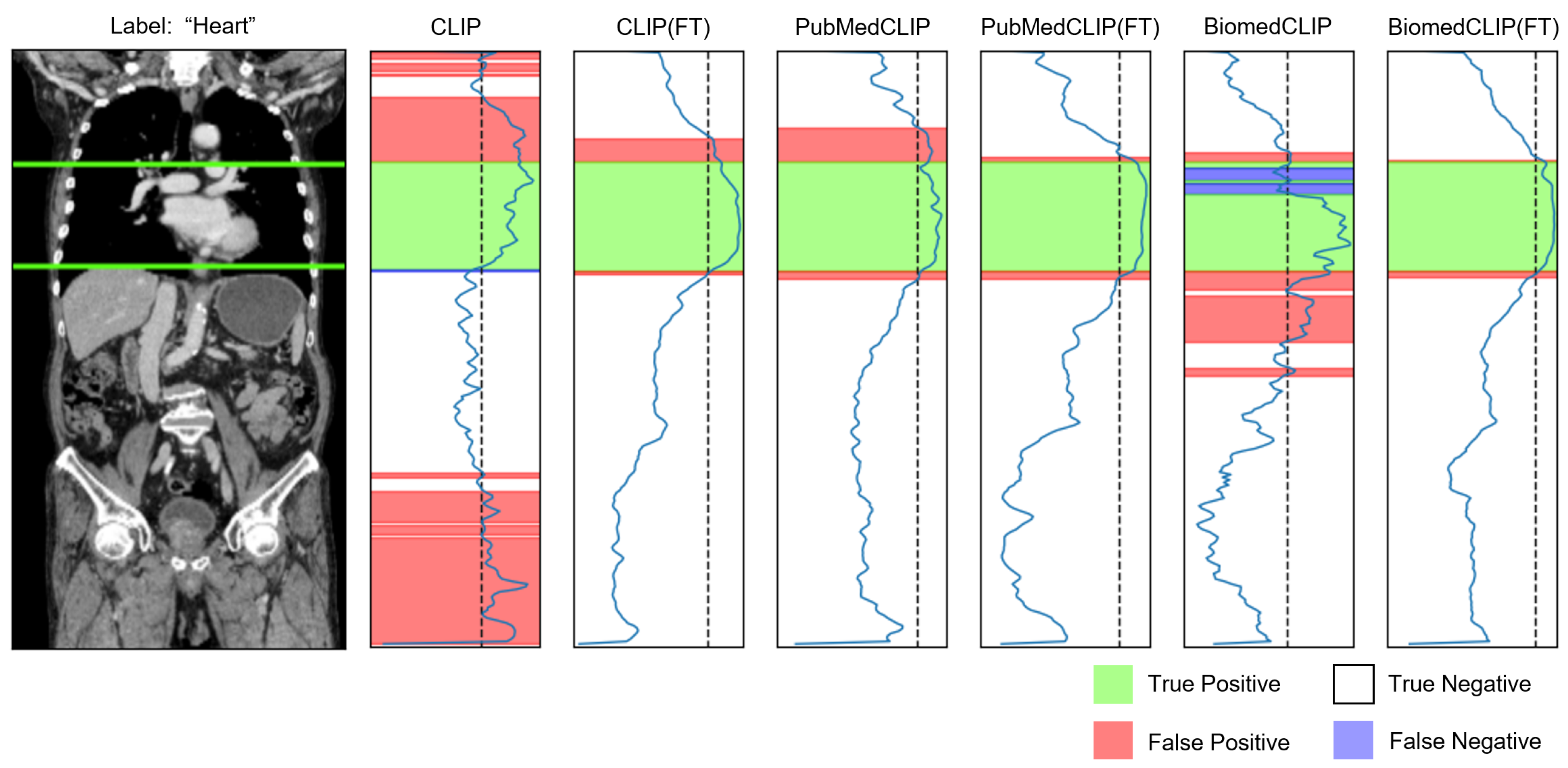
4.4. Visualization
5. Discussion
- Segmenting diagnostic reports into individual finding units;
- Acquiring the corresponding imaging series (including selection of contrast phases);
- Extracting key slices using the slice selector;
- Generating VQA pairs from each finding sentence through rule-based systems or LLMs.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CLIP | Contrastive Language–Image Pre-training |
| CT | Computed Tomography |
| VLM | Vision–Language Model |
| MedVQA | Medical Visual Question Answering |
| LLM | Large Language Model |
| SOTA | State of the Art |
| VQA | Visual Question Answering |
Appendix A. Finding Text Generation
Appendix A.1. Rule-Based
Appendix A.2. LLM-Based
Appendix B. Detailed Organ-Aware Results
| Organ | CLIP | CLIP(FT) | PubMedCLIP | PubMedCLIP(FT) | BiomedCLIP | BiomedCLIP(FT) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Word | Sentence | Word | Sentence | Word | Sentence | Word | Sentence | Word | Sentence | Word | Sentence | |
| aorta | 0.679 | 0.660 | 0.960 | 0.960 | 0.852 | 0.836 | 0.966 | 0.967 | 0.858 | 0.754 | 0.943 | 0.948 |
| colon | 0.676 | 0.747 | 0.877 | 0.877 | 0.817 | 0.859 | 0.880 | 0.878 | 0.878 | 0.886 | 0.878 | 0.878 |
| duodenum | 0.450 | 0.522 | 0.761 | 0.775 | 0.749 | 0.762 | 0.804 | 0.841 | 0.665 | 0.724 | 0.798 | 0.828 |
| esophagus | 0.614 | 0.731 | 0.932 | 0.931 | 0.936 | 0.918 | 0.958 | 0.958 | 0.873 | 0.895 | 0.973 | 0.972 |
| gallbladder | 0.250 | 0.235 | 0.494 | 0.495 | 0.450 | 0.472 | 0.547 | 0.547 | 0.432 | 0.451 | 0.543 | 0.537 |
| heart | 0.554 | 0.643 | 0.933 | 0.941 | 0.842 | 0.832 | 0.945 | 0.943 | 0.664 | 0.841 | 0.956 | 0.955 |
| kidney | 0.684 | 0.576 | 0.886 | 0.887 | 0.767 | 0.788 | 0.885 | 0.884 | 0.733 | 0.696 | 0.917 | 0.917 |
| liver | 0.600 | 0.570 | 0.900 | 0.901 | 0.791 | 0.832 | 0.928 | 0.921 | 0.821 | 0.750 | 0.914 | 0.912 |
| pancreas | 0.588 | 0.518 | 0.801 | 0.807 | 0.731 | 0.716 | 0.836 | 0.841 | 0.730 | 0.755 | 0.838 | 0.824 |
| stomach | 0.402 | 0.418 | 0.720 | 0.737 | 0.650 | 0.612 | 0.780 | 0.767 | 0.699 | 0.673 | 0.823 | 0.827 |
| spleen | 0.325 | 0.409 | 0.731 | 0.744 | 0.619 | 0.685 | 0.809 | 0.811 | 0.665 | 0.639 | 0.810 | 0.800 |
| thyroid gland | 0.363 | 0.227 | 0.744 | 0.744 | 0.552 | 0.576 | 0.648 | 0.647 | 0.219 | 0.643 | 0.727 | 0.728 |
| urinary bladder | 0.165 | 0.210 | 0.491 | 0.691 | 0.387 | 0.386 | 0.739 | 0.779 | 0.160 | 0.575 | 0.753 | 0.762 |
| trachea | 0.542 | 0.514 | 0.900 | 0.905 | 0.730 | 0.714 | 0.881 | 0.884 | 0.796 | 0.767 | 0.945 | 0.947 |
| lung | 0.816 | 0.827 | 0.964 | 0.963 | 0.954 | 0.904 | 0.976 | 0.975 | 0.906 | 0.907 | 0.980 | 0.981 |
Appendix C. Visualization
Appendix C.1. Lesion Awareness

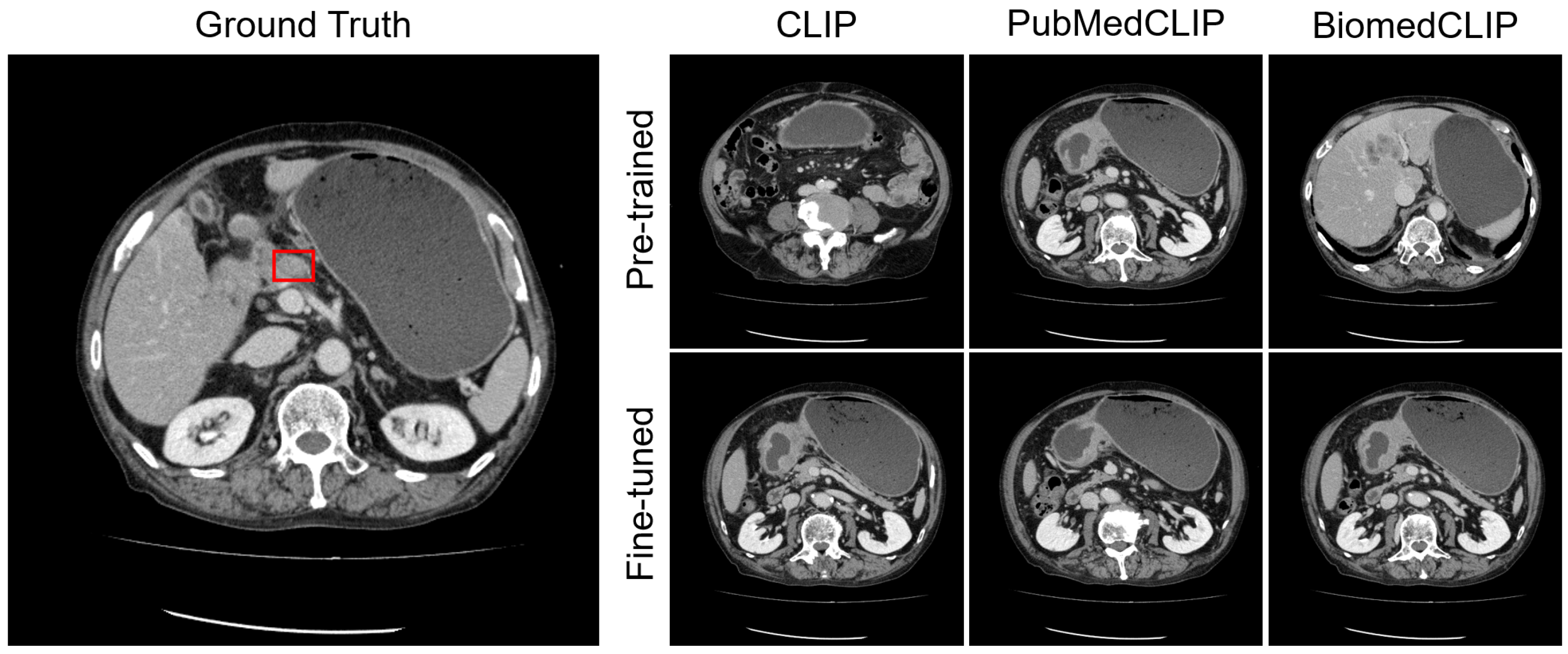
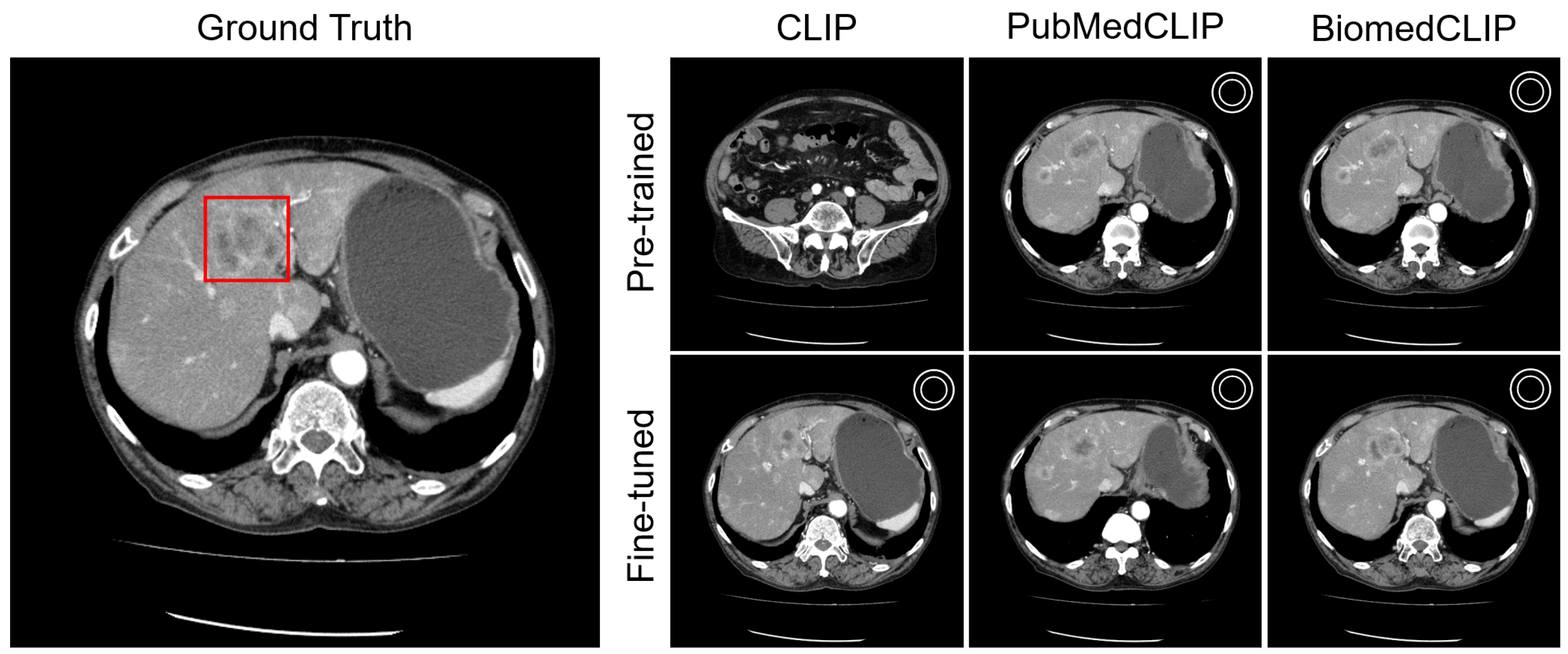

Appendix C.2. Organ Awareness
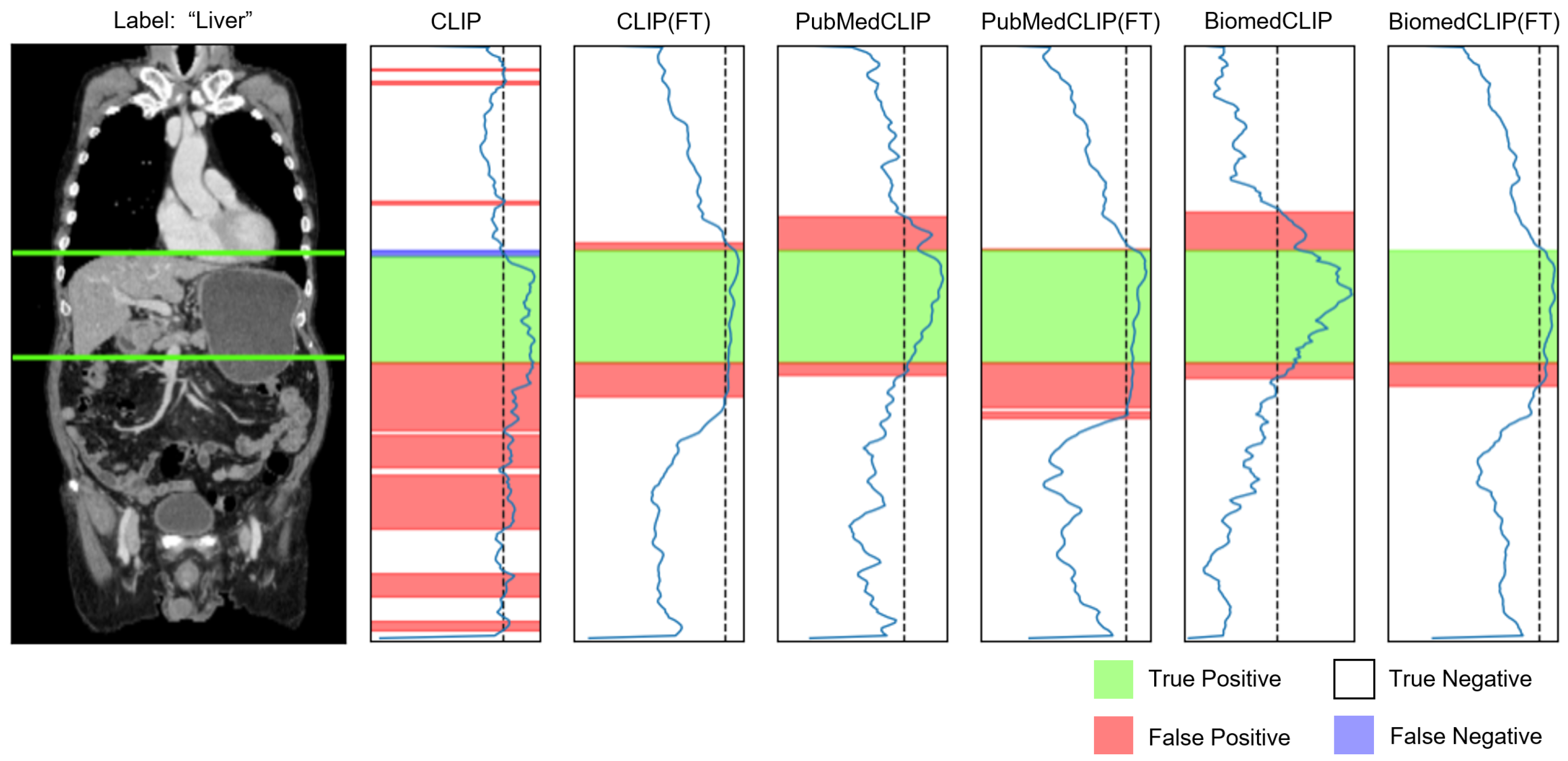
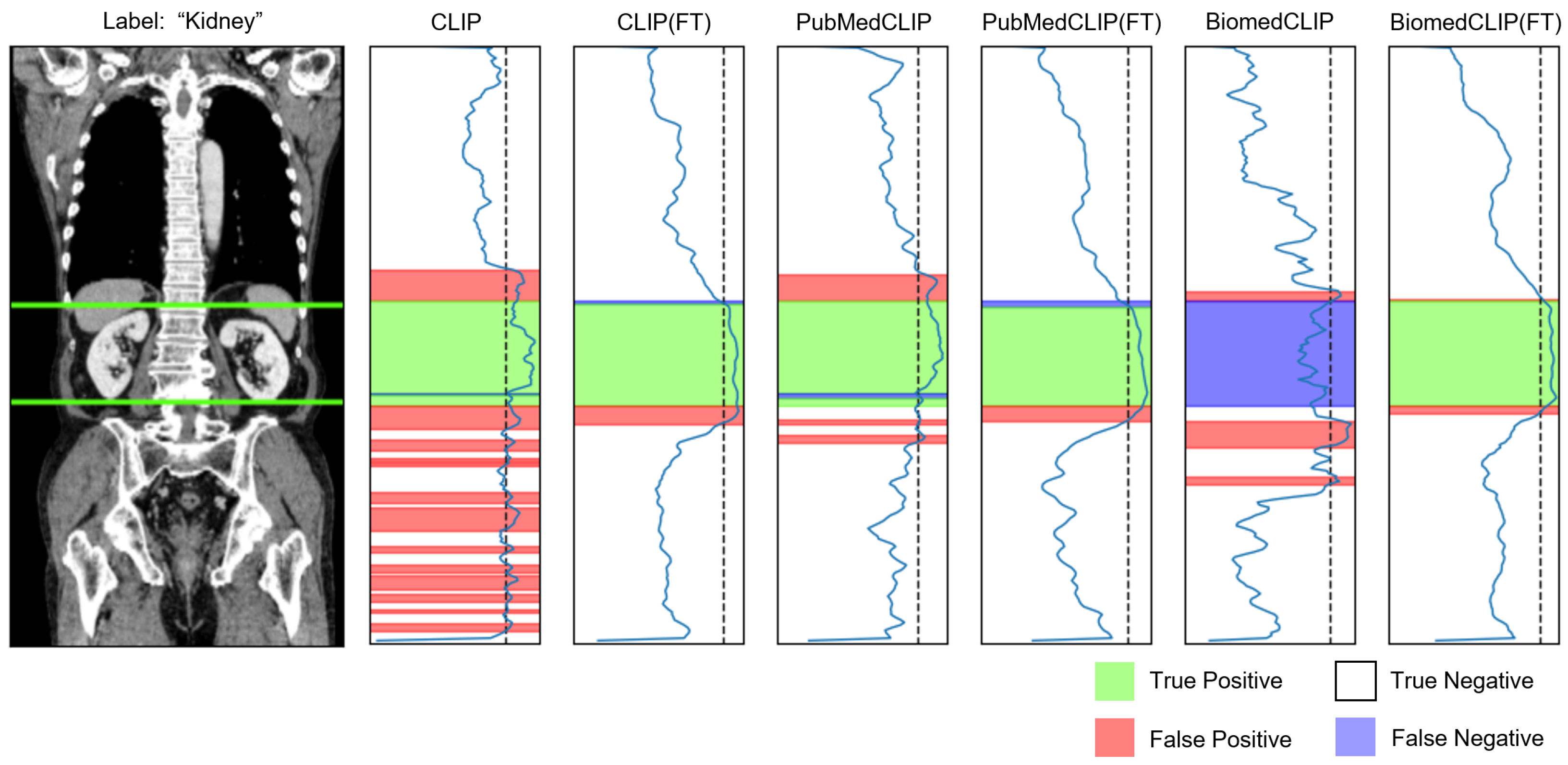
Appendix D. Impact of Organ-Aware Synthetic Data
| Method | Acc.@1 ↑ | Acc.@5 ↑ |
|---|---|---|
| BiomedCLIP | 44.83 | 60.92 |
| BiomedCLIP(FT) | 51.72 | 64.37 |
| BiomedCLIP(lesion-FT) | 45.98 | 63.22 |
References
- Li, F.; Zhang, R.; Zhang, H.; Zhang, Y.; Li, B.; Li, W.; Ma, Z.; Li, C. LLaVA-NeXT-Interleave: Tackling Multi-image, Video, and 3D in Large Multimodal Models. arXiv 2024, arXiv:2407.07895. [Google Scholar]
- Xiao, B.; Wu, H.; Xu, W.; Dai, X.; Hu, H.; Lu, Y.; Zeng, M.; Liu, C.; Yuan, L. Florence-2: Advancing a Unified Representation for a Variety of Vision Tasks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 16–22 June 2024; pp. 4818–4829. [Google Scholar]
- Chen, Z.; Wu, J.; Wang, W.; Su, W.; Chen, G.; Xing, S.; Zhong, M.; Zhang, Q.; Zhu, X.; Lu, L.; et al. InternVL: Scaling up Vision Foundation Models and Aligning for Generic Visual-Linguistic Tasks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 16–22 June 2024; pp. 24185–24198. [Google Scholar]
- Alayrac, J.B.; Donahue, J.; Luc, P.; Miech, A.; Barr, I.; Hasson, Y.; Lenc, K.; Mensch, A.; Millican, K.; Reynolds, M.; et al. Flamingo: A Visual Language Model for Few-Shot Learning. In Proceedings of the Advances in Neural Information Processing Systems, New Orleans, LA, USA, 28 November–9 December 2022; Koyejo, S., Mohamed, S., Agarwal, A., Belgrave, D., Cho, K., Oh, A., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2022; Volume 35, pp. 23716–23736. [Google Scholar]
- Lau, J.J.; Gayen, S.; Ben Abacha, A.; Demner-Fushman, D. A dataset of clinically generated visual questions and answers about radiology images. Sci. Data 2018, 5, 180251. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhan, L.M.; Xu, L.; Ma, L.; Yang, Y.; Wu, X.M. Slake: A Semantically-Labeled Knowledge-Enhanced Dataset For Medical Visual Question Answering. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1650–1654. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, C.; Zhao, Z.; Lin, W.; Zhang, Y.; Wang, Y.; Xie, W. PMC-VQA: Visual Instruction Tuning for Medical Visual Question Answering. arXiv 2024, arXiv:2305.10415. [Google Scholar]
- Li, C.; Wong, C.; Zhang, S.; Usuyama, N.; Liu, H.; Yang, J.; Naumann, T.; Poon, H.; Gao, J. LLaVA-Med: Training a Large Language-and-Vision Assistant for Biomedicine in One Day. In Proceedings of the Advances in Neural Information Processing Systems, New Orleans, LA, USA, 10–16 December 2023; Oh, A., Naumann, T., Globerson, A., Saenko, K., Hardt, M., Levine, S., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2023; Volume 36, pp. 28541–28564. [Google Scholar]
- Dong, W.; Shen, S.; Han, Y.; Tan, T.; Wu, J.; Xu, H. Generative Models in Medical Visual Question Answering: A Survey. Appl. Sci. 2025, 15, 2983. [Google Scholar] [CrossRef]
- Bae, S.; Kyung, D.; Ryu, J.; Cho, E.; Lee, G.; Kweon, S.; Oh, J.; Ji, L.; Chang, E.; Kim, T.; et al. EHRXQA: A multi-modal question answering dataset for electronic health records with chest x-ray images. Adv. Neural Inf. Process. Syst. 2024, 36, 3867–3880. [Google Scholar]
- Bai, F.; Du, Y.; Huang, T.; Meng, M.Q.H.; Zhao, B. M3D: Advancing 3D Medical Image Analysis with Multi-Modal Large Language Models. arXiv 2024, arXiv:2404.00578. [Google Scholar]
- Blankemeier, L.; Cohen, J.P.; Kumar, A.; Van Veen, D.; Gardezi, S.J.S.; Paschali, M.; Chen, Z.; Delbrouck, J.B.; Reis, E.; Truyts, C.; et al. Merlin: A Vision Language Foundation Model for 3D Computed Tomography. Res. Sq. 2024, rs.3.rs-4546309. [Google Scholar] [CrossRef]
- Hamamci, I.E.; Er, S.; Menze, B. CT2Rep: Automated Radiology Report Generation for 3D Medical Imaging. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2024, Marrakesh, Morocco, 6–10 October 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Springer: Cham, Switzerland, 2024; pp. 476–486. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning Transferable Visual Models From Natural Language Supervision. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Meila, M., Zhang, T., Eds.; PMLR: Washington, DC, USA, 2021; Volume 139, pp. 8748–8763. [Google Scholar]
- Eslami, S.; Meinel, C.; de Melo, G. PubMedCLIP: How Much Does CLIP Benefit Visual Question Answering in the Medical Domain? In Findings of the Association for Computational Linguistics: EACL 2023; Vlachos, A., Augenstein, I., Eds.; Association for Computational Linguistics: Dubrovnik, Croatia, 2023; pp. 1181–1193. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Y.; Usuyama, N.; Xu, H.; Bagga, J.; Tinn, R.; Preston, S.; Rao, R.; Wei, M.; Valluri, N.; et al. BiomedCLIP: A multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs. arXiv 2025, arXiv:2303.00915. [Google Scholar]
- Wang, Y.; Peng, J.; Dai, Y.; Jones, C.; Sair, H.; Shen, J.; Loizou, N.; Wu, J.; Hsu, W.C.; Imami, M.; et al. Enhancing vision-language models for medical imaging: Bridging the 3D gap with innovative slice selection. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 10–15 December 2024; Globerson, A., Mackey, L., Belgrave, D., Fan, A., Paquet, U., Tomczak, J., Zhang, C., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2024; Volume 37, pp. 99947–99964. [Google Scholar]
- Yuan, K.; Srivastav, V.; Navab, N.; Padoy, N. HecVL: Hierarchical Video-Language Pretraining for Zero-shot Surgical Phase Recognition. arXiv 2025, arXiv:2405.10075. [Google Scholar]
- Li, L.H.; Zhang, P.; Zhang, H.; Yang, J.; Li, C.; Zhong, Y.; Wang, L.; Yuan, L.; Zhang, L.; Hwang, J.N.; et al. Grounded Language-Image Pre-Training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 10965–10975. [Google Scholar]
- Zhong, Y.; Yang, J.; Zhang, P.; Li, C.; Codella, N.; Li, L.H.; Zhou, L.; Dai, X.; Yuan, L.; Li, Y.; et al. RegionCLIP: Region-Based Language-Image Pretraining. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 16793–16803. [Google Scholar]
- Zhai, X.; Mustafa, B.; Kolesnikov, A.; Beyer, L. Sigmoid Loss for Language Image Pre-Training. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 1–6 October 2023; pp. 11975–11986. [Google Scholar]
- Gao, P.; Geng, S.; Zhang, R.; Ma, T.; Fang, R.; Zhang, Y.; Li, H.; Qiao, Y. CLIP-Adapter: Better Vision-Language Models with Feature Adapters. Int. J. Comput. Vis. 2024, 132, 581–595. [Google Scholar] [CrossRef]
- Pelka, O.; Koitka, S.; Rückert, J.; Nensa, F.; Friedrich, C.M. Radiology Objects in COntext (ROCO): A Multimodal Image Dataset. In Proceedings of the Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis, Granada, Spain, 16 September 2018; Stoyanov, D., Taylor, Z., Balocco, S., Sznitman, R., Martel, A., Maier-Hein, L., Duong, L., Zahnd, G., Demirci, S., Albarqouni, S., et al., Eds.; Springer: Cham, Switzerland, 2018; pp. 180–189. [Google Scholar]
- Koleilat, T.; Asgariandehkordi, H.; Rivaz, H.; Xiao, Y. MedCLIP-SAM: Bridging Text and Image Towards Universal Medical Image Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2024, Marrakesh, Morocco, 6–10 October 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Springer: Cham, Switzerland, 2024; pp. 643–653. [Google Scholar]
- Zhao, T.; Gu, Y.; Yang, J.; Usuyama, N.; Lee, H.H.; Kiblawi, S.; Naumann, T.; Gao, J.; Crabtree, A.; Abel, J.; et al. A foundation model for joint segmentation, detection and recognition of biomedical objects across nine modalities. Nat. Methods 2024, 22, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Polis, B.; Zawadzka-Fabijan, A.; Fabijan, R.; Kosińska, R.; Nowosławska, E.; Fabijan, A. Exploring BiomedCLIP’s Capabilities in Medical Image Analysis: A Focus on Scoliosis Detection and Severity Assessment. Appl. Sci. 2025, 15, 398. [Google Scholar] [CrossRef]
- OpenAI; Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; et al. GPT-4 Technical Report. arXiv 2024, arXiv:2303.08774. [Google Scholar]
- Wasserthal, J.; Breit, H.C.; Meyer, M.T.; Pradella, M.; Hinck, D.; Sauter, A.W.; Heye, T.; Boll, D.T.; Cyriac, J.; Yang, S.; et al. TotalSegmentator: Robust Segmentation of 104 Anatomic Structures in CT Images. Radiol. Artif. Intell. 2023, 5, e230024. [Google Scholar] [CrossRef] [PubMed]
- DeVries, T.; Taylor, G.W. Improved Regularization of Convolutional Neural Networks with Cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Fixing weight decay regularization in adam. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Y.; Chen, J.N.; Xiao, J.; Lu, Y.; Landman, B.A.; Yuan, Y.; Yuille, A.; Tang, Y.; Zhou, Z. CLIP-Driven Universal Model for Organ Segmentation and Tumor Detection. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 1–6 October 2023; pp. 21095–21107. [Google Scholar] [CrossRef]
| Model | Baseline [%] | + Fine-Tuning [%] | [%] |
|---|---|---|---|
| CLIP | 24.14 | 40.23 | +16.09 |
| PubMedCLIP | 37.93 | 50.57 | +12.64 |
| BiomedCLIP | 50.57 | 56.32 | +5.75 |
| Method | Image Encoder | Text Encoder |
|---|---|---|
| CLIP | ViT-B/16 | Transformer |
| PubMedCLIP | ViT-B/32 | Transformer |
| BiomedCLIP | ViT-B/16 | PubMedBERT |
| Method | Acc.@1 ↑ | Acc.@5 ↑ |
|---|---|---|
| Radiologist2 | 78.16 | - |
| CLIP | 19.54 | 44.83 |
| CLIP(FT) | 40.23 | 49.43 |
| PubMedCLIP | 29.89 | 54.02 |
| PubMedCLIP(FT) | 42.53 | 59.77 |
| BiomedCLIP | 44.83 | 60.92 |
| BiomedCLIP(FT) | 51.72 | 64.37 |
| Method | Acceptance Rate ↑ |
|---|---|
| CLIP | 24.14 |
| CLIP(FT) | 40.23 |
| PubMedCLIP | 37.93 |
| PubMedCLIP(FT) | 50.57 |
| BiomedCLIP | 50.57 |
| BiomedCLIP(FT) | 56.32 |
| Method | Word | Sentence | ||
|---|---|---|---|---|
| Acc. ↑ | F1 ↑ | Acc. ↑ | F1 ↑ | |
| CLIP | ||||
| CLIP(FT) | ||||
| PubMedCLIP | ||||
| PubMedCLIP(FT) | ||||
| BiomedCLIP | ||||
| BiomedCLIP(FT) | ||||
| Method | Word | Sentence | ||
|---|---|---|---|---|
| Acc. ↑ | F1 ↑ | Acc. ↑ | F1 ↑ | |
| CLIP | ||||
| CLIP(FT) | ||||
| PubMedCLIP | ||||
| PubMedCLIP(FT) | ||||
| BiomedCLIP | ||||
| BiomedCLIP(FT) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, K.; Kikuchi, T. Feasibility Study of CLIP-Based Key Slice Selection in CT Images and Performance Enhancement via Lesion- and Organ-Aware Fine-Tuning. Bioengineering 2025, 12, 1093. https://doi.org/10.3390/bioengineering12101093
Yamamoto K, Kikuchi T. Feasibility Study of CLIP-Based Key Slice Selection in CT Images and Performance Enhancement via Lesion- and Organ-Aware Fine-Tuning. Bioengineering. 2025; 12(10):1093. https://doi.org/10.3390/bioengineering12101093
Chicago/Turabian StyleYamamoto, Kohei, and Tomohiro Kikuchi. 2025. "Feasibility Study of CLIP-Based Key Slice Selection in CT Images and Performance Enhancement via Lesion- and Organ-Aware Fine-Tuning" Bioengineering 12, no. 10: 1093. https://doi.org/10.3390/bioengineering12101093
APA StyleYamamoto, K., & Kikuchi, T. (2025). Feasibility Study of CLIP-Based Key Slice Selection in CT Images and Performance Enhancement via Lesion- and Organ-Aware Fine-Tuning. Bioengineering, 12(10), 1093. https://doi.org/10.3390/bioengineering12101093









