Abstract
Aquaporins (AQPs) are essential channel proteins that play central roles in maintaining water homeostasis. Here, a novel aquaporin gene, named KoPIP2;1, was cloned from the mangrove plant Kandelia obovata by RACE technology. The KoPIP2;1 gene was 1404 bp in length with an open reading frame (ORF) of 852 bp, encoded with 283 amino acids. Database comparisons revealed that KoPIP2;1 protein shared the highest identity (91.26%) with the aquaporin HbPIP2;2, which was isolated from Hevea brasiliensis. Gene expression analysis revealed that the KoPIP2;1 gene was induced higher in leaves than in stems and roots of K. obovata under cold stress. Transient expression of KoPIP2;1 in Nicotiana benthamiana epidermal cells revealed that the KoPIP2;1 protein was localized to the plasma membrane. Overexpressing KoPIP2;1 in Arabidopsis significantly enhanced the lateral root number of the transgenic lines. KoPIP2;1 transgenic Arabidopsis demonstrated better growth, elevated proline content, increased superoxide dismutase (SOD) and peroxidase (POD) activities, and reduced malondialdehyde (MDA) content compared with the wild-type Arabidopsis when exposed to cold stress. The findings suggest that overexpression of KoPIP2;1 probably conferred cold tolerance of transgenic Arabidopsis by enhancing osmoregulation and antioxidant capacity. This present data presents a valuable gene resource that contributes to the advancement of our understanding of aquaporins and their potential application in enhancing plant stress tolerance.
1. Introduction
The process of water movement plays a crucial role in facilitating optimal growth and reproductive functions in plants. Aquaporins (AQPs), a group of water channel proteins, can effectively enhance the permeability of water [1]. In addition to facilitating plant growth and development, AQPs play crucial roles in various aspects of plant water relations, including the regulation of hydraulic processes in roots and leaves, maintenance of cell turgor pressure, facilitation of root water uptake, and promotion of leaf transpiration [2]. AQPs regulate the facilitated diffusion of water and other uncharged solutes, including glycerol, hydrogen peroxide, carbon dioxide, ammonia, small organic acids, urea and metalloids. They also play a crucial role in maintaining water status, osmotic regulation, signal transduction, detoxification processes as well as the acquisition transportation of nutrients [1,3,4,5]. As water transporters, AQPs are involved in plant response to multiple external stimuli, including cold, drought, salt and waterlogging stress [6,7]. AQPs are classified into five subfamilies based on sequence similarity and subcellular localization: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin-26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs) and poorly characterized X intrinsic proteins (XIPs), of which PIPs are further subdivided into two phylogenetic subgroups: PIP1 and PIP2 [8,9]. All members of the AQP family contain highly conserved structural features and can form tetrameric quaternary structures in cell membranes [10]. Each AQP monomer contains six transmembrane domains (TM1–TM6), which are connected by five loops (loop A–loop E) [11]. The second structural characteristic is the aromatic/arginine (Ar/R) selectivity filter, which participates in substrate selection and prevents the ingress of other molecules into the aqueous pore [12]. The structural features of plant AQPs confer them with a wide range of selectivity for water and other solutes [2].
Studies have shown that some AQP genes can benefit plants under favorable and/or abiotic stress conditions [13,14], while others may have adverse effects [15,16]. However, our understanding of these proteins remains limited due to conflicting functions. Mangrove plants are important members of marine wetland ecosystems and mainly grow in the intertidal zone of tropical and subtropical regions [17]. An extraordinary cold or frost would significantly impede the growth of mangrove plants [18,19], and water imbalance is one of the main contributors to chilling injury [20]. The species Kandelia obovata exhibits the highest level of cold resistance among mangrove plants [21]. AQPs function as water channels and gated ion channels [22] and play important roles in water balance and water use efficiency [23]. Therefore, the AQPs of K. obovata may play positive roles in response to cold stress, and/or be useful genes for studying the stress response of mangroves. However, there is currently little literature about AQPs in K. obovata or other mangrove species [24,25,26,27].
In our previous study, we isolated an expressed sequence tag (EST) (Ko4002) from K. obovata under cold stress, which exhibited homology with other AQPs [28]. In order to gain a comprehensive understanding of the characteristics and functions of this gene, we conducted cloning and characterization of the full-length sequence of this AQP gene in this study, which was designated as KoPIP2;1. Furthermore, overexpression of KoPIP2;1 in Arabidopsis thaliana was performed to test its function under cold stress. This study will provide valuable clues for the function of AQP genes in K. obovata under cold stress, and also help to improve our understanding of the mechanism of stress resistance in mangrove plants.
2. Materials and Methods
2.1. Plant Material, Growth Conditions and Treatments
The seeds of K. obovata were collected from Guangdong Mangrove Ecological Technology Co. LTD (Zhuhai, Guangdong, China) and sown in clean sand at room temperature. The 3 months seedlings were transferred into growth chamber with normal condition (25 °C, relative humidity 75%, 14 h light/10 h dark cycle). After 7 days, these seedlings were treated under cold treatment (5 °C, relative humidity 75%, 14 h light/10 h dark cycle) for 0 h, 6 h, 12 h, 1 d, 2 d, 4 d, 7 d, 15 d and 20 d. The control group consisted of seedlings treated at 5 °C for 0 d. The leaves, stems and roots of treated seedlings were individually collected and rapidly frozen in liquid nitrogen before being stored at −80 °C until further use.
2.2. Isolation of Total RNA and cDNA Synthesis
Total RNA was extracted from K. obovata leaves using the Tiangen RNA plant Plus Reagent (Tiangen Biotech, Beijing, China) following a modified version of the described method [29]. Before the chloroform extraction step, we added a step wherein we extracted samples with phenol/chloroform/isoamyl (25:24:1 of volume ratio). The subsequent steps followed the reagent operating instructions. The integrity and purity of total RNA were assessed using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and confirmed by electrophoresis on a 1.0% agarose gel. The potential contamination of genomic DNA was eliminated through the use of RNase-free DNaseI (Promega, Madison, WI, USA). Subsequently, each RNA sample was equally mixed and subjected to SMARTTM reverse transcription Kit (Clontech, San Francisco, CA, USA) for synthesizing the first strand cDNA in accordance with the manufacturer’s instructions. The resulting cDNA samples were utilized to clone the full-length of the KoPIP2;1 gene and to conduct the RT-qPCR analysis.
2.3. Cloning the Full-Length cDNA of KoPIP2;1 Gene
The partial sequence of the KoPIP2;1 gene (GenBank accession number: JZ585815.1) has been mentioned in our previous study [28]. This EST sequence was used as the template for designing specific primers to clone the full-length cDNA sequence by rapid amplification of cDNA ends (RACE) technology in this study. The SMART RACE cDNA amplification kit (Clontech, San Francisco, CA, USA) was employed for PCR amplification according to the manufacturer’s protocol. Gene-specific primers (GSP1, 5′-GAGAGCCACAAAACAAAAGGAGGGGGT-3′, GSP2, 5′-ACCAGACCACAGCCACAGATCGCACCCA-3′) were designed for primary PCR amplification of the 5′ and 3′ end sequences of the KoPIP2;1 gene. Nested PCR reactions were performed using nested primers (NGSP1, 5′-GACTACCATGACCCACCTCCTGCTCCCT-3′, NGSP2, 5′-GGCCGGGTTAATGTGTCCTCCAGAGATA-3′) to obtain DNA fragments and RACE products which were purified by agarose gel electrophoresis and confirmed by sequencing. The 3′- and 5′- nucleotide sequences obtained were assembled using DNAMAN 6.0 software to obtain the full-length sequence of KoPIP2;1 with overlapping regions. Subsequently, primers were designed based on the assembled sequence for cloning the complete length of KoPIP2;1. The newly obtained KoPIP2;1 sequence was then submitted to the company (BGI, Shenzhen, Guangdong, China) for sequencing, confirming the full-length cDNA sequence of KoPIP2;1, which was subsequently submitted to GenBank and assigned accession number KP267759.1.
2.4. Bioinformatics Analysis of KoPIP2;1 Gene
The deduced amino acid sequence of KoPIP2;1 (GenBank accession number: KP267759.1) was predicted using the ORF Finder tool provided by the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 24 June 2023). Sequence comparisons with known sequences were conducted utilizing the NCBI BLAST tools (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 24 June 2023). The molecular weight and theoretical pI were predicted using the Compute pI/MW tool (http://web.expasy.org/compute_pi/, accessed on 24 June 2023), while TMHMM-2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/, accessed on 24 June 2023) was utilized to predict the transmembrane domain. Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan, accessed on 24 June 2023) was employed to detect motif sequences, and subcellular localization was predicted by combining PSORT (http://www.psort.org/, accessed on 24 June 2023) and Softberry ProComp v. 9.0 (http://www.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc, accessed on 24 June 2023). The ClustalX 1.83 software was utilized to perform multiple sequence alignments. A total of 40 homologues of AQPs from the NCBI database were employed to investigate the evolutionary relationship of the deduced KoPIP2;1 protein. The phylogenetic tree was constructed using the Neighbor-Joining method in MEGA 5.0 software. The three-dimensional (3D) structure of KoPIP2;1 was generated using the SWISS-MODEL tool (http://www.swissmodel.expasy.org/, accessed on 24 June 2023).
2.5. Expression Analysis by RT-qPCR
The transcriptional levels of the KoPIP2;1 gene under cold stress were determined using a real-time quantitative PCR (RT-qPCR) method. Seedlings were subjected to 5 °C for different durations (0 h, 6 h, 12 h, 1 d, 2 d, 4 d, 7 d, 15 d and 20 d), with seedlings treated at 0 day serving as control. The leaves, stems and roots were harvested separately under different conditions. Total RNA was extracted from samples and cDNA synthesis was performed following the methods according to the above Section 2.2. The K. obova’s 18S rRNA was used as reference [30,31]. Each treatment was conducted with three independent biological replicates. The primers for KoPIP2;1 (forward primer, CTCGGCGAAGGACTACCA; reverse primer, TACCCAGAATGTCAACACCAG) were designed for RT-qPCR analysis. The SYBR Premix Ex TaqTM II (Takara, Dalian, Liaoning, China) reagents were used in the RT-qPCR experiment and analyzed by iCycler iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The cycling parameters included an initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. Each RT-qPCR reaction was performed in triplicate. Transcripts were quantified using 2−∆∆CT method [32]. The data were expressed as mean ± standard deviation (x ± SD). Statistical analysis was conducted using the Student’s t-test, and graphical representation was generated with GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA).
2.6. Subcellular Localization Analysis
The KoPIP2;1 gene was cloned into the pFGC5941-35S-GFP vector [33] with N-terminal fusion using the Hieff Clone® Plus One Step Cloning Kit (Yeasen Biotech, Shanghai, China) following the manufacturer’s instructions. Subsequently, the resulting fluorescence-tagged 35S-KoPIP2;1-GFP vector was introduced into Nicotiana benthamiana epidermal cells via agrobacterium-mediated transformation. The preparation of N. benthamiana leaves and transformation procedures were carried out as previously described [28]. Briefly, recombinant plasmid 35S-KoPIP2;1-GFP was transiently introduced into the leaf epidermis of N. benthamiana via Agrobacterium tumefaciens EHA105-mediated transformation, while the vector containing only 35S-GFP served as a control. The fluorescence signal in leaves was visualized using Zeiss LSM710 laser scanning confocal microscopy (×63), with excitation and emission wavelengths of 489 nm and 510 nm for GFP signal detection, respectively. Fluorescence intensity was measured at 493 to 542 nm for GFP.
2.7. Generation of KoPIP2;1 Transgenic Arabidopsis Plants
The KoPIP2;1 gene was cloned into the binary vector pGW505-1 using Invitrogen Company’s Gateway technology, following the manufacturer’s instructions, to investigate its function in a heterologous expression system. The primers TOPO-F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGCAAAGGACGTTGAAGTTCAAG-3′) and TOPO-R (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGCATTGCTCCTGAAGGATCCGA-3′) were designed for directional cloning in accordance with the specified requirements. Subsequently, the amplified PCR products were cloned into an entry vector using the pENTR™/D-TOPO™ cloning Kit (Invitrogen, Carlsbad, CA, USA) and subjected to sequencing analysis. Confirmed recombinant entry vectors were subsequently transferred into the plant binary vector pGWB505 through LR Clonase™ II Enzyme Mix (Invitrogen, Carlsbad, CA, USA) for construction of plant Gateway® expression vector and sequenced. The pGWB505 vector containing KoPIP2;1 recombinant clones was introduced into A. tumefaciens strain EHA105 using the freeze–thaw method for transformation. Positive colonies were then subjected to floral dip-mediated transformation in Arabidopsis plants [34]. Positive transgenic lines of A. thaliana were subsequently obtained and selected by culturing on MS medium agar plates supplemented with 50 mg/L kanamycin. The integration of transgenes in these plants was confirmed through RT-PCR (reverse transcription-PCR) analysis using gene-specific primers F2 (5′-ATGGCAAAGGACGTTGAAGTTCAAG-3′) and R2 (5′-TTAAGCATTGCTCCTGAAGGATCCG-3′). The transgenic lines were advanced through self-pollination until T3 transgenic plants were obtained. Finally, the T3 or T4 homozygous lines were utilized for functional analysis.
2.8. Physiological Analysis of Transgenic A. thaliana Lines Exposed to Cold Stress
Wild-type (WT) A. thaliana and transgenic Arabidopsis lines overexpressing KoPIP2;1 (Lines 2, 3, and 5) were subjected to cold treatments. Cold stress was applied to 12-day-old seedlings by incubating them at 5 °C for 10 days. Seedlings grown under normal conditions at 22 °C were used as controls. The fresh weight and number of lateral roots in these seedlings were quantified. The levels of proline and malondialdehyde (MDA), as well as the activities of superoxide dismutase (SOD) and peroxidase (POD), were quantified using the corresponding assay kits (Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China) following the manufacturer’s instructions. All experiments were conducted in triplicate. Statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, San Diego, CA, USA). One-way ANOVA followed by Duncan’s test was employed to determine significant differences using SPSS statistics 25.
3. Results
3.1. Characterization and Sequence Analysis of KoPIP2;1
The full-length cDNA sequence of this aquaporin gene with 1404 base pairs (bp) was generated and named KoPIP2;1 (GenBank ID: KP267759.1). The KoPIP2;1 sequence comprises a 136 bp 5′-untranslated region (UTR), a 416 bp 3′-UTR, and an 852 bp complete open reading frame (ORF) that encodes a protein of 283 amino acids with a calculated molecular weight (MW) of 30.41 kDa and an isoelectric point (pI) of 6.05. Bioinformatics analysis revealed that the KoPIP2;1 protein contains 24 positively charged residues (Arg and Lys) and 21 negatively charged residues (Asp and Glu) and is predicted to be localized in the plasma membrane. Similar to other PIPs, KoPIP2;1 contains six transmembrane domains (TM1–TM6), five loops (Loop A–Loop E) and conserved dual NPA motifs (NPA Ⅰ and NPA Ⅱ) (Figure 1), which are typical domains of the AQP family. Additionally, the KoPIP2;1 protein harbors two highly conversed sequences (signature GGGANXXXXGY and signature TGI/TNPARSL/FGAAI/VI/VF/YN in Loop C and Loop E, respectively) within its central region, as well as a distinctive conserved sequence (P/K)/DYX(E/D)PP(P/R)X3-4(E/D)XXELXXWSF(Y/W)R at its N-terminal, all of which are typical features of the PIP subfamily [35]. Furthermore, sequence analysis showed that KoPIP2;1 exhibited the highest sequence identity (91.26%) with Hevea brasiliensis aquaporin HbPIP2;2 (GenBank accession number: XP_021662216.1). Sequence alignment also demonstrated that KoPIP2;1 displayed greater similarity to PIP2s than PIP1s in both the N-terminus and C-terminus regions (Figure 1). These findings suggest that KoPIP2;1 belongs to the PIP2 family.
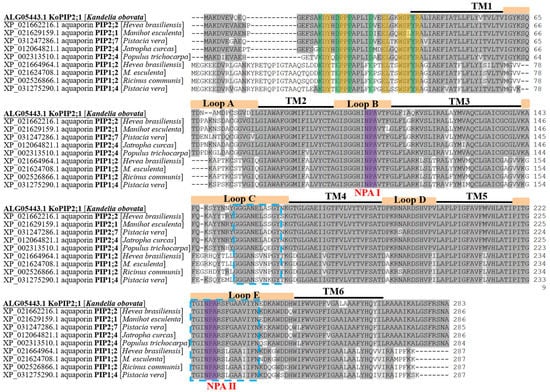
Figure 1.
Sequence alignment of KoPIP2;1 and other plant PIP1s and PIP2s. The amino acids that are identical among proteins are shaded in dark gray, while the similar acids are shaded in light gray. The conserved amino acids of PIPs at N-terminal (P/K)/DYX(E/D)PP(P/R)X3-4(E/D)XXELXXWSF(Y/W)R are highlighted with green and yellow shading. The six transmembrane domains (TM1–TM6) and five loops (Loop A–Loop E) of AQPs are represented by black lines and orange boxes above the sequences, respectively. The two conserved NPA motifs (NPA I and NPA II) are shaded in purple. The blue dotted boxed sequences depict the two conserved sequences (GGGANXXXXGY and TGI/TNPARSL/FGAAI/VI/VF/YN) present in all of the PIPs.
The SWISS-MODEL online tool was utilized to construct the 3D model of KoPIP2;1 (Figure S1). A sequence similarity comparison between KoPIP2;1 and its template (SoPIP2-1, Spinacia oleracea aquaporin PIP2;1, SMTL id: 4jc6.2.A) [36] revealed a similarity of 78.42%, indicating that the 3D model of KoPIP2;1 was acceptable. Homology modeling demonstrated that the 3D model structure of KoPIP2;1 consisted of a homo-tetramer, with each monomer containing eleven α-helixes, two β-strands, some random coils, three octyl-β-glucanpyranoside molecules, seven cadmium ions and four mercury ions. It is suggested that the presence of cadmium and mercury ions may impact the gating mechanism of AQPs [36].
3.2. Phylogenic Analysis at the Protein Level
We have downloaded representative amino acid sequences of different types of Arabidopsis aquaporins, as well as some aquaporins that exhibit high homology with the KoPIP2;1 sequence through NCBI blast analysis. According to phylogenetic tree (Figure 2), KoPIP2;1 was distinctly classified in the aquaporin PIP2 subgroup, demonstrating that KoPIP2;1 is a member of the PIP2 subfamily. Furthermore, KoPIP2;1 was on the same PIP2 branch as its highest homologue (Hevea brasiliensis aquaporin PIP2;2, HbPIP2;2) and its 3D modeling template (Spinacia oleracea aquaporin PIP2;1, SoPIP2;1). According to these data, we named the aquaqporin KoPIP2;1 in this study.
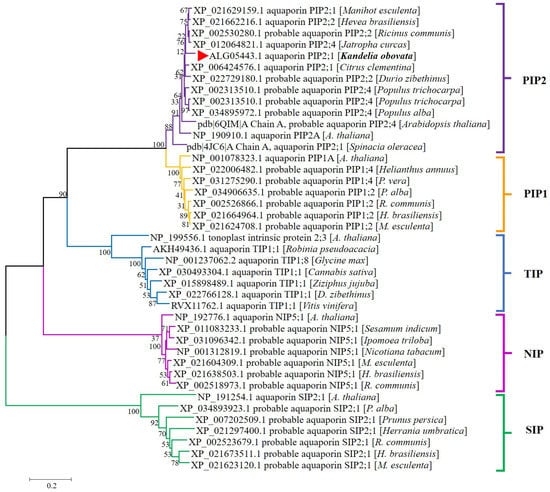
Figure 2.
Phylogenetic relationship between KoPIP2;1 and other plant AQPs. A total of 40 plant amino acid sequences, obtained from the NCBI database with accession numbers, were included in the tree. The KoPIP2;1 protein is represented by a red triangle. This tree was constructed using the Neighbor-Joining method with 1000 bootstrap replications. The scale indicates the branch length.
3.3. Expression Patterns of KoPIP2;1 in Response to Cold Stress
Under cold stress, the expression of KoPIP2;1 is significantly induced in various tissues compared with normal conditions (Figure 3). The results show that the KoPIP2;1 gene were highly expressed in the leaves, but lesser expressed in stems and roots of K. obovata under cold stress. During the experimental period, the expression levels of KoPIP2;1 initially increased and then decreased in different tissues. However, the time points of their peak expression levels varied, with leaves peaking at 6 h (11.79-fold) (Figure 3A), stems at 12 h (5.33-fold) (Figure 3B), and roots at 6 h (5.14-fold) (Figure 3C). It can be stated that KoPIP2;1 was predominantly induced during the early stage (6 h–1 d) in K. obovata under cold stress. Notably, significant wilting and dehydration of K. obovata leaves were observed after 15 days under cold stress in our previous study [37]. These results suggest that KoPIP2;1 is an early responsive gene and may play crucial roles in the response to cold stress in K. obovata.
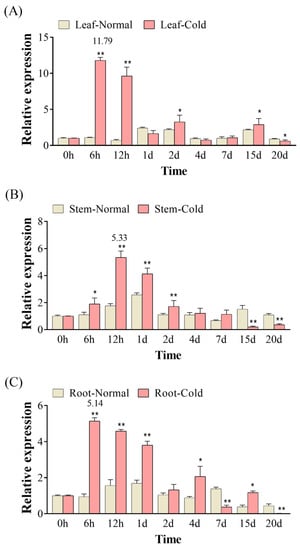
Figure 3.
Gene expression of KoPIP2;1 in K. obovata under cold stress and normal conditions. The relative gene expression levels of KoPIP2;1 in leaf (A), stem (B) and root (C) were normalized using the reference gene Ko18S. The data represented here are the average values obtained from three biological repetitions, with error bars indicating standard deviations (p values were calculated using Student’s t-test. * p < 0.05; ** p < 0.01).
3.4. Subcellular Localization of KoPIP2;1 in Tobacco Epidermal Cells
The 35S-KoPIP2;1-GFP vector, labeled with fluorescence, was introduced into the epidermal cells of N. benthamiana and examined using confocal laser-scanning microscopy. As depicted in Figure 4, the GFP fluorescence signal of 35S-KoPIP2;1-GFP was predominantly accumulated in the plasma membrane, whereas the control GFP protein was distributed throughout the entire cell, including the nucleus. These findings indicate that KoPIP2;1 is a plasma membrane protein and are consistent with bioinformatics predictions.
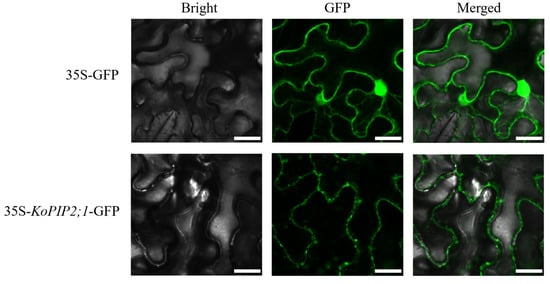
Figure 4.
Subcellular localization of KoPIP2;1 in N. benthamiana epidermal cells. GFP fluorescence was used to observe the transient expression of KoPIP2;1. The leaves with overexpression of 35S-KoPIP2;1-GFP and 35S-GFP (used as the control) were imaged by confocal microscopy. Scale bar = 100 µm.
3.5. Overexpression of KoPIP2;1 Enhanced Cold Tolerance of Transgenic Arabidopsis
As shown in Figure 5, the WT Arabidopsis and three KoPIP2;1 transgenic Arabidopsis lines (Line 2, 3, 5) showed no significant difference in fresh weight under normal growth conditions (CK) and cold stress, respectively (Figure 5A–C). Nevertheless, KoPIP2;1 transgenic lines exhibited superior growth and significantly higher lateral root numbers than WT plants under both CK and cold conditions (Figure 5A,B,D). A well-developed root system typically results in more efficient absorption of nutrients and water, indicating that overexpression of KoPIP2;1 could enhance the cold resistance of transgenic A. thaliana. To explore the involvement of KoPIP2;1 in osmoregulation, the proline content was measured in both WT and transgenic plants. As shown in Figure 5E, transgenic lines exhibited significantly higher proline content than WT under cold stress, indicating that overexpression of KoPIP2;1 may enhance osmoregulatory capacity by increasing the proline content of plant cells. In addition, to investigate the involvement of KoPIP2;1 in antioxidant function, MDA content and SOD and POD activities were examined. As depicted in Figure 5F–H, the KoPIP2;1 transgenic lines exhibited significantly lower MDA content and higher SOD and POD activities than WT plants under cold stress, indicating that overexpression of KoPIP2;1 improved the efficiency of antioxidant systems and reduced membrane damage in transgenic Arabidopsis under cold stress. These findings suggest that overexpression of KoPIP2;1 may enhance osmoregulation and antioxidant capacity in transgenic Arabidopsis plants, thereby conferring cold tolerance.
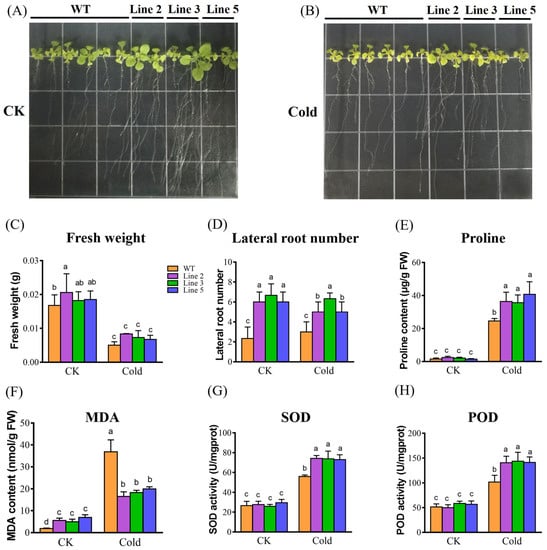
Figure 5.
Cold tolerance analysis of KoPIP2;1 transgenic Arabidopsis under cold stress. Twelve-day-old seedlings were grown in MS agar medium and subjected to normal growth condition (A) or cold stress condition (B) for 10 days, respectively. The fresh weight (C), lateral root number (D), proline content (E), MDA content (F), SOD activity (G), and POD activity (H) were measured in both the KoPIP2;1 transgenic lines and wild-type plants. Line 2, Line 3, and Line 5 represent KoPIP2;1 transgenic lines 2, 3 and 5, respectively. WT represents the wild type. The data were subjected to one-way analysis of variance, followed by Duncan’s test. Error bars with dissimilar letters indicate statistically significant differences (p < 0.05, Duncan’s test).
4. Discussion
The KoPIP2;1 was located on the plasma membrane by the subcellular localization analysis, similar results of PIPs localization on the plasma membrane have been reported [38,39]. The RT-qPCR results reveal that the expression of KoPIP2;1 exhibited tissue-specific patterns, which were also observed in other AQPs across various plant organs, including leaves, stems, roots, flowers, fruits and seeds [40]. Expression levels of most Saussurea involucrata PIPs were decreased in both roots and leaves under salt stress and involved in mediating water transport in tomato [41]. Tissue-specific expression of KoPIP2;1 implies that the KoPIP2;1 protein may play positive roles in K. obovata, participating in abiotic stress responses.
AQPs have been reported to mediate transcellular (across membranes) root water transport and hydraulic conductance [2]. Many PIPs have been observed to facilitate water exchange between intracellular and extracellular compartments, playing a crucial role in the maintenance of water balance within the cytoplasm [4,42,43]. Research has reported that overexpression of aquaporin genes confers benefits to transgenic plants under various stress conditions, such as cold, drought, salt, and nitrate reduction stresses [10,44,45]. In this study, it was found that the transgenic plants exhibited a higher number of lateral roots compared with WT plants under favorable conditions. This observation suggests that the KoPIP2;1 gene has an impact on plant morphology under normal conditions in transgenic plants. Similar phenotypes have been observed in transgenic rice overexpressing OsPIP2;3 (Oryza sativa PIP) and in transgenic A. thaliana overexpressing PgTIP1 (Panax ginseng TIP) [46,47]. This is likely due to a compensation mechanism that increases lateral roots or enhanced water transport resulting from the overexpression of KoPIP2;1, leading to morphological changes.
Furthermore, our findings indicate that plants overexpressing KoPIP2;1 exhibit superior growth compared with WT plants when subjected to cold stress. This suggests that KoPIP2;1 confers tolerance to cold stress in transgenic Arabidopsis plants. Similarly, the overexpression of SiPIP1;5A (Saussurea involucrata PIP) enhances the cold tolerance of tomato by regulating cell water balance [48]. Conversely, some AQP members have been shown to be negatively associated with abiotic stress. The overexpression of AtPIP1;4 and AtPIP2;5 in plants has been seen to result in accelerated water loss during drought stress [16]. Overexpressing AtPIP1;b in tobacco has led to rapid wilting under drought stress [15]. Therefore, members of AQP family exhibit differential cellular functions.
Abiotic stresses always disturb osmotic balance in plants. In general, proline is widely distributed in many organisms, and the accumulation of this substance can improve cellular protection and reduce the cellular osmotic potential [49]. In this study, KoPIP2;1 overexpressing plants exhibited higher proline level compared with WT plants, indicating that KoPIP2;1 may regulate osmotic imbalance induced by cold stress. This finding is consistent with a previous study demonstrating that overexpression of TsPIP1;1 (Thellungiella salsuginea PIP) improved cellular proline accumulation in rice under salt stress [44].
Abiotic stress induces the rapid accumulation of reactive oxygen species (ROS), which can cause severe damage to cell membranes by oxidizing DNA, proteins, and lipids [50]. MDA is commonly used to assess ROS-mediated cellular injuries and membrane damage [51]. Our findings indicate that transgenic plants exhibit lower MDA levels than the WT under cold stress, indicating that KoPIP2;1 can mitigate membrane damage in overexpressing plants. This finding is consistent with previous studies, which have demonstrated that overexpression of TsPIP1;1 and MaPIP1;1 also reduces cellular MDA levels [44,52]. SOD and POD serve as ROS scavengers [53]. The overexpression of KoPIP2;1 in plants has resulted in increased levels of both SOD and POD activities compared with WT plants. Taken together, these findings suggest that the overexpression of KoPIP2;1 can enhance plant osmotic regulation and antioxidant capacity, which in turn reduces membrane injury. Certainly, this is only the start in terms of the study of the function of KoPIP2;1, further investigations, including detailed analysis in transgenic plants, are necessary to fully elucidate its role.
5. Conclusions
In summary, we successfully cloned and characterized the aquaporin gene KoPIP2;1 from the mangrove plant K. obovata. Overexpression of KoPIP2;1 improved the cold tolerance of transgenic A. thaliana by regulating solute accumulation and antioxidant capacity, suggesting an important role for KoPIP2;1 in K. obovata under cold stress. Future investigations will focus on identifying upstream regulators of KoPIP2;1 and evaluating the growth performance of these transgenic lines exposed to diverse stresses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bioengineering10070878/s1, Figure S1: Prediction of 3D structure of KoPIP2;1 and comparison with its template.
Author Contributions
Conceptualization, J.F., Y.W. and H.C.; Data curation, F.S.; Formal analysis, J.F.; Funding acquisition, J.F., Y.W. and H.C.; Investigation, J.F.; Methodology, J.F. and F.S.; Resources, H.C., M.W. and C.S.; Software, H.W. and M.W.; Supervision, Y.W.; Validation, H.W. and C.S.; Visualization, Y.W., H.C. and H.W.; Writing—original draft, J.F.; Writing—review and editing, Y.W., H.C., H.W., M.W., F.S. and C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 41706118, No. 41676086 and No. U1901211), the Basic and Applied Basic Research Project of Guangzhou Basic Research Program (No. 2023A04J0898), the Marine Economy Development Project of Guangdong Province (No. GDNRC[2023]43), the International Partnership Program of Chinese Academy of Sciences (No. 133244KYSB20180012), the National Key Research and Development Plan (No. 2017FY100700), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA23050200, No. XDA13010500 and No. XDA13020503), and the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. GML2019ZD0305).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequence data of KoPIP2;1 can be obtained from the NCBI database with accession number: KP267759.1 (https://www.ncbi.nlm.nih.gov/nucleotide/KP267759.1/, accessed on 24 June 2023). All analyzed or generated data is included in this article. The data analyzed or generated in this study can be obtained from the corresponding author upon reasonable request.
Acknowledgments
We are very grateful to Yong-Jia Zhong and Lei Zheng of Fujian Agriculture and Forestry University for their assistance in experiment of subcellular localization.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shivaraj, S.M.; Sharma, Y.; Chaudhary, J.; Rajora, N.; Sharma, S.; Thakral, V.; Ram, H.; Sonah, H.; Singla-Pareek, S.L.; Sharma, T.R.; et al. Dynamic role of aquaporin transport system under drought stress in plants. Environ. Exp. Bot. 2021, 184, 104367. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Z.J.; Liu, F.; Sun, L.R.; Hao, F.S. Versatile roles of aquaporins in plant growth and development. Int. J. Mol. Sci. 2020, 21, 9485. [Google Scholar] [CrossRef] [PubMed]
- Noronha, H.; Silva, A.; Mitani-Ueno, N.; Conde, C.; Sabir, F.; Prista, C.; Soveral, G.; Isenring, P.; Ma, J.F.; Belanger, R.R.; et al. The grapevine NIP2;1 aquaporin is a silicon channel. J. Exp. Bot. 2020, 71, 6789–6798. [Google Scholar] [CrossRef] [PubMed]
- Scharwies, J.D.; Dinneny, J.R. Water transport, perception, and response in plants. J. Plant Res. 2019, 132, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Q.; You, S.Y.; Wang, Y.B.; Huang, L.; Wang, M. Influence of frost on nutrient resorption during leaf senescence in a mangrove at its latitudinal limit of distribution. Plant Soil 2011, 342, 105–115. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wu, H.H.; Zou, Y.N.; Wu, Q.S.; Kuča, K. Mycorrhizal response strategies of trifoliate orange under well-watered, salt stress, and waterlogging stress by regulating leaf aquaporin expression. Plant Physiol. Bioch. 2021, 162, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, W.; Liu, J.H.; Song, S.; Hou, X.W.; Jia, C.H.; Li, J.Y.; Miao, H.X.; Wang, Z.; Tie, W.W.; et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.). Plant Physiol. Biochem. 2020, 147, 66–76. [Google Scholar] [CrossRef]
- Hussain, A.; Tanveer, R.; Mustafa, G.; Farooq, M.; Amin, I.; Mansoor, S. Comparative phylogenetic analysis of aquaporins provides insight into the gene family expansion and evolution in plants and their role in drought tolerant and susceptible chickpea cultivars. Genomics 2020, 112, 263–275. [Google Scholar] [CrossRef]
- Kaldenhoff, R.; Fischer, M. Functional aquaporin diversity in plants. BBA Biomembr. 2006, 1758, 1134–1141. [Google Scholar] [CrossRef]
- Heymann, J.B.; Engel, A. Structural clues in the sequences of the aquaporins. J. Mol. Biol. 2000, 295, 1039–1053. [Google Scholar] [CrossRef]
- Chowdhury, R.; Ren, T.; Shankla, M.; Decker, K.; Grisewood, M.; Prabhakar, J.; Baker, C.; Golbeck, J.H.; Aksimentiev, A.; Kumar, M.; et al. PoreDesigner for tuning solute selectivity in a robust and highly permeable outer membrane pore. Nat. Commun. 2018, 9, 3661. [Google Scholar] [CrossRef]
- Törnroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef]
- Zhang, W.E.; Hu, J.M.; Li, F.; Chen, E.J.; Zhao, T.; Pan, X.J. Cloning and expression of tonoplast membrane intrinsic protein genes in leaves of Vitis heyneana and overexpression of VhTIP2;1 in Arabidopsis confer drought tolerance. Acta Physiol. Plant 2023, 45, 44. [Google Scholar] [CrossRef]
- Yang, X.M.; Li, J.W.; Ji, C.C.; Wei, Z.X.; Zhao, T.; Pang, Q.Y. Overexpression of an aquaporin gene EsPIP1;4 enhances abiotic stress tolerance and promotes flowering in Arabidopsis thaliana. Plant Physiol. Bioch. 2022, 193, 25–35. [Google Scholar] [CrossRef]
- Aharon, R.; Shahak, Y.; Wininger, S.; Bendov, R.; Kapulnik, Y.; Galili, G. Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 2003, 15, 439–447. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, S.H.; Rhee, J.Y.; Chung, G.C.; Ahn, S.J.; Kang, H. Transgenic Arabidopsis and tobacco plants overexpressing an aquaporin respond differently to various abiotic stresses. Plant Mol. Biol. 2007, 64, 621–632. [Google Scholar] [CrossRef]
- Tregarot, E.; Caillaud, A.; Cornet, C.C.; Taureau, F.; Catry, T.; Cragg, S.M.; Failler, P. Mangrove ecological services at the forefront of coastal change in the French overseas territories. Sci. Total Environ. 2021, 763, 143004. [Google Scholar] [CrossRef]
- Chen, L.Z.; Wang, W.Q.; Zhang, Y.H.; De, Y.H.; Huang, L.; Zhao, C.L.; Yang, S.C.; Yang, Z.W.; Chen, Y.C.; Xu, H.L.; et al. Damage to mangroves from extreme cold in early 2008 in Southern China. Acta Phytoecol. Sin. 2010, 34, 186–194. [Google Scholar]
- Lu, W.X.; Zhang, B.H.; Yang, S.C. Survive the north: Transplantation for conservation of mangrove forests requires consideration of influences of low temperature, mating system and their joint effects on effective size of the reforested populations. Front. Ecol. Evol. 2023, 11, 1160468. [Google Scholar] [CrossRef]
- Sohag, A.M.; Tahjib-Ul-Arif, M.; Afrin, S.; Khan, M.K.; Hannan, A.; Skalicky, M.; Mortuza, M.G.; Brestic, M.; Hossain, M.A.; Murata, Y. Insights into nitric oxide-mediated water balance, antioxidant defence and mineral homeostasis in rice (Oryza sativa L.) under chilling stress. Nitric Oxide-Biol. Chem. 2020, 100, 7–16. [Google Scholar] [CrossRef]
- Tian, K.; Li, Q.; Zhang, X.M.; Guo, H.Y.; Wang, Y.H.; Cao, P.L.; Xu, S.Y.; Li, W.Y. Analysis of the expression and function of the CBL-CIPK network and MAPK cascade genes in Kandelia obovata seedlings under cold stress. Front. Mar. Sci. 2023, 10, 1113278. [Google Scholar] [CrossRef]
- Ozu, M.; Alvear-Arias, J.J.; Fernandez, M.; Caviglia, A.; Pena-Pichicoi, A.; Carrillo, C.; Carmona, E.; Otero-Gonzalez, A.; Garate, J.A.; Amodeo, G.; et al. Aquaporin gating: A new twist to unravel permeation through water channels. Int. J. Mol. Sci. 2022, 23, 12317. [Google Scholar] [CrossRef]
- Zhao, C.X.; Shao, H.B.; Chu, L.Y. Aquaporin structure-function relationships, water flow through plant living cells. Colloid Surf. B 2008, 62, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Fang, X.D.; Lin, Q.F.; Li, G.Y.; Zhao, W.M. Identification and expression analysis of a full-length cDNA encoding a Kandelia candel tonoplast intrinsic protein. Act. J. Pharm. Biotech. 2003, 19, 147–152. [Google Scholar]
- Lu, Y.J.; Han, Y.S.; Li, N.N.; Hou, P.C.; Huang, X.X.; Deng, S.R.; Zhao, R.; Shen, X.; Chen, S.L. Molecular cloning and salt-tolerance of gene KcTIP1 from Kandelia candel. J. Northwest Agric. For. Univ. China 2013, 41, 162–171. [Google Scholar]
- Reef, R.; Schmitz, N.; Rogers, B.A.; Ball, M.C.; Lovelock, C.E. Differential responses of the mangrove Avicennia marina to salinity and abscisic acid. Funct. Plant Biol. 2012, 39, 1038–1046. [Google Scholar] [CrossRef]
- Tan, W.K.; Lin, Q.; Lim, T.M.; Kumar, P.; Loh, C.S. Dynamic secretion changes in the salt glands of the mangrove tree species Avicennia officinalis, in response to a changing saline environment. Plant Cell Environ. 2013, 36, 1410–1422. [Google Scholar] [CrossRef]
- Fei, J.; Wang, Y.S.; Jiang, Z.Y.; Cheng, H.; Zhang, J.D. Identification of cold tolerance genes from leaves of mangrove plant Kandelia obovata by suppression subtractive hybridization. Ecotoxicology 2015, 24, 1686–1696. [Google Scholar] [CrossRef]
- Song, H.; Wang, Y.S. Analysis and improvement of high-quality RNA extraction in leaves of mangrove plants. Ecol. Sci. 2011, 30, 201–206. [Google Scholar]
- Fei, J.; Wang, Y.S.; Cheng, H.; Su, Y.B.; Zhong, Y.J.; Zheng, L. Cloning and characterization of KoOsmotin from mangrove plant Kandelia obovata under cold stress. BMC Plant Biol. 2021, 21, 10. [Google Scholar] [CrossRef]
- Peng, Y.L.; Wang, Y.S.; Cheng, H.; Sun, C.C.; Wu, P.; Wang, L.Y.; Fei, J. Characterization and expression analysis of three CBF/DREB1 transcriptional factor genes from mangrove Avicennia marina. Aquat. Toxicol. 2013, 140, 68–76. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lei, X.J.; Tan, B.; Liu, Z.Y.; Wu, J.; Lv, J.X.; Gao, C.Q. ThCOL2 improves the salt stress tolerance of Tamarix hispida. Front. Plant Sci. 2021, 12, 653791. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar]
- Weig, A.; Deswarte, C.; Chrispeels, M.J. The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 1997, 114, 1347–1357. [Google Scholar] [CrossRef]
- Frick, A.; Jarva, M.; Ekvall, M.; Uzdavinys, P.; Nyblom, M.; Tornroth-Horsefield, S. Mercury increases water permeability of a plant aquaporin through a non-cysteine-related mechanism. Biochem. J. 2013, 454, 491–499. [Google Scholar] [CrossRef]
- Fei, J.; Wang, Y.S.; Cheng, H.; Su, Y.B.; Zhong, Y.J.; Zheng, L. The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2022, 22, 274. [Google Scholar] [CrossRef]
- Bienert, M.D.; Diehn, T.A.; Richet, N.; Chaumont, F.; Bienert, G.P. Heterotetramerization of plant PIP1 and PIP2 aquaporins is an evolutionary ancient feature to guide PIP1 plasma membrane localization and function. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Shi, J.; Wang, J.F.; Li, R.; Li, D.B.; Xu, F.F.; Sun, Q.Q.; Zhao, B.; Mao, A.J.; Guo, Y.D. Expression patterns of genes encoding plasma membrane aquaporins during fruit development in cucumber (Cucumis sativus L.). Plant Physiol. Bioch. 2015, 96, 329–336. [Google Scholar] [CrossRef]
- Kumawat, S.; Khatri, P.; Ahmed, A.; Vats, S.; Kumar, V.; Jaswal, R.; Wang, Y.; Xu, P.; Mandlik, R.; Shivaraj, S.M.; et al. Understanding aquaporin transport system, silicon and other metalloids uptake and deposition in bottle gourd (Lagenaria siceraria). J. Hazard Mater. 2021, 409, 124598. [Google Scholar] [CrossRef]
- Jia, J.H.; Liang, Y.F.; Gou, T.Y.; Hu, Y.H.; Zhu, Y.X.; Hu, H.Q.; Guo, J.; Gong, H.J. The expression response of plasma membrane aquaporins to salt stress in tomato plants. Environ. Exp. Bot. 2020, 178, 104190. [Google Scholar] [CrossRef]
- Tailor, A.; Bhatla, S.C. Polyamine homeostasis modulates plasma membrane- and tonoplast-associated aquaporin expression in etiolated salt-stressed sunflower (Helianthus annuus L.) seedlings. Protoplasma 2021, 258, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.F.; Xu, H.; Khan, S.; Equiza, M.A.; Lee, S.H.; Vaziriyeganeh, M.; Zwiazek, J.J. Plant water transport and aquaporins in oxygen-deprived environments. J. Plant Physiol. 2018, 227, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qiang, X.J.; Han, X.R.; Jiang, L.L.; Zhang, S.H.; Han, J.; He, R.; Cheng, X.G. Ectopic expression of a Thellungiella salsuginea aquaporin gene, TsPIP1;1, increased the salt tolerance of rice. Int. J. Mol. Sci. 2018, 19, 2229. [Google Scholar] [CrossRef]
- Zhuo, C.; Wang, T.; Guo, Z.; Lu, S. Overexpression of MfPIP2-7 from Medicago falcata promotes cold tolerance and growth under NO3− deficiency in transgenic tobacco plants. BMC Plant Biol. 2016, 16, 138. [Google Scholar] [CrossRef]
- Sun, J.Y.; Liu, X.S.; Khan, I.U.; Wu, X.C.; Yang, Z.M. OsPIP2;3 as an aquaporin contributes to rice resistance to water deficit but not to salt stress. Environ. Exp. Bot. 2021, 183, 104342. [Google Scholar] [CrossRef]
- Peng, Y.H.; Lin, W.L.; Cai, W.M.; Arora, R. Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 2007, 226, 729–740. [Google Scholar] [CrossRef]
- Li, J.; Xia, W.W.; Zang, H.X.; Dai, B.; Zhang, Y.; Feng, Y.J.; Wang, A.Y.; Lin, Z.P.; Liu, H.L.; Zhu, J.B. Expression analysis of aquaporin genes in Saussurea involucrata rosette leaves and functional analysis of upregulated SiPIP1;5A under low-temperature stress. Environ. Exp. Bot. 2020, 171, 103958. [Google Scholar] [CrossRef]
- Koenigshofer, H.; Loeppert, H.G. The up-regulation of proline synthesis in the meristematic tissues of wheat seedlings upon short-term exposure to osmotic stress. J. Plant Physiol. 2019, 237, 21–29. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Chomkitichai, W.; Chumyam, A.; Rachtanapun, P.; Uthaibutra, J.; Saengnil, K. Reduction of reactive oxygen species production and membrane damage during storage of ‘Daw’ longan fruit by chlorine dioxide. Sci. Hortic. 2014, 170, 143–149. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Zhang, J.; Jia, C.; Miao, H.; Xu, B.; Jin, Z. A banana aquaporin gene, MaPIP1;1, is involved in tolerance to drought and salt stresses. BMC Plant Biol. 2014, 14, 59. [Google Scholar] [CrossRef]
- Wen, M.; Lin, X.; Yu, Y.S.; Wu, J.J.; Xu, Y.J.; Xiao, G.S. Natamycin treatment reduces the quality changes of postharvest mulberry fruit during storage. J. Food Biochem. 2019, 43, e12934. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).