Predictive Model for Listeria monocytogenes in RTE Meats Using Exclusive Food Matrix Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Growth Rate Estimation
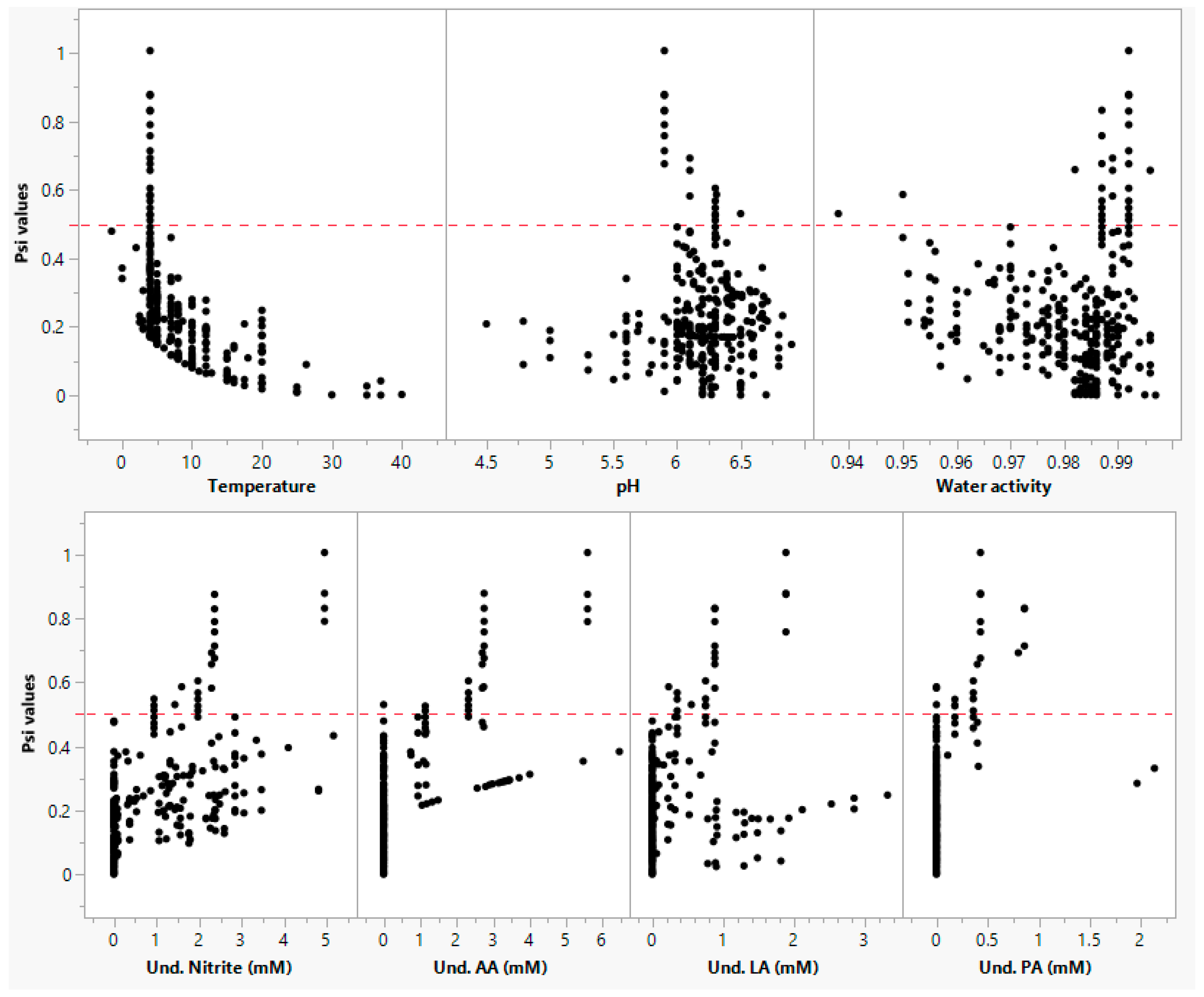
2.2. Data Selection Criteria and Assumptions
2.3. Development of Secondary Growth Rate Model
2.4. Secondary Modeling Approach
2.5. Secondary Growth Rate Model Validation
2.6. Development of Lag Time Model and Its Validation
3. Results
3.1. Development of a Secondary Model for Listeria Growth Prediction
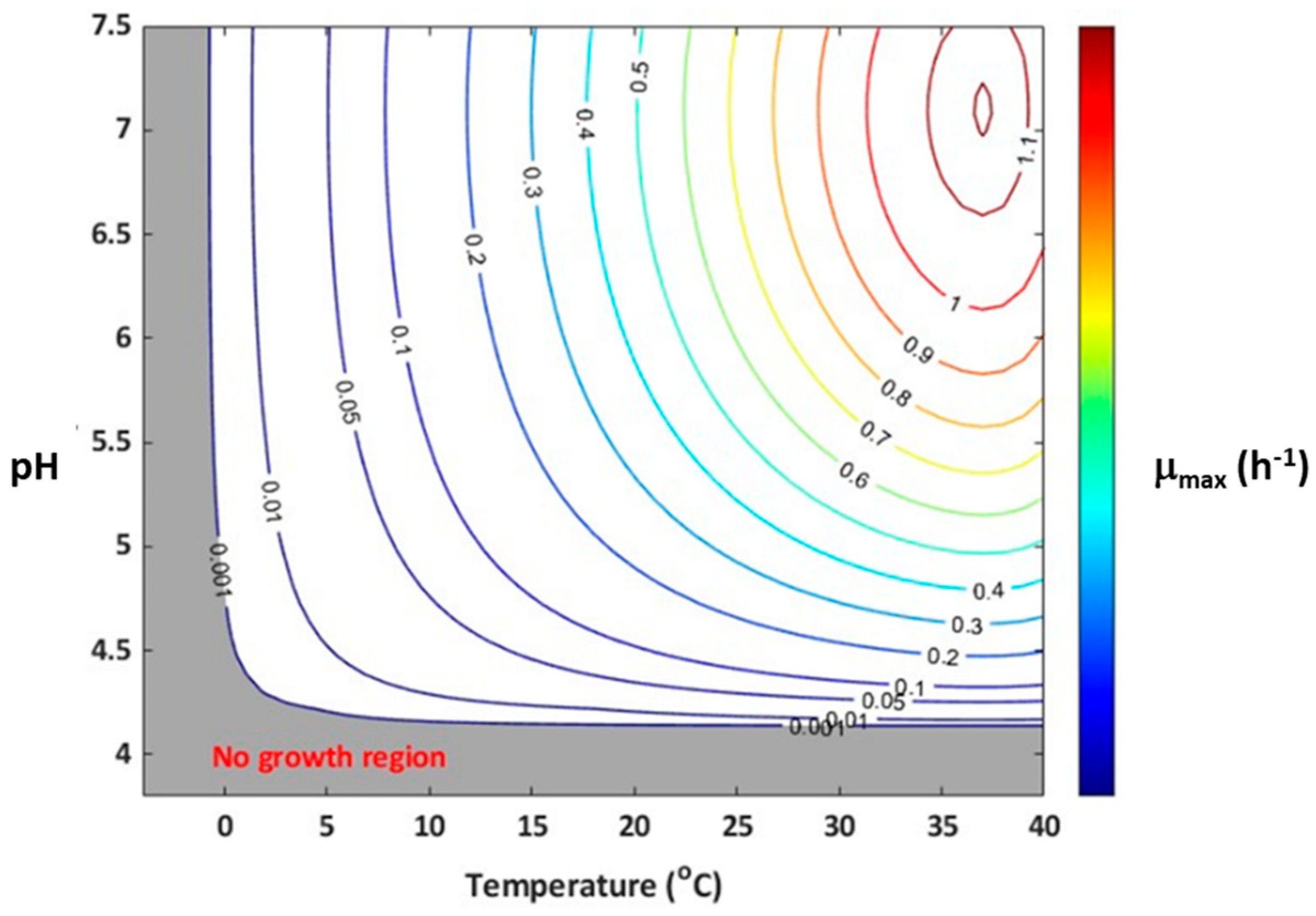
3.1.1. Estimation of Gamma Model Parameters
3.1.2. Estimation of MICs of Inhibitory Compounds
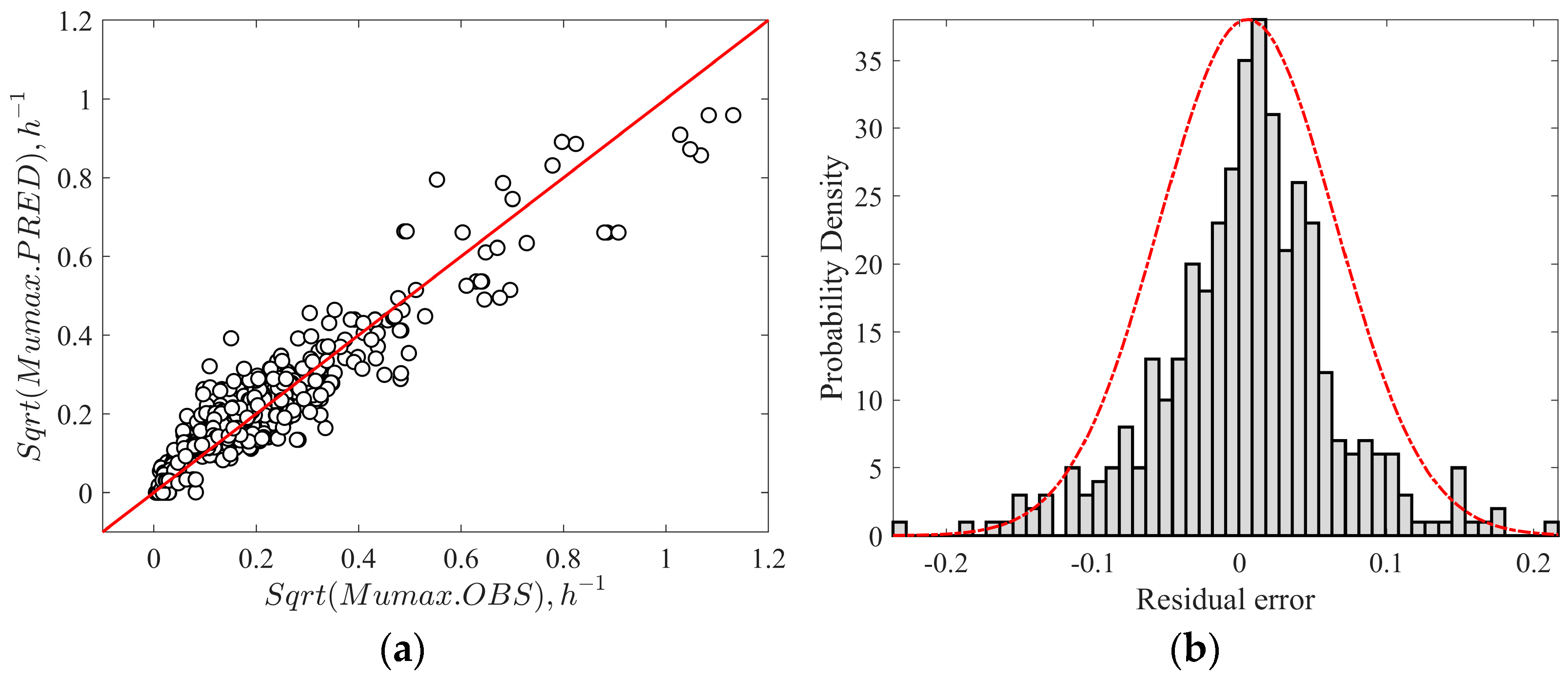
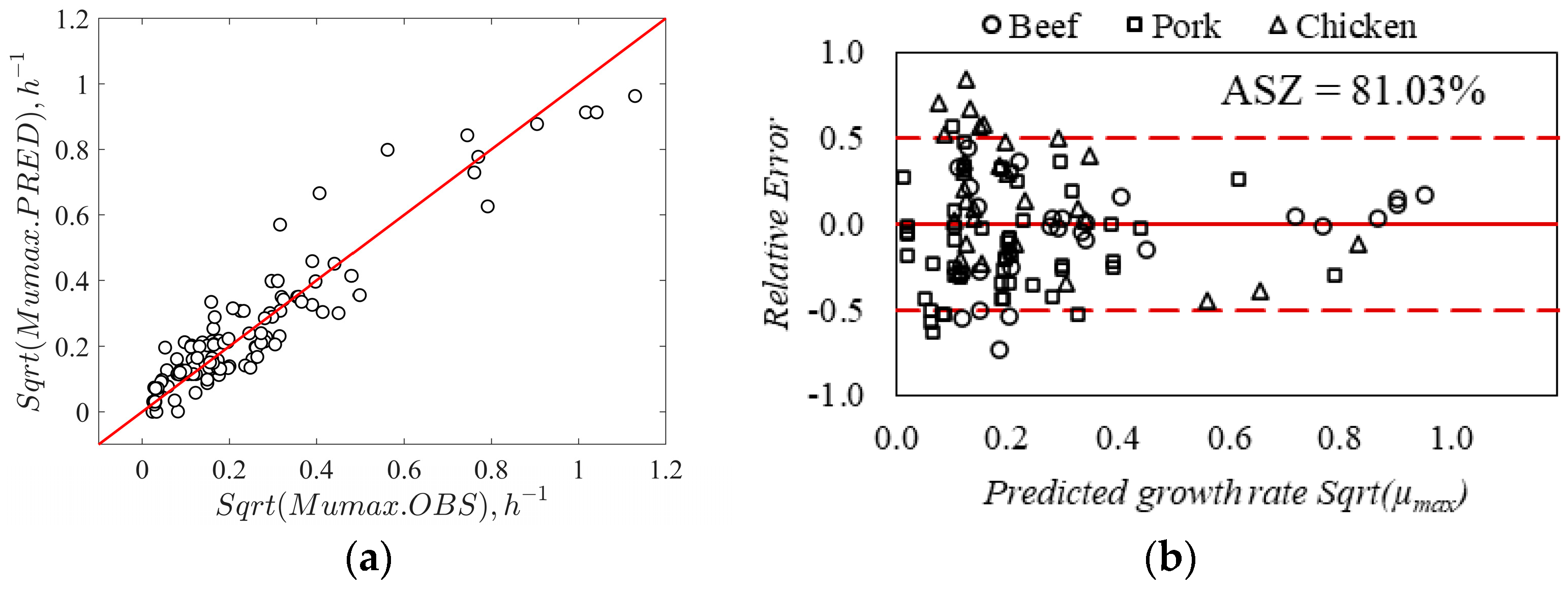
3.2. Validation of the Secondary Growth Model
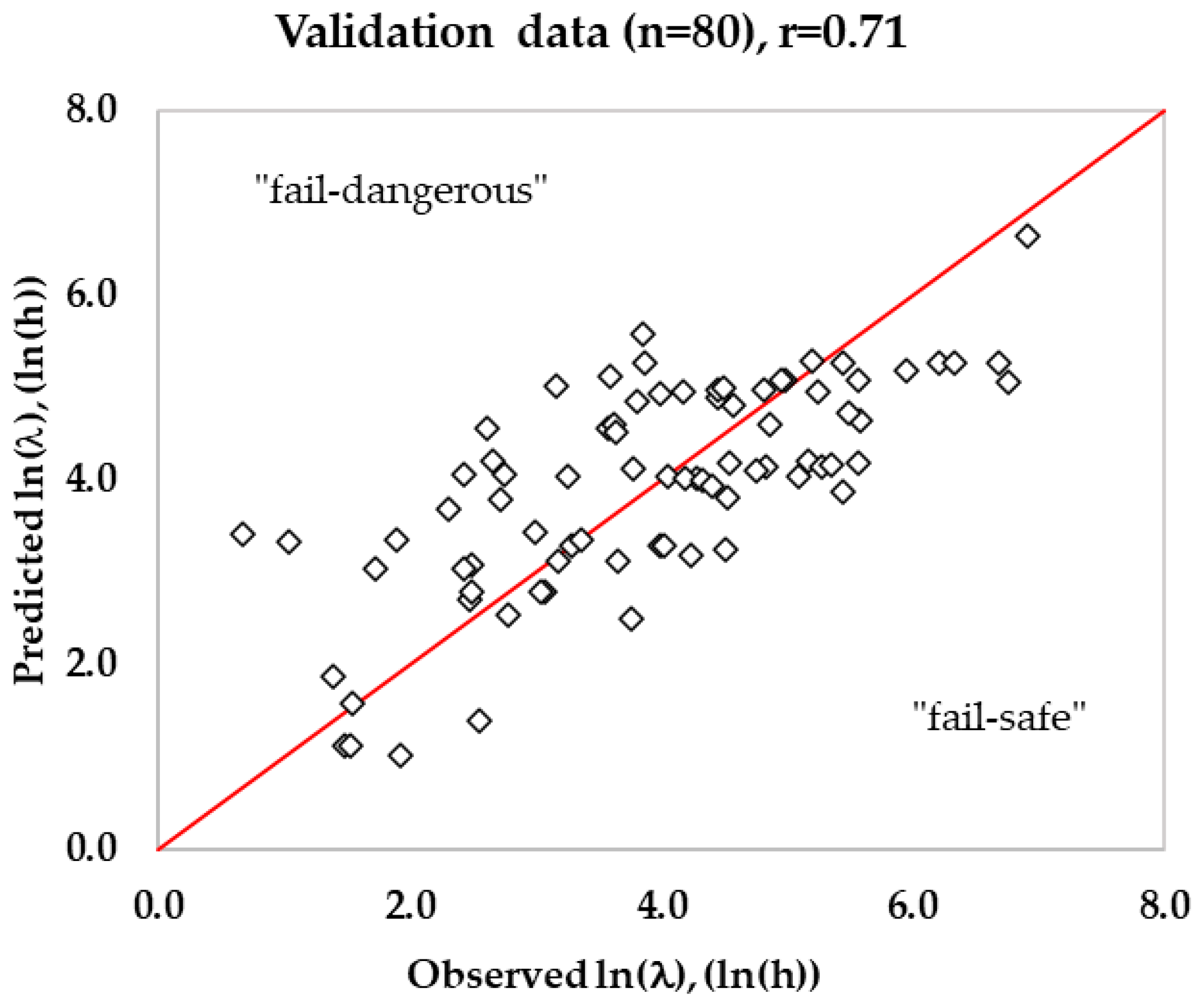
3.3. Validation of Predicted Lag Time in RTE Meats
4. Discussion
| Products | No. of Strains | n | T (°C) | pH | NaCl | aw | Acetate (%) | Lactate (%) | Propionate (%) | Nitrite (ppm) | Data Source | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wiener pork and Bratwurst | 5 | 4 | 3–7 | 5.9–6.3 | 1.5–2 | 0.97–0.98 | 1–6 | 156 | Pub | [14] | ||
| Ground cooked ham | 5 | 36 | 4–10 | 5.7–6.1 | 2.4 | 0.986 | 1–2 | 1 | Cb | [90] | ||
| Cooked Pork liver sausage | 1 | 24 | 5–20 | 6.01–6.12 | 2–4 | 0.97–0.98 | 0–4 | 98.5 | Pub | [96] | ||
| Sliced cooked ham | 6 | 9 | 7 | 6.22 | 3 | 0.978 | 0–3 | Cb | [97] | |||
| RTE Products | 5 | 4 | 4 | 6.13–6.2 | 2 | 0.983–0.99 | 0–2.5 | 97 | Pub | [87] | ||
| Cooked Cured sliced ham | 3 | 18 | 4–12 | 6.2 | 2.8 | 0.979 | 0–2 | 190 | Pub | [86] | ||
| Ham | 8 | 3 | 4 | 6.27–6.42 | 2.2 | 0.984 | 0.108–0.12 | Pub | [65] | |||
| Pork-beef frankfurter | 3 | 18 | 4–12 | 6.02–6.17 | 2.2 | 0.979 | 0.2 | 156 | Pub | [98] | ||
| Pork-beef bologna | 5 | 1 | 4 | 6.07–6.14 | 2.13–2.16 | 0.979–0.98 | 0–0.05 | 156 | Pub | [88] | ||
| Cooked ham and Mortadella | 1 | 12 | 4–12 | 6.1–6.3 | 2.5–2.8 | 0.976–0.979 | 0.43–0.7 | 102 | Pub | [31] | ||
| Pork Bologna | 10 | 4 | 4–10 | 6.3–6.5 | 2 | 0.98 | 0–1.8 | 156 | Pub | [99] | ||
| Servelat sausage and cooked ham | 3 | 4 | 4–9 | 6–6.3 | 2–2.5 | 0.98 | 0–2.5 | Pub | [100] | |||
| Sliced Cooked Ham | 5 | 5 | 4 | 6.39 | 2.59 | 0.967 | 1.6 | 156 | Pub | [101] | ||
| RTE ham model | 5 | 90 | 4 | 5.5–6.6 | 0.5–2.5 | 0.98–0.99 | 0–0.74 | 0–3.06 | 0.05–0.3 | 0–200 | Pub | [81] |
| Sliced cooked Cured ham | 3 | 8 | 5–10 | 6.2 | 2 | 0.985 | 0-0.2 | 98.5 | Pub | [102] | ||
| Cooked sliced ham | 3 | 5 | 8 | 6.2 | 3 | 0.978 | Cb | [103] | ||||
| Pack Slice Cook Pork | 1 | 2 | 4 | 5.99–6.05 | 2 | 0.985 | Cb | [104] | ||||
| Ham | 1 | 14 | 0–15 | 6.1–6.4 | 2.2–2.4 | 0.982–0.984 | 84–110 | Cb | [105] | |||
| Pork ham | 1 | 8 | 0–15 | 6.6 | 2.7–4 | 0.97–0.98 | 11–170 | Cb | [73] | |||
| Ham | 1 | 10 | 47.2 | 6.4 | 2 | 0.985 | Cb | [106] | ||||
| Mortadella (bologna ham) | 2 | 10 | 8 | 6.19 | 2 | 0.981 | 60 | Cb | [107] | |||
| Cooked cured pork shoulder | 1 | 12 | 0–16 | 6.26 | 2 | 0.985 | 150 | Cb | [108] | |||
| RTE ham and sausages | 3 | 8 | 4–35 | 6.2 | 1.8–2.2 | 0.984–0.986 | Pub | [109] | ||||
| Cooked Cured Pork Sausage | 5 | 30 | 7 | 6.59 | 1.84 | 0.985 | 50–300 | Pub | [110] | |||
| Cooked ham | 1 | 2 | 7 | 6.1–6.2 | 2.2 | 0.984 | Cb | [71] | ||||
| Pork Live pate | 5 | 16 | 4–10 | 6–6.15 | 1–3 | 0973–0.991 | 0–200 | Pub | [111] | |||
| Sliced Cooked ham | 1 | 5 | 2–15 | 6.07 | 2.72 | 0.98 | 100 | Pub | [85] | |||
| Processed meats- bologna | 5 | 7 | 4.4 | 4.8–6.3 | 2.3–3 | 0.95–0.97 | 0–48 | Pub | [79] | |||
| Pork Chorizo | 1 | 13 | 5–30 | 4.79–6.5 | 1.84 | 0.984 | Pub | [112] | ||||
| Meatballs and Sundae | 7 | 10 | 5–37 | 5.6–6.9 | 0.5–2.1 | 0.997–0.989 | Pub | [113] |
| Products | No. of Strains | n | T (°C) | pH | NaCl (%) | aw | Acetate (%) | Lactate (%) | Propionate (%) | Nitrite (ppm) | Data Source | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Turkey slurry | 4 | 6 | 4–25 | 5.2–6.2 | 1.3–2.1 | 0.98–0.99 | 0–2.5 | 200 | Pub | [114] | ||
| RTE sliced turkey breast | 3 | 18 | 4–12 | 6.2 | 2.2 | 0.98 | 0–2 | Pub | [86] | |||
| Uncured Turkey | 8 | 7 | 4 | 6.1–6.4 | 2 | 0.98 | 98.5 | Pub | [65] | |||
| Uncured turkey | 5 | 1 | 4 | 6.19 | 2 | 0.98 | 0.05 | Pub | [101] | |||
| Comminuted Chicken | 3 | 9 | 5–35 | 6.5 | 2 | 0.98 | 0–4 | Pub | [66] | |||
| Chicken salad | 5 | 5 | 4–12.8 | 5.6–6 | 0.61 | 0.99 | 0.045–0.051 | Cb | [115] | |||
| Sliced cooked Turkey bologna | 7 | 3 | 4 | 6.5–6.7 | 2 | 0.98 | 0–0.5 | 0–2 | 156 | Pub | [116] | |
| Sliced Cooked Ham | 5 | 5 | 4 | 6.42 | 1.7 | 0.972 | 3.2 | 0.05–0.3 | Pub | [88] | ||
| Cured Deli Style Turkey | 5 | 8 | 4–7 | 6.1–6.4 | 1.7–1.8 | 0.97 | 1.8 | 0.2–0.5 | Pub | [89] | ||
| Turkey bologna | 5 | 2 | 4–7 | 6.17 | 2.2 | 0.973 | 1.6 | 60 | Pub | [117] | ||
| RTE turkey meat | 1 | 2 | 10 | 6.2 | 2.5 | 0.98 | Pub | [118] | ||||
| Sliced roasted turkey | 3 | 3 | 5–10 | 6.2 | 2.5 | 0.98 | Pub | [102] | ||||
| Chicken liver pate or minced chicken breast | 1 | 2 | 6.8–30.4 | 5.6 | 0.8 | 0.99 | Cb | [119] | ||||
| Precooked chicken nuggets | 1 | 3 | 3–11 | 6.5 | 1.5 | 0.98 | Pub | [120] | ||||
| Chicken breast | 2 | 30 | 0–15 | 5.6 | 2–4 | 0.97–0.98 | Cb | [76] | ||||
| Dark-meat chicken nuggets | 1 | 3 | 3–11 | 6.5 | 1.5 | 0.98 | Cb | [121] | ||||
| Sliced Chicken Breast | 1 | 2 | 7 | 6.2 | 2.8 | 0.97 | Cb | [71] | ||||
| RTE chicken salad | 3 | 9 | 4–16 | 5.93 | 1.3 | 0.99 | Pub | [122] | ||||
| Chicken nuggets | 6 | 5 | 4–16 | 6.21 | 1.5 | 0.98 | Pub | [123] | ||||
| Chicken salad | 3 | 9 | 5–25 | 5.9 | 0.612 | 0.99 | Pub | [124] | ||||
| Processed meats- Sliced chicken | 5 | 3 | 4.4 | 6.3–6.5 | 1.7–2.7 | 0.97–0.98 | Pub | [79] | ||||
| Cooked deli turkey breast | 5 | 31 | 5 | 6.1–6.83 | 0.6–2.5 | 0.996–0.982 | 0–1.35 | Pub/PC | [125] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A Review of Listeria Monocytogenes: An Update on Outbreaks, Virulence, Dose-Response, Ecology, and Risk Assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- CDC Listeira (Listeriosis). Available online: https://www.cdc.gov/listeria/index.html (accessed on 10 October 2022).
- Hwang, C.-A.; Sheen, S. Growth Characteristics of Listeria Monocytogenes as Affected by a Native Microflora in Cooked Ham under Refrigerated and Temperature Abuse Conditions. Food Microbiol. 2011, 28, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, S.; Chen, X.; Qu, C. Review Controlling Listeria Monocytogenes in Ready-to-Eat Meat and Poultry Products: An Overview of Outbreaks, Current Legislations, Challenges, and Future Prospects. Trends Food Sci. Technol. 2021, 116, 24–35. [Google Scholar] [CrossRef]
- Potter, A.; Murray, J.; Lawson, B.; Graham, S. Trends in Product Recalls within the Agri-Food Industry: Empirical Evidence from the USA, UK and the Republic of Ireland. Trends Food Sci. Technol. 2012, 28, 77–86. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Drosinos, E.H.; Skandamis, P.N. Food Recalls and Warnings Due to the Presence of Foodborne Pathogens—A Focus on Fresh Fruits, Vegetables, Dairy and Eggs. Curr. Opin. Food Sci. 2017, 18, 71–75. [Google Scholar] [CrossRef]
- Soon, J.M.; Brazier, A.K.M.; Wallace, C.A. Determining Common Contributory Factors in Food Safety Incidents—A Review of Global Outbreaks and Recalls 2008–2018. Trends Food Sci. Technol. 2020, 97, 76–87. [Google Scholar] [CrossRef]
- EFSA-ECDPC. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e7666. [Google Scholar] [CrossRef]
- FSIS-USDA. 9 CFR Part 430: Control of Listeria Monocytogenes in Ready-to-Eat Meat and Poultry Products. Food Saf. Insp. Serv. Fed. Regist. 2015, 80, 35178–35188. [Google Scholar]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs; European Commission: Brussels, Belgium, 2005; pp. 1–26.
- Health-Canada Policy on Listeria Monocytogenes in Ready-to-Eat Foods. Available online: https://www.canada.ca/en/health-canada/services/food-nutrition/legislation-guidelines/policies/policy-listeria-monocytogenes-ready-eat-foods-2011.html (accessed on 10 October 2022).
- Australian-Government. Australia New Zealand Food Standards Code—Standard 1.6.1–Microbiological Limits in Food, Schedule 27; Australian-Government: Canberra, Australia, 2017.
- de Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The Global Burden of Listeriosis: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef]
- Glass, K.A.; Granberg, D.A.; Smith, A.L.; Mcnamara, A.M.; Hardin, M.; Mattias, J.; Ladwig, K.; Johnson, E.A. Inhibition of Listeria Monocytogenes by Sodium Diacetate and Sodium Lactate on Wieners and Cooked Bratwurst. J. Food Prot. 2002, 65, 116–123. [Google Scholar] [CrossRef]
- Lu, Z.; Sebranek, J.G.; Dickson, J.S.; Mendonca, A.F.; Bailey, T.B. Inhibitory Effects of Organic Acid Salts for Control of Listeria Monocytogenes on Frankfurters. J. Food Prot. 2005, 68, 499–506. [Google Scholar] [CrossRef]
- Sansawat, T.; Zhang, L.; Jeong, J.Y.; Xu, Y.; Hessell, G.; Ryser, E.T.; Harte, J.B.; Tempelman, R.; Kang, I. Inhibition of Listeria Monocytogenes in Full- and Low-Sodium Frankfurters at 4, 7, or 10°C Using Spray-Dried Mixtures of Organic Acid Salts. J. Food Prot. 2013, 76, 1557–1567. [Google Scholar] [CrossRef]
- European-Commission. Regulation (EC) No 1333/2008 of the European Parliament and of the Council on Food Additives. Off. J. Eur. Union 2008, 354, 16e33. [Google Scholar]
- FDA 21CFR172.135—Food Additives Permitted for Direct Addition to Food for Human Consumption; Food & Drug Administration: Silver Spring, MD, USA, 1994; p. 21.
- Samelis, J.; Sofos, J.N.; Kain, M.L.; Scanga, J.A.; Belk, K.E.; Smith, G.C. Organic Acids and Their Salts as Dipping Solutions to Control Listeria Monocytogenes Inoculated Following Processing of Sliced Pork Bologna Stored at 4 °C in Vacuum Packages. J. Food Prot. 2001, 64, 1722–1729. [Google Scholar] [CrossRef]
- Mani-López, E.; García, H.S.; López-Malo, A. Organic Acids as Antimicrobials to Control Salmonella in Meat and Poultry Products. Food Res. Int. 2012, 45, 713–721. [Google Scholar] [CrossRef]
- Skjerdal, T.; Gangsei, L.E.; Alvseike, O.; Kausrud, K.; De Cesare, A.; Alexa, E.-A.; Alvarez-Ordóñez, A.; Moen, L.H.; Osland, A.M.; From, C.; et al. Development and Validation of a Regression Model for Listeria Monocytogenes Growth in Roast Beefs. Food Microbiol. 2021, 98, 103770. [Google Scholar] [CrossRef]
- Giffel, M.C.; Zwietering, M.H. Validation of Predictive Models Describing the Growth of Listeria Monocytogenes. Int. J. Food Microbiol. 1999, 46, 135–149. [Google Scholar] [CrossRef]
- Silva, B.N.; Cadavez, V.; Teixeira, J.A.; Ellouze, M.; Gonzales-Barron, U. Cardinal Parameter Meta-Regression Models Describing Listeria Monocytogenes Growth in Broth. Food Res. Int. 2020, 136, 109476. [Google Scholar] [CrossRef]
- Augustin, J.-C.; Carlier, V. Modelling the Growth Rate of Listeria Monocytogenes with a Multiplicative Type Model Including Interactions between Environmental Factors. Int. J. Food Microbiol. 2000, 56, 53–70. [Google Scholar] [CrossRef]
- Augustin, J.-C.; Carlier, V. Mathematical Modelling of the Growth Rate and Lag Time for Listeria Monocytogenes. Int. J. Food Microbiol. 2000, 56, 29–51. [Google Scholar] [CrossRef]
- LeMarc, Y.; Pin, C.; Baranyi, J. Methods to Determine the Growth Domain in a Multidimensional Environmental Space. Int. J. Food Microbiol. 2005, 100, 3–12. [Google Scholar] [CrossRef]
- Zuliani, V.; Lebert, I.; Augustin, J.-C.; Garry, P.; Vendeuvre, J.-L.; Lebert, A. Modelling the Behaviour of Listeria Monocytogenes in Ground Pork as a Function of PH, Water Activity, Nature and Concentration of Organic Acid Salts. J. Appl. Microbiol. 2007, 103, 536–550. [Google Scholar] [CrossRef]
- Pujol, L.; Kan-King-Yu, D.; Le Marc, Y.; Johnston, M.D.; Rama-Heuzard, F.; Guillou, S.; McClure, P.; Membré, J.-M. Establishing Equivalence for Microbial-Growth-Inhibitory Effects (“Iso-Hurdle Rules”) by Analyzing Disparate Listeria Monocytogenes Data with a Gamma-Type Predictive Model. Appl. Environ. Microbiol. 2012, 78, 1069–1080. [Google Scholar] [CrossRef]
- Coroller, L.; Kan-King-Yu, D.; Leguerinel, I.; Mafart, P.; Membré, J.-M. Modelling of Growth, Growth/No-Growth Interface and Nonthermal Inactivation Areas of Listeria in Foods. Int. J. Food Microbiol. 2012, 152, 139–152. [Google Scholar] [CrossRef]
- Martinez-Rios, V.; Gkogka, E.; Dalgaard, P. New Term to Quantify the Effect of Temperature on PHmin-Values Used in Cardinal Parameter Growth Models for Listeria Monocytogenes. Front. Microbiol. 2019, 10, 1510. [Google Scholar] [CrossRef]
- Hereu, A.; Dalgaard, P.; Garriga, M.; Aymerich, T.; Bover-Cid, S. Analysing and Modelling the Growth Behaviour of Listeria Monocytogenes on RTE Cooked Meat Products after a High Pressure Treatment at 400 MPa. Int. J. Food Microbiol. 2014, 186, 84–94. [Google Scholar] [CrossRef]
- Couvert, O.; Guégan, S.; Hézard, B.; Huchet, V.; Lintz, A.; Thuault, D.; Stahl, V. Modeling Carbon Dioxide Effect in a Controlled Atmosphere and Its Interactions with Temperature and PH on the Growth of L. Monocytogenes and P. Fluorescens. Food Microbiol. 2017, 68, 89–96. [Google Scholar] [CrossRef]
- McClure, P.J.; Cole, M.B.; Davies, K.W. An Example of the Stages in the Development of a Predictive Mathematical Model for Microbial Growth: The Effects of NaCl, PH and Temperature on the Growth of Aeromonas Hydrophila. Int. J. Food Microbiol. 1994, 23, 359–375. [Google Scholar] [CrossRef]
- Oscar, T.E. Validation of Lag Time and Growth Rate Models for Salmonella Typhimurium: Acceptable Prediction Zone Method. J. Food Sci. 2005, 70, M129–M137. [Google Scholar] [CrossRef]
- Fakruddin, M.; Mazumdar, R.M.; Mannan, K.S. Bin Predictive Microbiology: Modeling Microbial Responses in Food. Ceylon J. Sci. Biol. Sci. 2011, 40, 121–131. [Google Scholar] [CrossRef]
- Koutsoumanis, K.P.; Kendall, P.A.; Sofos, J.N. A Comparative Study on Growth Limits of Listeria Monocytogenes as Affected by Temperature, PH and Aw When Grown in Suspension or on a Solid Surface. Food Microbiol. 2004, 21, 415–422. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, F.; Valero, A. Predictive Microbiology in Foods; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Skandamis, P.N.; Jeanson, S. Colonial vs. Planktonic Type of Growth: Mathematical Modeling of Microbial Dynamics on Surfaces and in Liquid, Semi-Liquid and Solid Foods. Front. Microbiol. 2015, 6, 1178. [Google Scholar] [CrossRef]
- McMeekin, T.A.; Baranyi, J.; Bowman, J.; Dalgaard, P.; Kirk, M.; Ross, T.; Schmid, S.; Zwietering, M.H. Information Systems in Food Safety Management. Int. J. Food Microbiol. 2006, 112, 181–194. [Google Scholar] [CrossRef]
- Gill, C.O.; Greer, G.G.; Dilts, B.D. The Aerobic Growth of Aeromonas Hydrophila and Listeria Monocytogenes in Broths and on Pork. Int. J. Food Microbiol. 1997, 35, 67–74. [Google Scholar] [CrossRef]
- Lobete, M.M.; Fernandez, E.N.; Impe, J.F.M. Van Recent Trends in Non-Invasive in Situ Techniques to Monitor Bacterial Colonies in Solid (Model) Food. Front. Microbiol. 2015, 6, 148. [Google Scholar] [CrossRef]
- Coroller, L.; Guerrot, V.; Huchet, V.; LeMarc, Y.; Mafart, P.; Sohier, D.; Thuault, D. Modelling the Influence of Single Acid and Mixture on Bacterial Growth. Int. J. Food Microbiol. 2005, 100, 167–178. [Google Scholar] [CrossRef]
- Nyhan, L.; Begley, M.; Mutel, A.; Qu, Y.; Johnson, N.; Callanan, M. Predicting the Combinatorial Effects of Water Activity, PH and Organic Acids on Listeria Growth in Media and Complex Food Matrices. Food Microbiol. 2018, 74, 75–85. [Google Scholar] [CrossRef]
- Martinez-Rios, V.; Gkogka, E.; Dalgaard, P. Predicting Growth of Listeria Monocytogenes at Dynamic Conditions during Manufacturing, Ripening and Storage of Cheeses—Evaluation and Application of Models. Food Microbiol. 2020, 92, 103578. [Google Scholar] [CrossRef]
- Rosso, L.; Bajard, S.; Flandrois, J.P.; Lahellec, C.; Fournaud, J.; Veit, P. Differential Growth of Listeria Monocytogenes at 4 and 8°C: Consequences for the Shelf Life of Chilled Products. J. Food Prot. 1996, 59, 944–949. [Google Scholar] [CrossRef]
- Augustin, J.-C.; Zuliani, V.; Cornu, M.; Guillier, L. Growth Rate and Growth Probability of Listeria Monocytogenes in Dairy, Meat and Seafood Products in Suboptimal Conditions. J. Appl. Microbiol. 2005, 99, 1019–1042. [Google Scholar] [CrossRef]
- Le Marc, Y.; Huchet, V.; Bourgeois, C.M.; Guyonnet, J.P.; Mafart, P.; Thuault, D. Modelling the Growth Kinetics of Listeria as a Function of Temperature, PH and Organic Acid Concentration. Int. J. Food Microbiol. 2002, 73, 219–237. [Google Scholar] [CrossRef]
- Wemmenhove, E.; van Valenberg, H.J.F.; Zwietering, M.H.; van Hooijdonk, T.C.M.; Wells-Bennik, M.H.J. Minimal Inhibitory Concentrations of Undissociated Lactic, Acetic, Citric and Propionic Acid for Listeria Monocytogenes under Conditions Relevant to Cheese. Food Microbiol. 2016, 58, 63–67. [Google Scholar] [CrossRef]
- Mejlholm, O.; Gunvig, A.; Borggaard, C.; Blom-Hanssen, J.; Mellefont, L.; Ross, T.; Leroi, F.; Else, T.; Visser, D.; Dalgaard, P. Predicting Growth Rates and Growth Boundary of Listeria Monocytogenes—An International Validation Study with Focus on Processed and Ready-to-Eat Meat and Seafood. Int. J. Food Microbiol. 2010, 141, 137–150. [Google Scholar] [CrossRef]
- Chirife, J.; Resnik, S.L. Unsaturated Solutions of Sodium Chloride as Reference Sources of Water Activity at Various Temperatures. J. Food Sci. 1984, 49, 1486–1488. [Google Scholar] [CrossRef]
- Frida.fooddata.dk Food Data. Available online: https://frida.fooddata.dk (accessed on 10 November 2022).
- USDA-ARS. FoodData Central. Available online: https://fdc.nal.usda.gov/ (accessed on 25 November 2022).
- Ross, T.; Dalgaard, P. Secondary Models. In Modeling Microbial Responses in Food; McKellar, R.C., Lu, X., Eds.; CRC Press: Mississauga, ON, Canada, 2004; p. 360. [Google Scholar]
- Lambert, R.J.; Stratford, M. Weak-Acid Preservatives: Modelling Microbial Inhibition and Response. J. Appl. Microbiol. 1999, 86, 157–164. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; Monteoliva García, G.; Bodea, I.; Cotter, P.D.; Iguaz, A.; Garre, A. A Secondary Model for the Effect of PH on the Variability in Growth Fitness of Listeria Innocua Strains. Food Res. Int. 2024, 186, 114314. [Google Scholar] [CrossRef]
- Cornu, M.; Beaufort, A.; Rudelle, S.; Laloux, L.; Bergis, H.; Miconnet, N.; Serot, T.; Delignette-Muller, M.L. Effect of Temperature, Water-Phase Salt and Phenolic Contents on Listeria Monocytogenes Growth Rates on Cold-Smoked Salmon and Evaluation of Secondary Models. Int. J. Food Microbiol. 2006, 106, 159–168. [Google Scholar] [CrossRef]
- Moller, C.O.A.O.A.; Ilg, Y.; Aabo, S.; Christensen, B.B.B.; Dalgaard, P.; Hansen, T.B.B. Effect of Natural Microbiota on Growth of Salmonella Spp. in Fresh Pork—A Predictive Microbiology Approach. Food Microbiol. 2013, 34, 284–295. [Google Scholar] [CrossRef]
- Ross, T.; Dalgaard, P.; Tienungoon, S. Predictive Modelling of the Growth and Survival of Listeria in Fishery Products. Int. J. Food Microbiol. 2000, 62, 231–245. [Google Scholar] [CrossRef]
- ICMSF. Microorganisms in Foods: Microbiological Specifications of Food Pathogens; Blackie Academic and Professional: London, UK, 1996. [Google Scholar]
- Anyasi, T.; Jideani, A.I.O.; Edokpayi, J.N.; Anokwuru, C.P. Application of Organic Acids in Food Preservation. In Organic Acids, Characteristics, Properties and Synthesis; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2017; pp. 1–47. [Google Scholar]
- Hills, B.P.; Manning, C.E.; Ridge, Y.; Brocklehurst, T. Water Availability and the Survival of Salmonella Typhimurium in Porous Systems. Int. J. Food Microbiol. 1997, 36, 187–198. [Google Scholar] [CrossRef]
- Mejlholm, O.; Dalgaard, P. Development and Validation of an Extensive Growth and Growth Boundary Model for Listeria Monocytogenes in Lightly Preserved and Ready-to-Eat Shrimp. J. Food Prot. 2009, 72, 2132–2143. [Google Scholar] [CrossRef]
- Mbandi, E.; Shelef, L.A. Enhanced Inhibition of Listeria Monocytogenes and Salmonella Enteritidis in Meat by Combinations of Sodium Lactate and Diacetate. J. Food Prot. 2001, 64, 640–644. [Google Scholar] [CrossRef]
- Mbandi, E.; Shelef, L.A.A. Enhanced Antimicrobial Effects of Combination of Lactate and Diacetate on Listeria Monocytogenes and Salmonella Spp. in Beef Bologna. Int. J. Food Microbiol. 2002, 76, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.E.I.; Moosekian, S.R.; Todd, E.C.D.; Ryser, E.T. Growth of Listeria Monocytogenes in Different Retail Delicatessen Meats during Simulated Home Storage. J. Food Prot. 2012, 75, 896–905. [Google Scholar] [CrossRef]
- Shelef, L.A.; Yang, Q. Growth Suppression of Listeria Monocytogenes by Lactates in Broth, Chicken, and Beef. J. Food Prot. 1991, 54, 283–287. [Google Scholar] [CrossRef]
- Stekelenburg, F.K. Enhanced Inhibition of Listeria Monocytogenes in Frankfurter Sausage by the Addition of Potassium Lactate and Sodium Diacetate Mixtures. Food Microbiol. 2003, 20, 133–137. [Google Scholar] [CrossRef]
- Porto, A.C.S.; Franco, B.D.G.M.; Sant’anna, E.S.; Call, J.E.; Piva, A.; Luchansky, J.B. Viability of a Five-Strain Mixture of Listeria Monocytogenes in Vacuum-Sealed Packages of Frankfurters, Commercially Prepared with and without 2.0 or 3.0% Added Potassium Lactate, during Extended Storage at 4 and 10 °C. J. Food Prot. 2002, 65, 308–316. [Google Scholar] [CrossRef]
- Morey, A.; Bowers, J.W.J.; Bauermeister, L.J.; Singh, M.; Huang, T.; McKee, S.R. Effect of Salts of Organic Acids on Listeria Monocytogene, Shelf Life, Meat Quality, and Consumer Acceptability of Beef Frankfurters. J. Food Sci. 2014, 79, M54–M60. [Google Scholar] [CrossRef] [PubMed]
- Grant, I.R.; Nixon, C.R.; Patterson, M.F. Comparison of the Growth of Listeria Monocytogenes in Unirradiated and Irradiated Cook-Chill Roast Beef and Gravy at Refrigeration Temperatures. Lett. Appl. Microbiol. 1993, 17, 55–57. [Google Scholar] [CrossRef]
- Beumer, R. Growth of Listeria Monocytogenes on Sliced Cooked Meat Products. Food Microbiol. 1996, 13, 333–340. [Google Scholar] [CrossRef]
- Huang, L. Growth Kinetics of Listeria Monocytogenes in Broth and Beef Frankfurters- Determination of Lag Phase Duration and Exponential Growth Rate under Isothermal Conditions. J. Food Sci. 2008, 73, E235–E242. [Google Scholar] [CrossRef]
- Grau, F.H.; Vanderlinde, P.B. Occurrence, Numbers, and Growth of Listeria Monocytogenes on Some Vacuum-Packaged Processed Meats. J. Food Prot. 1992, 55, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.A.; Mott, S.J. Growth of Listeria Monocytogenes, Aeromonas Hydrophila and Yersinia Enterocolitica on Cooked Beef under Refrigeration and Mild Temperature Abuse. Food Microbiol. 1993, 10, 429–437. [Google Scholar] [CrossRef]
- Hudson, J.A.; Mott, S.J.; Penney, N. Growth of Listeria Monocytogenes, Aeromonas Hydrophila, and Yersinia Enterocolitica on Vacuum and Saturated Carbon Dioxide Controlled Atmosphere-Packaged Sliced Roast Beef. J. Food Prot. 1994, 57, 204–208. [Google Scholar] [CrossRef]
- Nyati, H. Survival Characteristics and the Applicability of Predictive Mathematical Modelling to Listeria Monocytogenes Growth in Sous Vide Products. Int. J. Food Microbiol. 2000, 56, 123–132. [Google Scholar] [CrossRef]
- Diez-Gonzalez, F.; Belina, D.; Labuza, T.P.; Pal, A. Modeling the Growth of Listeria Monocytogenes Based on a Time to Detect Model in Culture Media and Frankfurters. Int. J. Food Microbiol. 2007, 113, 277–283. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Liu, B.; Dong, Q. One-Step Analysis for Listeria Monocytogenes Growth in Ready-to-Eat Braised Beef at Dynamic and Static Conditions. J. Food Prot. 2019, 82, 1820–1827. [Google Scholar] [CrossRef]
- Glass, K.A.; Doyle, M.P. Fate of Listeria Monocytogenes in Processed Meat Products during Refrigerated Storage. Appl. Environ. Microbiol. 1989, 55, 1565–1569. [Google Scholar] [CrossRef]
- Bucur, F.I.; Borda, D.; Neagu, C.; Grigore-Gurgu, L.; Nicolau, A.I. Deterministic Approach and Monte Carlo Simulation to Predict Listeria Monocytogenes Time to Grow on Refrigerated Ham: A Study Supporting Risk-Based Decisions for Consumers’ Health. J. Food Prot. 2023, 86, 100026. [Google Scholar] [CrossRef]
- Dussault, D.; Vu, K.D.; Lacroix, M. Development of a Model Describing the Inhibitory Effect of Selected Preservatives on the Growth of Listeria Monocytogenes in a Meat Model System. Food Microbiol. 2016, 53, 115–121. [Google Scholar] [CrossRef]
- Augustin, J.-C.; Bergis, H.; Midelet-Bourdin, G.; Cornu, M.; Couvert, O.; Denis, C.; Huchet, V.; Lemonnier, S.; Pinon, A.; Vialette, M.; et al. Design of Challenge Testing Experiments to Assess the Variability of Listeria Monocytogenes Growth in Foods. Food Microbiol. 2011, 28, 746–754. [Google Scholar] [CrossRef]
- Aryani, D.C.; den Besten, H.M.W.; Hazeleger, W.C.; Zwietering, M.H. Quantifying Strain Variability in Modeling Growth of Listeria Monocytogenes. Int. J. Food Microbiol. 2015, 208, 19–29. [Google Scholar] [CrossRef]
- van der Veen, S.; Moezelaar, R.; Abee, T.; Wells-Bennik, M.H.J. The Growth Limits of a Large Number of Listeria Monocytogenes Strains at Combinations of Stresses Show Serotype- and Niche-Specific Traits. J. Appl. Microbiol. 2008, 105, 1246–1258. [Google Scholar] [CrossRef]
- Serra-Castelló, C.; Costa, J.C.C.P.; Jofré, A.; Bolívar, A.; Pérez-Rodríguez, F.; Bover-Cid, S. A Mathematical Model to Predict the Antilisteria Bioprotective Effect of Latilactobacillus Sakei CTC494 in Vacuum Packaged Cooked Ham. Int. J. Food Microbiol. 2022, 363, 109491. [Google Scholar] [CrossRef]
- Pal, A.; Labuza, T.; Diezgonzalez, F. Shelf Life Evaluation for Ready-to-Eat Sliced Uncured Turkey Breast and Cured Ham under Probable Storage Conditions Based on Listeria Monocytogenes and Psychrotroph Growth. Int. J. Food Microbiol. 2008, 126, 49–56. [Google Scholar] [CrossRef]
- Seman, D.L.; Borger, A.C.; Meyer, J.D.; Hall, P.A.; Milkowski, A.L. Modeling the Growth of Listeria Monocytogenes in Cured Ready-to-Eat Processed Meat Products by Manipulation of Sodium Chloride, Sodium Diacetate, Potassium Lactate, and Product Moisture Content. J. Food Prot. 2002, 65, 651–658. [Google Scholar] [CrossRef]
- Glass, K.; Preston, D.; Veesenmeyer, J. Inhibition of Listeria Monocytogenes in Turkey and Pork-Beef Bologna by Combinations of Sorbate, Benzoate, and Propionate. J. Food Prot. 2007, 70, 214–217. [Google Scholar] [CrossRef]
- Glass, K.A.; Mcdonnell, L.M.; Vontayson, R.; Wanless, B.; Badvela, M. Inhibition of Listeria Monocytogenes by Propionic Acid–Based Ingredients in Cured Deli-Style Turkey. J. Food Prot. 2013, 76, 2074–2078. [Google Scholar] [CrossRef]
- Hwang, C.-A.; Sheen, S.; Juneja, V. Effects of Sodium Lactate on the Survival of Listeria Monocytogenes, Escherichia Coli O157:H7, and Salmonella Spp. in Cooked Ham at Refrigerated and Abuse Temperature. Food Nutr. Sci. 2011, 2, 464–470. [Google Scholar] [CrossRef][Green Version]
- Delignette-Muller, M.L. Relation between the Generation Time and the Lag Time of Bacterial Growth Kinetics. Int. J. Food Microbiol. 1998, 43, 97–104. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Klawitter, L.A. Effect of Temperature History on the Growth of Listeria Monocytogenes Scott A at Refrigeration Temperatures. Int. J. Food Microbiol. 1991, 12, 235–245. [Google Scholar] [CrossRef]
- Pla, M.-L.; Oltra, S.; Esteban, M.-D.; Andreu, S.; Palop, A. Comparison of Primary Models to Predict Microbial Growth by the Plate Count and Absorbance Methods. BioMed Res. Int. 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Baka, M.; Noriega, E.; Stamati, I.; Logist, F.; Van Impe, J.F.M. Critical Assessment of the Time-to-Detection Method for Accurate Estimation of Microbial Growth Parameters. J. Food Saf. 2015, 35, 179–192. [Google Scholar] [CrossRef]
- Begot, C.; Lebert, I.; Lebert, A. Variability of the Response of 66Listeria MonocytogenesandListeria Innocuastrains to Different Growth Conditions. Food Microbiol. 1997, 14, 403–412. [Google Scholar] [CrossRef]
- Weaver, R.A.; Shelef, L.A.; Seman, D.L.; Borger, A.C.; Meyer, J.D.; Hall, P.A.; Milkowski, A.L.; Serra-Castelló, C.; Costa, J.C.C.P.; Jofré, A.; et al. Antilisterial Activity of Sodium, Potassium or Calcium Lactate in Pork Liver Sausage. J. Food Saf. 1993, 13, 133–146. [Google Scholar] [CrossRef]
- Uyttendaele, M.; Rajkovic, A.; Benos, G.; Franc ois, K.; Devlieghere, K.; Debevere, J. Evaluation of a Challenge Testing Protocol to Assess the Stability of Ready-to-Eat Cooked Meat Products against Growth of Listeria Monocytogenes. Int. J. Food Microbiol. 2004, 90, 219–236. [Google Scholar] [CrossRef]
- Pal, A.; Labuza, T.P.; Diez-Gonzalez, F. Evaluating the Growth of Listeria Monocytogenes in Refrigerated Ready-to-Eat Frankfurters: Influence of Strain, Temperature, Packaging, Lactate and Diacetate, and Background Microflora. J. Food Prot. 2008, 71, 1806–1816. [Google Scholar] [CrossRef]
- Barmpalia, I.M.; Koutsoumanis, K.P.; Geornaras, I.; Belk, K.E.; Scanga, J.A.; Kendall, P.A.; Smith, G.C.; Sofos, J.N. Effect of Antimicrobials as Ingredients of Pork Bologna for Listeria Monocytogenes Control during Storage at 4 or 10 °C. Food Microbiol. 2005, 22, 205–211. [Google Scholar] [CrossRef]
- Blom, H.; Nerbrink, E.; Dainty, R.; Hagtvedt, T.; Borch, E.; Nissen, H.; Nesbakken, T. Addition of 2.5% Lactate and 0.25% Acetate Controls Growth of Listeria Monocytogenes in Vacuum-Packed, Sensory-Acceptable Servelat Sausage and Cooked Ham Stored at 4 °C. Int. J. Food Microbiol. 1997, 38, 71–76. [Google Scholar] [CrossRef]
- Glass, K.A.; Mcdonnell, L.M.; Rassel, R.C.; Zierke, K.L. Controlling Listeria Monocytogenes on Sliced Ham and Turkey Products Using Benzoate, Propionate, and Sorbate. J. Food Prot. 2007, 70, 2306–2312. [Google Scholar] [CrossRef]
- Burnett, S.L.; Mertz, E.L.; Bennie, B.; Ford, T.; Starobin, A. Growth or Survival of Listeria Monocytogenes in Ready-to-Eat Meat Products and Combination Deli Salads During Refrigerated Storage. J. Food Sci. 2005, 70, m301–m304. [Google Scholar] [CrossRef]
- Bredholt, S.; Nesbakken, T.; Holck, A. Protective Cultures Inhibit Growth of Listeria Monocytogenes and Escherichia Coli O157:H7 in Cooked, Sliced, Vacuum- and Gas-Packaged Meat. Int. J. Food Microbiol. 1999, 53, 43–52. [Google Scholar] [CrossRef]
- Sym’Previus Ecole Nationale Vétérinaire d’Alfort, Maisons Alfort, France. Sym’Previus Partner. Available online: https://combase.errc.ars.usda.gov/ (accessed on 25 November 2022).
- Food Research Association (FRA), UK. FRA Food Standards Agency Funded Data Generated at Leatherhead. Available online: https://frida.fooddata.dk/food/lists/grouped/56/65?lang=en&#group65 (accessed on 25 November 2022).
- TIA Tasmanian Institute of Agriculture (TIA), University of Tasmania, Australia. Available online: https://combase.errc.ars.usda.gov/ (accessed on 25 November 2022).
- Daminelli, P.; Dalzini, E.; Cosciani-Cunico, E.; Finazzi, G.; D’Amico, S.; Losio, M.N. Prediction of the Maximal Growth Rate of Listeria Monocytogenes in Sliced Mortadella by the Square Root Type Model. Ital. J. Food Sci. 2014, 26, 261–267. [Google Scholar]
- Mataragas, M.; Drosinos, E.H.; Siana, P.; Skandamis, P.; Metaxopoulos, I. Determination of the Growth Limits and Kinetic Behavior of Listeria Monocytogenes in a Sliced Cooked Cured Meat Product: Validation of the Predictive Growth Model under Constant and Dynamic Temperature Storage Conditions. J. Food Prot. 2006, 69, 1312–1321. [Google Scholar] [CrossRef]
- Luo, K.; Hong, S.-S.; Oh, D.-H. Modeling the Effect of Storage Temperatures on the Growth of Listeria Monocytogenes on Ready-to-Eat Ham and Sausage. J. Food Prot. 2015, 78, 1675–1681. [Google Scholar] [CrossRef]
- King, A.M.; Glass, K.A.; Milkowski, A.L.; Seman, D.L.; Sindelar, J.J. Modeling the Impact of Ingoing Sodium Nitrite, Sodium Ascorbate, and Residual Nitrite Concentrations on Growth Parameters of Listeria Monocytogenes in Cooked, Cured Pork Sausage. J. Food Prot. 2016, 79, 184–193. [Google Scholar] [CrossRef]
- Farber, J.M.; McKellar, R.C.; Ross, W.H. Modelling the Effects of Various Parameters on the Growth of Listeria Monocytogenes on Liver Pâté. Food Microbiol. 1995, 12, 447–453. [Google Scholar] [CrossRef]
- Gutiérrez-Chocoza, M.A.; López-Romero, J.C.; García-Galaz, A.; González-Ríos, H.; Peña-Ramos, A.; Juneja, V.K.; Pérez-Báez, A.J.; Valenzuela-Melendres, M. Modeling the Effects of Temperature and PH on Listeria Monocytogenes Growth in Mexican-Style Pork Chorizo. Appl. Food Res. 2023, 3, 100336. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, H.J. Modeling and Scenario-Based Risk Estimation of Listeria Monocytogenes in Meatballs and Sundae, a Korean Black Pudding, under Different Packaging Conditions. Food Control 2023, 152, 109886. [Google Scholar] [CrossRef]
- Schlyter, J.H.; Glass, K.A.; Loeffelholz, J.; Degnan, A.J.; Luchansky, J.B. The Effects of Diacetate with Nitrite, Lactate, or Pediocin on the Viability of Listeria Monocytegenes in Turkey Slurries. Int. J. Food Microbiol. 1993, 19, 271–281. [Google Scholar] [CrossRef]
- Erickson, J.P.; Mckenna, D.N.; Woodruff, M.A.; Bloom, J.S. Fate of Salmonella Spp., Listeria Monocytogenes, and Indigenous Spoilage Microorganisms in Home-Style Salads Prepared with Commercial Real Mayonnaise or Reduced Calorie Mayonnaise Dressings. J. Food Prot. 1993, 56, 1015–1021. [Google Scholar] [CrossRef]
- Wederquist, H.J.; Sofos, J.N.; Schmidt, G.R. Listeria Monocytogenes Inhibition in Refrigerated Vacuum Packaged Turkey Bologna by Chemical Additives. J. Food Sci. 1994, 59, 498–500. [Google Scholar] [CrossRef]
- Glass, K.; Lindsey, M.; Sawyer, C.; Claus, J. Minimum Nitrite Levels Required to Control Listeria Monocytogenes on Ready-to-Eat Poultry Products Manufactured with Lactate and Diacetate; University of Wisconsin-Madison: Madison, WI, USA, 2008. [Google Scholar]
- Peterson, L.; Faith, N.; Czuprynski, C. Growth of L. Monocytogenes Strain F2365 on Ready-to-Eat Turkey Meat Does Not Enhance Gastrointestinal Listeriosis in Intragastrically Inoculated A/J Mice. Int. J. Food Microbiol. 2008, 126, 112–115. [Google Scholar] [CrossRef]
- Bovill, R.; Bew, J.; Cook, N.; D’Agostino, M.; Wilkinson, N.; Baranyi, J. Predictions of Growth for Listeria Monocytogenes and Salmonella during Fluctuating Temperature. Int. J. Food Microbiol. 2000, 59, 157–165. [Google Scholar] [CrossRef]
- Marshall, D.L.; Wiese-Lehigh, P.L.; Wells, J.H.; Farr, A.J. Comparative Growth of Listeria Monocytogenes and Pseudomonas Fluorescens on Precooked Chicken Nuggets Stored under Modified Atmospheres. Food Prot. 1991, 54, 841–844. [Google Scholar] [CrossRef]
- Zhao, Y.; Wells, J.; Marshall, D.L. Description of Log Phase Growth for Selected Microorganisms during Modified Atmosphere Storage. J. Food Process Eng. 1992, 15, 299–317. [Google Scholar] [CrossRef]
- Bernardo, R.; Barreto, A.S.; Nunes, T.; Henriques, A.R. Estimating Listeria Monocytogenes Growth in Ready-to-Eat Chicken Salad Using a Challenge Test for Quantitative Microbial Risk Assessment. Risk Anal. 2020, 40, 2427–2441. [Google Scholar] [CrossRef]
- Lianou, A.; Raftopoulou, O.; Spyrelli, E.; Nychas, G.-J.E. Growth of Listeria Monocytogenes in Partially Cooked Battered Chicken Nuggets as a Function of Storage Temperature. Foods 2021, 10, 533. [Google Scholar] [CrossRef]
- Sahu, S.N.; Kim, B.; Ferguson, M.S.; Zink, D.L.; Datta, A.R. Growth Potential of Listeria Monocytogenes in Artificially Contaminated Celery and Chicken Salad. Food Control 2017, 73, 1229–1236. [Google Scholar] [CrossRef]
- Shrestha, S.; Erdmann, J.J.; Smith, S.A. Predicting the Growth of Listeria Monocytogenes in Cooked, Sliced Deli Turkey Breast as a Function of Clean-Label Antimicrobials, PH, Moisture, and Salt. J. Food Prot. 2022, 85, 945–955. [Google Scholar] [CrossRef]


| Factors | Criteria to Exclude |
|---|---|
| Growth curves | Curves having less than three points in the exponential phase |
| Treatments | Irradiation and high-pressure processed samples |
| Growth rate | The estimated growth rate (μmax) equal to zero |
| Antimicrobials | Products with surface treatment (only products with antimicrobials incorporated into the food formulation were considered) |
| Poor data | Curves with poor model fitting (R2 < 0.9) |
| Storage atmosphere | Modified Atmospheric Package or CO2. |
| Acetic | Lactic | Propionic | Nitrite | References | |
|---|---|---|---|---|---|
| pKa | 4.76 | 3.8 | 4.87 | 3.37 | [46,47] |
| MICU (mM) | 20.3 | 8.0 | 8.8 | 25 |
| Parameters | Estimated Value | 95% CI Values | Literature Range | References | |
|---|---|---|---|---|---|
| LCI | UCI | ||||
| µopt (h−1) | 1.126 | 0.65 | 1.66 | 0.85 to 1.33 | [27,43] |
| Topt (°C) | 37.0 | 34.83 | 39.38 | 35.9 to 39.7 | [23,43] |
| Tmin (°C) | −1.57 | −2.14 | −1.0 | −4.5 to 1.16 | [43,47] |
| pHmin | 4.19 | 3.56 | 4.79 | 4.03 to 4.57 | [29] |
| awmin | 0.932 | 0.904 | 0.938 | 0.92 to 0.93 | [43] |
| awopt | 0.998 | 0.995 | 1.00 | 0.997 to 1.0 | [25] |
| MICU NIT | 22.12 | 2.36 | 41.8 | 11.4 to 25 | [27,43,47,48] |
| MICU AA | 18.33 | 0.98 | 37.6 | 17.8 to 22.8 | |
| MICU LA | 6.88 | 4.04 | 9.73 | 1.7 to 9.8 | |
| MICU PA | 9.88 | −9.0 | 28.8 | 7.6 to 9.9 | |
| Model | Bf | Af | ASZ (%) | %B | %D | FS (%) | FD (%) | AIC |
|---|---|---|---|---|---|---|---|---|
| Without interaction | 1.12 | 1.48 | 76.7 | 12.7 | 48.1 | 15.5 | 7.7 | −228.7 |
| With interaction | 0.96 | 1.50 | 81.0 | −4.0 | 50.4 | 8.6 | 10.3 | −230.7 |
| Products | No. of Strains | n | T (°C) | pH | NaCl | aw | Acetate (%) | Lactate (%) | Propionate (%) | Nitrite (ppm) | Data Source | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comminuted beef emulsion | 1 | 11 | 5–10 | 6.30 | 2.00 | 0.986 | 0.1–0.2 | 1.8–2.5 | Cb | [63] | ||
| Beef Bologna | 5 | 8 | 5–10 | 5.9–6.3 | 2.50 | 0.973 | 2.50 | Cb | [64] | |||
| Roasted beef | 4 | 18 | 4–12 | 5.80 | 0.30 | 0.998 | 0–0.1 | 0.2–0.4 | 98.5 | Pub | [21] | |
| Roasted beef | 8 | 3 | 4 | 5.6–6.19 | 1.15 | 0.990 | 0.12 | Pub | [65] | |||
| Frankfurters | 6 | 21 | 4–10 | 6.15–6.4 | 1.80 | 0.983 | 0.25–0.8 | 0.14–0.25 | 112.5 | Pub | [16] | |
| Comminuted meat | 3 | 25 | 5–35 | 6.27 | 2.00 | 0.985 | 0–4 | 140 | Pub | [66] | ||
| Frankfurter sausage | 1 | 2 | 4 | 6.1–6.3 | 2.04–2.11 | 0.981 | 0.12–0.18 | 0.66–2.26 | 11–19 | Pub | [67] | |
| Frankfurter | 5 | 10 | 4–10 | 5.68–6.18 | 1.73–2 | 0.980–0.983 | 0–3 | 4.1–4.8 | Pub | [68] | ||
| Frankfurter | 1 | 3 | 4 | 6.20 | 1.80 | 0.982 | 0–2 | 98.5 | Pub | [69] | ||
| Beef gravy | 2 | 4 | 5–10 | 6.00 | 1.00 | 0.994 | Pub | [70] | ||||
| Luncheon meat | 1 | 2 | 7 | 6–6.3 | 1.10 | 0.992 | Pub | [71] | ||||
| Frankfurter | 4 | 5 | 15–40 | 6.30 | 1.80 | 0.982 | Pub | [72] | ||||
| Corned beef | 1 | 5 | 0–15 | 6.20 | 3.25 | 0.973 | 5.00 | Cb | [73] | |||
| Cooked beef | 2 | 4 | 5–10 | 5.80 | 1.00 | 0.996 | Cb | [74] | ||||
| Sliced roast meat | 2 | 4 | −1.5–3 | 6.10 | 1.15 | 0.990 | Cb | [75] | ||||
| Beef sirloin | 1 | 5 | 0–15 | 6.00 | 1.80 | 0.982 | Cb | [76] | ||||
| Frankfurter | 1 | 3 | 4–18 | 6.20 | 1.80 | 0.982 | 98.50 | Pub | [77] | |||
| Ready-to-eat braised beef | 4 | 24 | 4–40 | 6.20 | 1.00 | 0.990 | PC | [78] | ||||
| Ham and roasted meat | 5 | 4 | 4.4 | 5.4–6.4 | 0.6–3 | 0.976–0.992 | 28–42 | Pub | [79] | |||
| Ham | 1 | 12 | 4–7 | 6.67 | 1.9 | 0.986 | 85 | Pub | [80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gowda, N.A.N.; Singh, M.; Lommerse, G.; Kumar, S.; Heintz, E.; Subbiah, J. Predictive Model for Listeria monocytogenes in RTE Meats Using Exclusive Food Matrix Data. Foods 2024, 13, 3948. https://doi.org/10.3390/foods13233948
Gowda NAN, Singh M, Lommerse G, Kumar S, Heintz E, Subbiah J. Predictive Model for Listeria monocytogenes in RTE Meats Using Exclusive Food Matrix Data. Foods. 2024; 13(23):3948. https://doi.org/10.3390/foods13233948
Chicago/Turabian StyleGowda, N. A. Nanje, Manjari Singh, Gijs Lommerse, Saurabh Kumar, Eelco Heintz, and Jeyamkondan Subbiah. 2024. "Predictive Model for Listeria monocytogenes in RTE Meats Using Exclusive Food Matrix Data" Foods 13, no. 23: 3948. https://doi.org/10.3390/foods13233948
APA StyleGowda, N. A. N., Singh, M., Lommerse, G., Kumar, S., Heintz, E., & Subbiah, J. (2024). Predictive Model for Listeria monocytogenes in RTE Meats Using Exclusive Food Matrix Data. Foods, 13(23), 3948. https://doi.org/10.3390/foods13233948





