Metagenomic Study on Chinese Homemade Paocai: The Effects of Raw Materials and Fermentation Periods on the Microbial Ecology and Volatile Components
Abstract
:1. Introduction
2. Materials and Methods
2.1. Paocai Preparations
2.2. DNA Extraction, Library Construction and Metagenomic Sequencing
2.3. Metagenome Data Integration
2.4. Species Annotation
2.5. Determination of VOCs Using GC–MS
2.6. Statistical Analysis
3. Results and Discussion
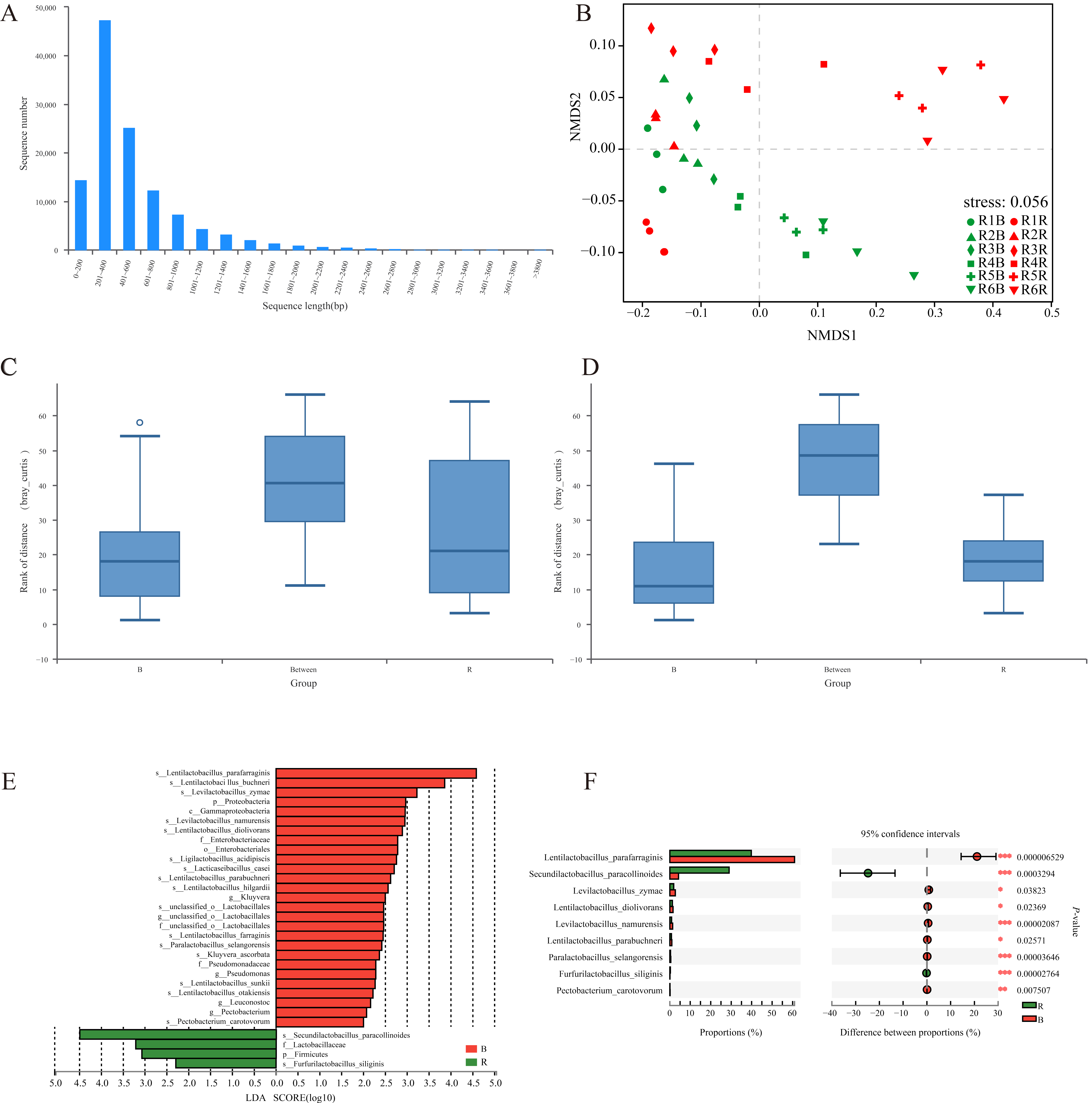
3.1. Metagenomic Sequencing Statistics and Quality Control
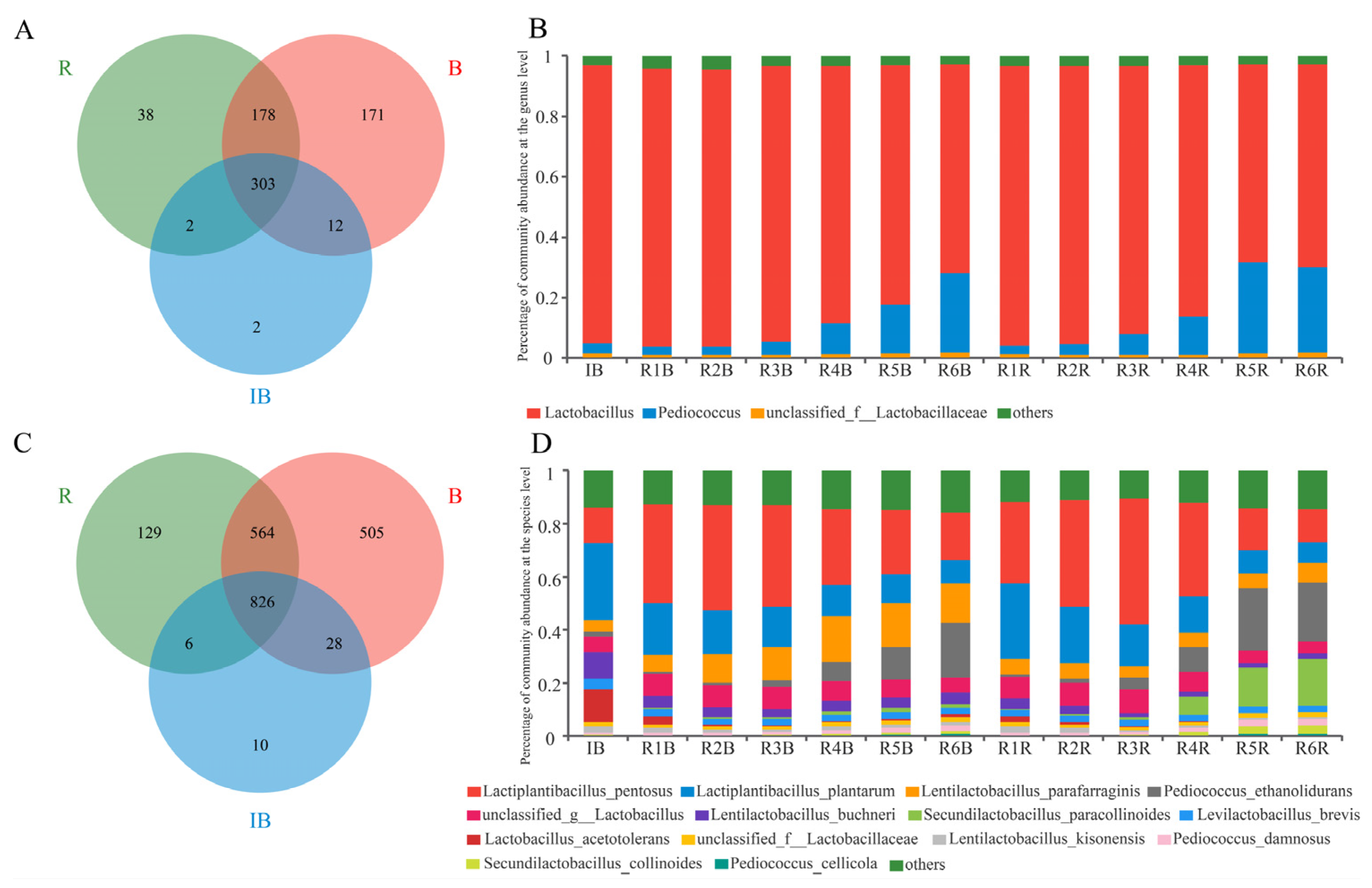
3.2. Microbial Structure
3.3. Microbial Composition Comparison
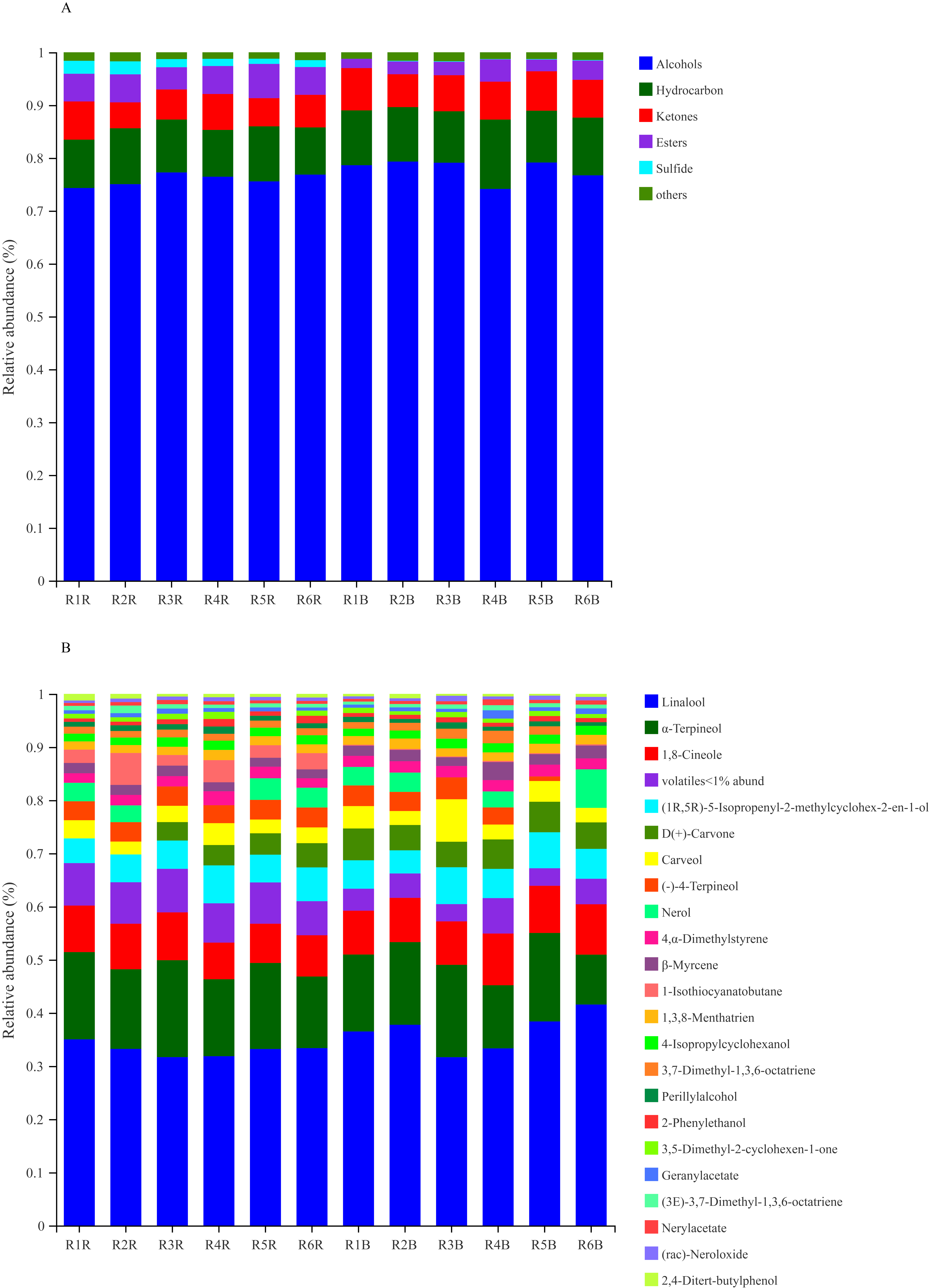
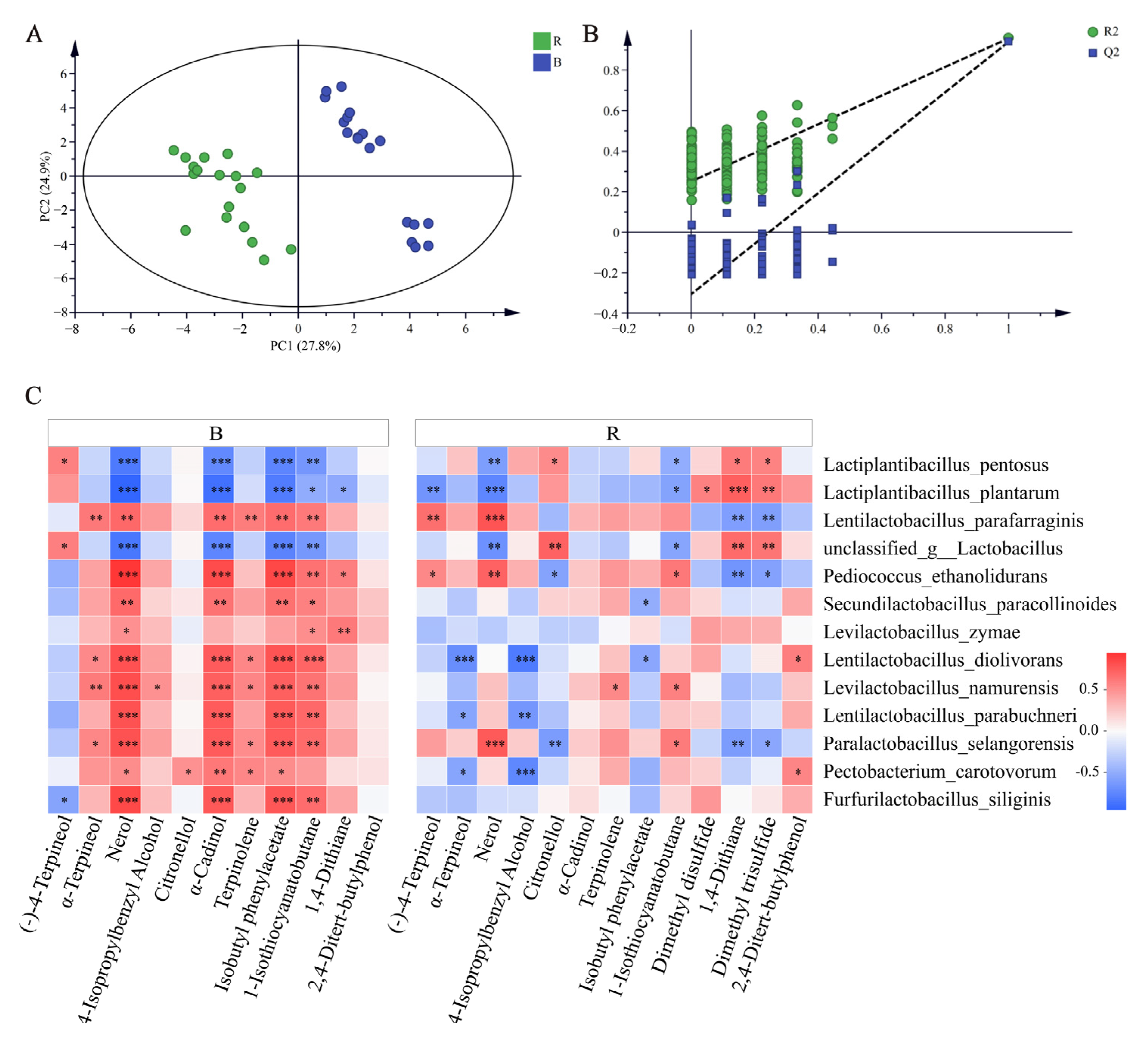
3.4. Profiles of VOCs during Different Paocai Fermentations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, D.Q.; Tong, C. Bacterial community diversity of traditional fermented vegetables in China. LWT—Food Sci. Technol. 2017, 86, 40–48. [Google Scholar] [CrossRef]
- Chen, G.; Yu, W.H.; Zhang, Q.S.; Song, P.; Zhang, B.B.; Liu, Z.; You, J.G.; Li, H. Research of Sichuan Paocai and Lactic Acid Bacteria. Adv. J. Food Sci. Technol. 2014, 6, 1–5. [Google Scholar] [CrossRef]
- Liang, H.P.; Chen, H.Y.; Ji, C.F.; Lin, X.P.; Zhang, W.X.; Li, L. Dynamic and Functional Characteristics of Predominant Species in Industrial Paocai as Revealed by Combined DGGE and Metagenomic Sequencing. Front. Microbiol. 2018, 9, 2416. [Google Scholar] [CrossRef]
- Zhang, S.S. Chinese Pickles; Higher Education Press: Beijing, China, 1994. [Google Scholar]
- Yan, P.-M.; Xue, W.-T.; Tan, S.-S.; Zhang, H.; Chang, X.-H. Effect of inoculating lactic acid bacteria starter cultures on the nitrite concentration of fermenting Chinese paocai. Food Control 2008, 19, 50–55. [Google Scholar] [CrossRef]
- Song, H.S.; Whon, T.W.; Kim, J.; Lee, S.H.; Kim, J.Y.; Kim, Y.B.; Choi, H.-J.; Rhee, J.-K.; Roh, S.W. Microbial niches in raw ingredients determine microbial community assembly during kimchi fermentation. Food Chem. 2020, 318, 126481. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, B.; Zhao, W.; Lao, F.; Chen, F.; Liao, X.; Wu, J. Shifts in autochthonous microbial diversity and volatile metabolites during the fermentation of chili pepper (Capsicum frutescens L.). Food Chem. 2021, 335, 127512. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Xiong, T.; Peng, Z.; Liu, C.; Huang, T.; Yu, H.; Xie, M. Correlation between microbiota and flavours in fermentation of Chinese Sichuan Paocai. Food Res. Int. 2018, 114, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Yang, B.; Lu, W.; Liu, X.; Zhao, J.; Ge, L.; Zhu, Y.; Lai, H.; Paul Ross, R.; Chen, W.; et al. Divergent role of abiotic factors in shaping microbial community assembly of paocai brine during aging process. Food Res. Int. 2020, 137, 109559. [Google Scholar] [CrossRef]
- Chen, A.J.; Luo, W.; Peng, Y.T.; Niu, K.L.; Liu, X.Y.; Shen, G.H.; Zhang, Z.Q.; Wan, H.; Luo, Q.Y.; Li, S.S. Quality and microbial flora changes of radish paocai during multiple fermentation rounds. Food Control 2019, 106, 106733. [Google Scholar] [CrossRef]
- Yang, J.; Cao, J.; Xu, H.; Hou, Q.; Yu, Z.; Zhang, H.; Sun, Z. Bacterial diversity and community structure in Chongqing radish paocai brines revealed using PacBio single-molecule real-time sequencing technology. J. Sci. Food Agric. 2018, 98, 3234–3245. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yang, J.; Hou, Q.; Xu, H.; Zheng, Y.; Zhang, H.; Zhang, L. Assessment of bacterial profiles in aged, home-made Sichuan paocai brine with varying titratable acidity by PacBio SMRT sequencing technology. Food Control 2017, 78, 14–23. [Google Scholar] [CrossRef]
- Wang, D.D.; Chen, G.; Tang, Y.; Li, H.; Shen, W.; Wang, M.; Liu, S.; Qin, W.; Zhang, Q. Effects of temperature on paocai bacterial succession revealed by culture-dependent and culture-independent methods. Int. J. Food Microbiol. 2020, 317, 108463. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved Selection of Internal Transcribed Spacer-Specific Primers Enables Quantitative, Ultra-High-Throughput Profiling of Fungal Communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [Green Version]
- Agyirifo, D.S.; Wamalwa, M.; Otwe, E.P.; Galyuon, I.; Runo, S.; Takrama, J.; Ngeranwa, J. Metagenomics analysis of cocoa bean fermentation microbiome identifying species diversity and putative functional capabilities. Heliyon 2019, 5, e02170. [Google Scholar] [CrossRef] [Green Version]
- Verce, M.; De Vuyst, L.; Weckx, S. Shotgun Metagenomics of a Water Kefir Fermentation Ecosystem Reveals a Novel Oenococcus Species. Front. Microbiol. 2019, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Parente, E.; Ercolini, D. Metagenomics insights into food fermentations. Microb. Biotechnol. 2017, 10, 91–102. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K. SOAP: Short Oligonucleotide Alignment Program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.S.; Huang, T.; Huang, C.L.; Hardie, J.; Peng, Z.; Xie, M.Y.; Xiong, T. The microbial communities and flavour compounds of Jiangxi yancai, Sichuan paocai and Dongbei suancai: Three major types of traditional Chinese fermented vegetables. LWT—Food Sci. Technol. 2020, 121, 108865. [Google Scholar] [CrossRef]
- Liu, Z.; Peng, Z.; Huang, T.; Xiao, Y.; Li, J.; Xie, M.; Xiong, T. Comparison of bacterial diversity in traditionally homemade paocai and Chinese spicy cabbage. Food Microbiol. 2019, 83, 141–149. [Google Scholar] [CrossRef]
- Xiong, T.; Guan, Q.; Song, S.; Hao, M.; Xie, M. Dynamic changes of lactic acid bacteria flora during Chinese sauerkraut fermentation. Food Control 2012, 26, 178–181. [Google Scholar] [CrossRef]
- Liu, A.; Liu, G.; Huang, C.; Shen, L.; Li, C.; Liu, Y.; Liu, S.; Hu, B.; Chen, H. The bacterial diversity of ripened Guang’yuan Suancai and In Vitro evaluation of potential probiotic lactic acid bacteria isolated from Suancai. LWT—Food Sci. Technol. 2017, 85, 175–180. [Google Scholar] [CrossRef]
- Kim, M.; Chun, J. Bacterial community structure in kimchi, a Korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 2005, 103, 91–96. [Google Scholar] [CrossRef]
- An, F.; Sun, H.; Wu, J.; Zhao, C.; Li, T.; Huang, H.; Fang, Q.; Mu, E.; Wu, R. Investigating the core microbiota and its influencing factors in traditional Chinese pickles. Food Res. Int. 2021, 147, 110543. [Google Scholar] [CrossRef]
- Lee, K.; Lee, Y. Effect of Lactobacillus plantarum as a starter on the food quality and microbiota of kimchi. Food Sci. Biotechnol. 2010, 19, 641–646. [Google Scholar] [CrossRef]
- Wang, Z.; Shao, Y. Effects of microbial diversity on nitrite concentration in pao cai, a naturally fermented cabbage product from China. Food Microbiol. 2018, 72, 185–192. [Google Scholar] [CrossRef]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Conlan, S.; Kong, H.H.; Segre, J.A. Species-Level Analysis of DNA Sequence Data from the NIH Human Microbiome Project. PLoS ONE 2012, 7, e47075. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Pereira, I.; Severino, P.; Santos, A.C.; Silva, A.M.; Souto, E.B. Linalool bioactive properties and potential applicability in drug delivery systems. Colloids Surf. B 2018, 171, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Kupska, M.; Chmiel, T.; Jedrkiewicz, R.; Wardencki, W.; Namiesnik, J. Comprehensive two-dimensional gas chromatography for determination of the terpenes profile of blue honeysuckle berries. Food Chem. 2014, 152, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Verzera, A.; Dima, G.; Tripodi, G.; Ziino, M.; Lanza, C.M.; Mazzaglia, A. Fast Quantitative Determination of Aroma Volatile Constituents in Melon Fruits by Headspace-Solid-Phase Microextraction and Gas Chromatography-Mass Spectrometry. Food Anal. Meth. 2011, 4, 141–149. [Google Scholar] [CrossRef]
- Hanschen, F.S.; Rohn, S.; Mewis, I.; Schreiner, M.; Kroh, L.W. Influence of the chemical structure on the thermal degradation of the glucosinolates in broccoli sprouts. Food Chem. 2012, 130, 1–8. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Xian, S.; Liu, X.; Shen, G.; Zhang, Z.; Hou, X.; Chen, A. Metagenomic Study on Chinese Homemade Paocai: The Effects of Raw Materials and Fermentation Periods on the Microbial Ecology and Volatile Components. Foods 2022, 11, 62. https://doi.org/10.3390/foods11010062
Jiang L, Xian S, Liu X, Shen G, Zhang Z, Hou X, Chen A. Metagenomic Study on Chinese Homemade Paocai: The Effects of Raw Materials and Fermentation Periods on the Microbial Ecology and Volatile Components. Foods. 2022; 11(1):62. https://doi.org/10.3390/foods11010062
Chicago/Turabian StyleJiang, Linjun, Shuang Xian, Xingyan Liu, Guanghui Shen, Zhiqing Zhang, Xiaoyan Hou, and Anjun Chen. 2022. "Metagenomic Study on Chinese Homemade Paocai: The Effects of Raw Materials and Fermentation Periods on the Microbial Ecology and Volatile Components" Foods 11, no. 1: 62. https://doi.org/10.3390/foods11010062
APA StyleJiang, L., Xian, S., Liu, X., Shen, G., Zhang, Z., Hou, X., & Chen, A. (2022). Metagenomic Study on Chinese Homemade Paocai: The Effects of Raw Materials and Fermentation Periods on the Microbial Ecology and Volatile Components. Foods, 11(1), 62. https://doi.org/10.3390/foods11010062






