In Vitro and Reactive Metabolites Investigation of Metabolic Profiling of Tyrosine Kinase Inhibitors Dubermatinib in HLMs by LC–MS/MS
Abstract
1. Introduction
2. Chemicals and Methods
2.1. Chemicals
2.2. Chromatographic Conditions
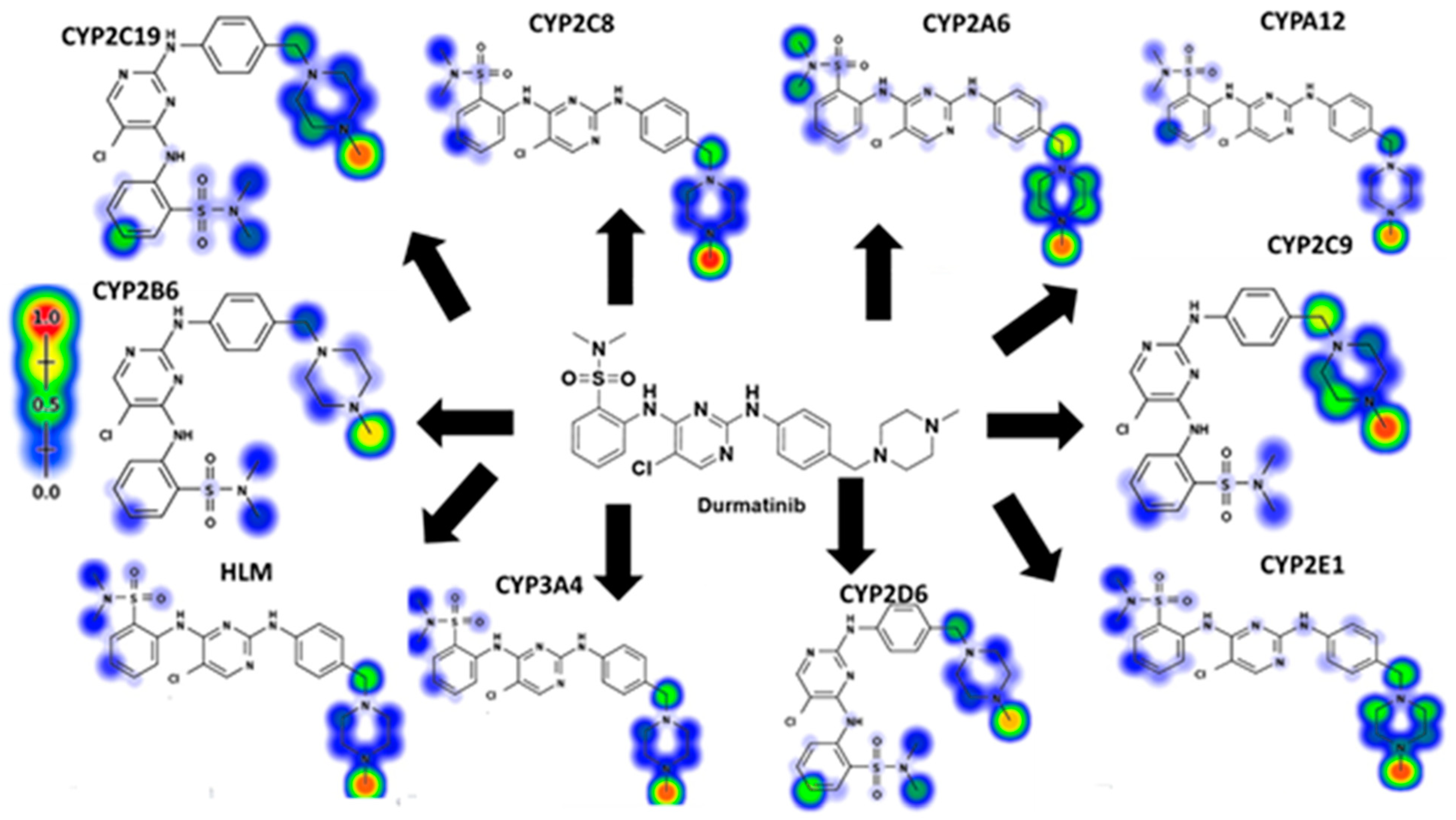
2.3. Xenosite Web Predictor for In Silico Prediction
2.4. In Vitro Metabolism of DMB
3. Results and Discussion
3.1. DMB Metabolism and Reactivity (In Silico Prediction)
3.2. DMB Fragmentation
3.3. Phase I Metabolites of DMB Identified In Vitro
- M1 and M2 (N-demethylation metabolites) at (m/z: 502).
- M3 (m/z = 474) was found to be the only metabolite with triple N-demethylation.
- M4 (m/z = 532) was shown to be a hydroxylated metabolite.
- M5 at (m/z = 498), a product of dechlorination and hydroxylation, has been isolated.
- The metabolite M6 at (m/z: 484) has been shown to undergo N-demethylation, dechlorination, and hydroxylation.
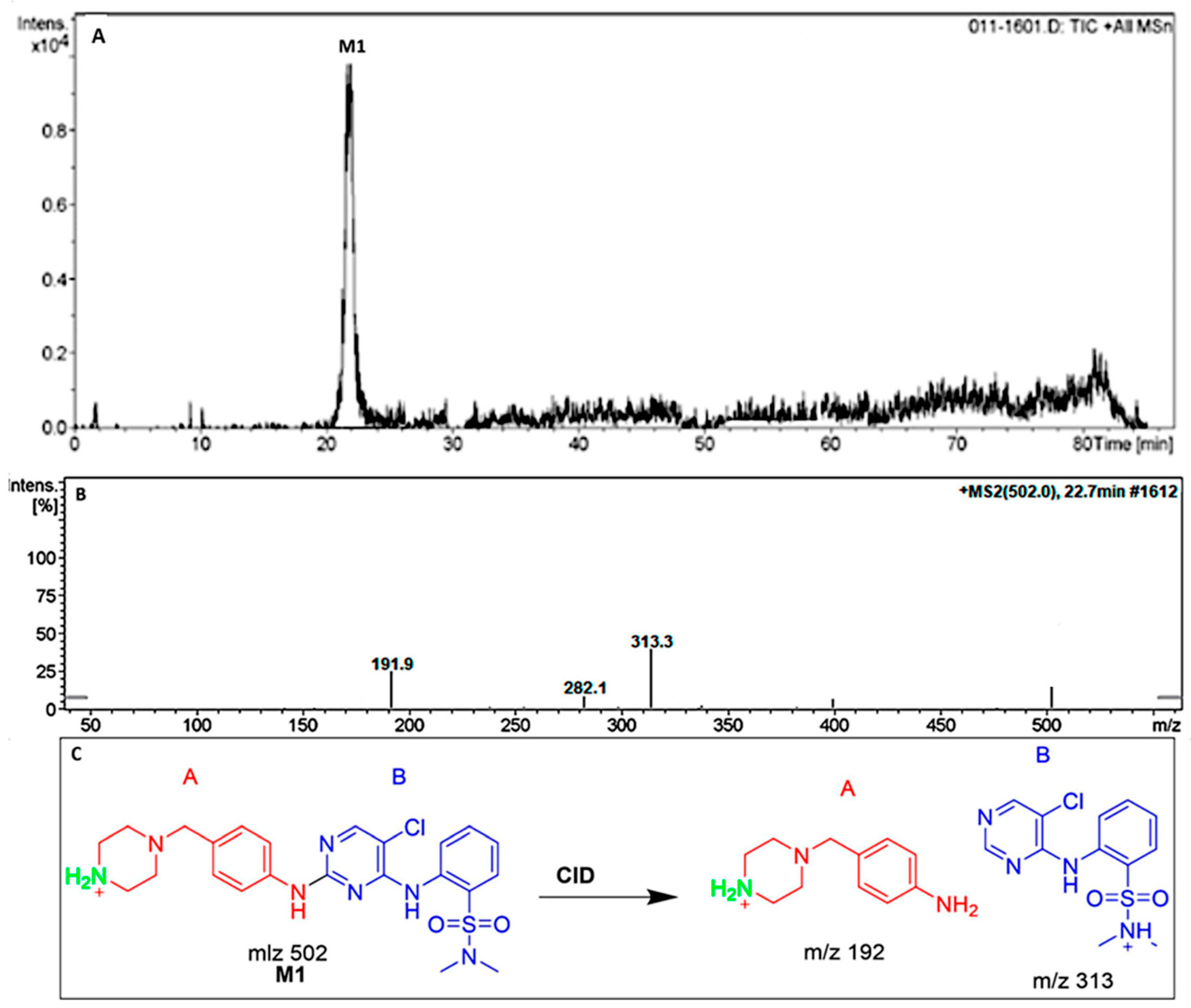
3.3.1. The DMB M1 Metabolites Identification
3.3.2. Identification of the DMB M2 Metabolites
3.3.3. Identification of the DMB M3 Metabolite
3.3.4. Identification of the DMB M4 Metabolite
3.3.5. Identification of the DMB M5 Metabolite
3.3.6. Identification of the DMB M6 Metabolite
3.3.7. Identification of the M7 DMB Metabolite
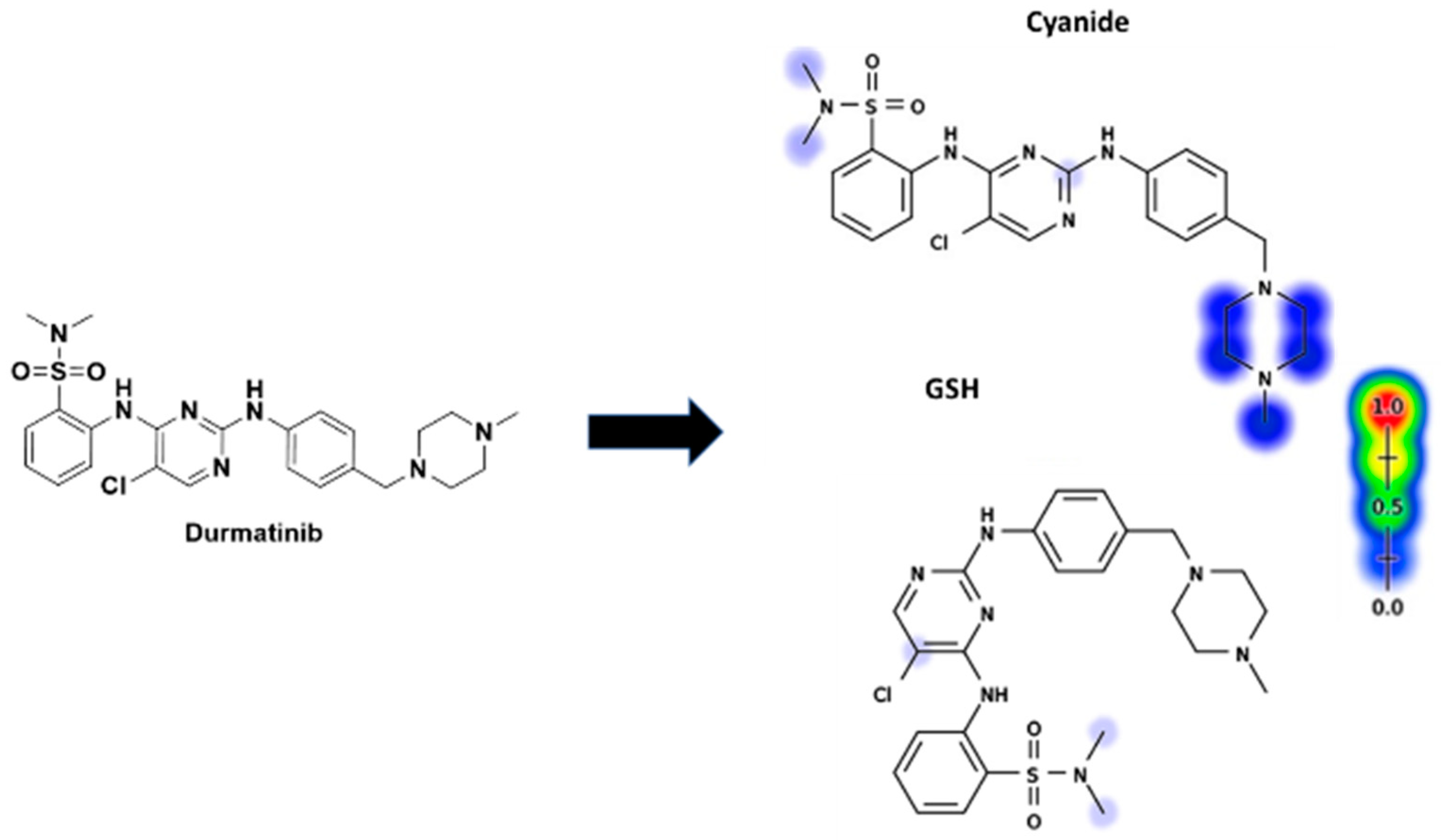
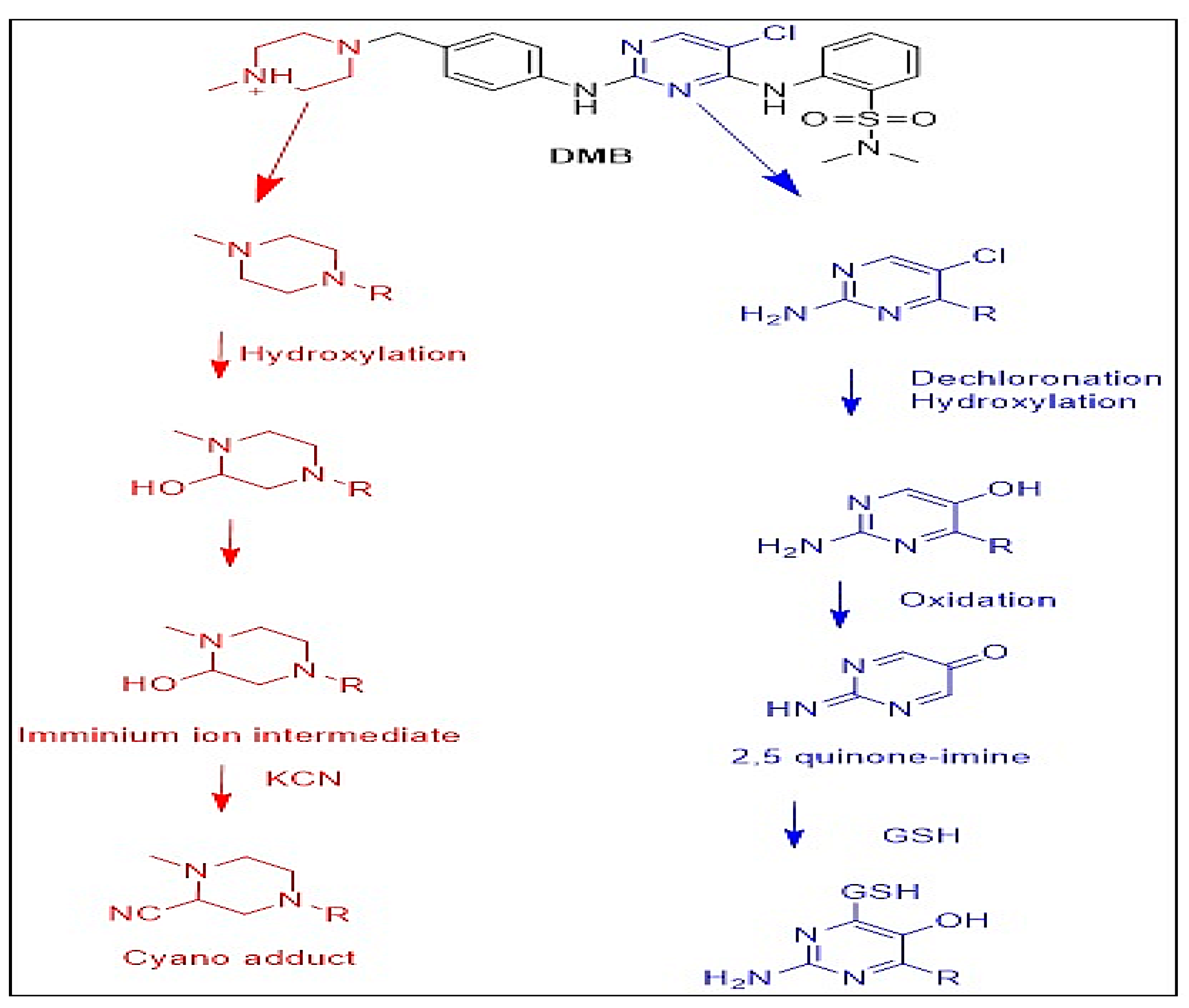
3.4. Identification of In Vitro DMB Reactive Metabolites
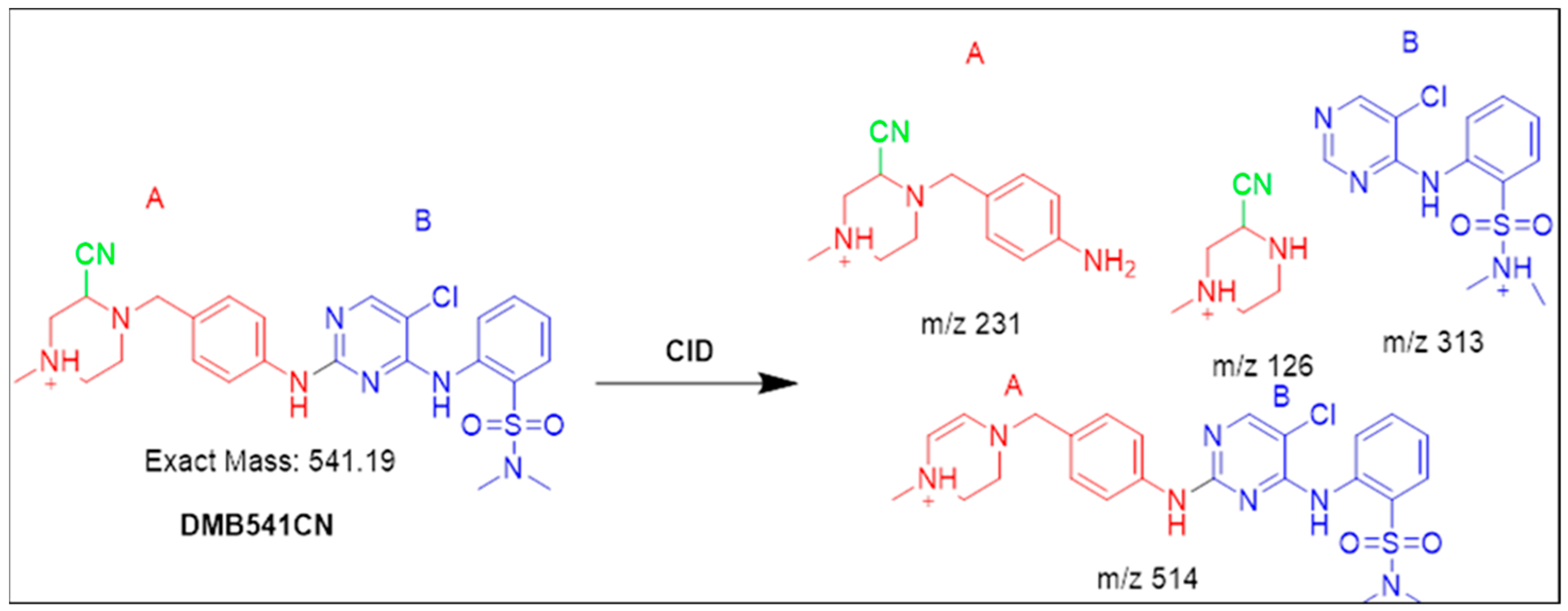
3.4.1. Identification of the DMB541CN Cyano Adduct Reactive Metabolite of DMB
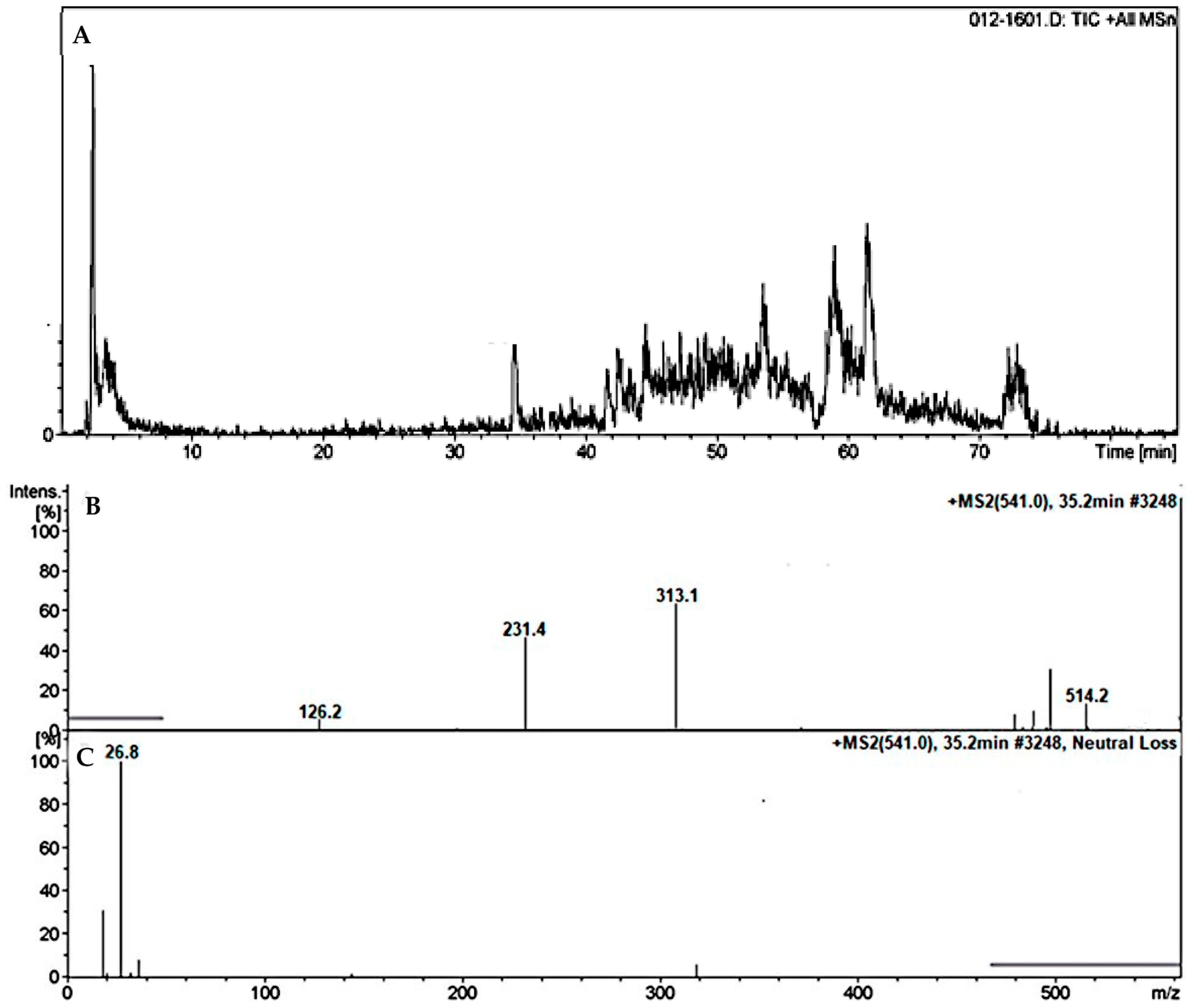
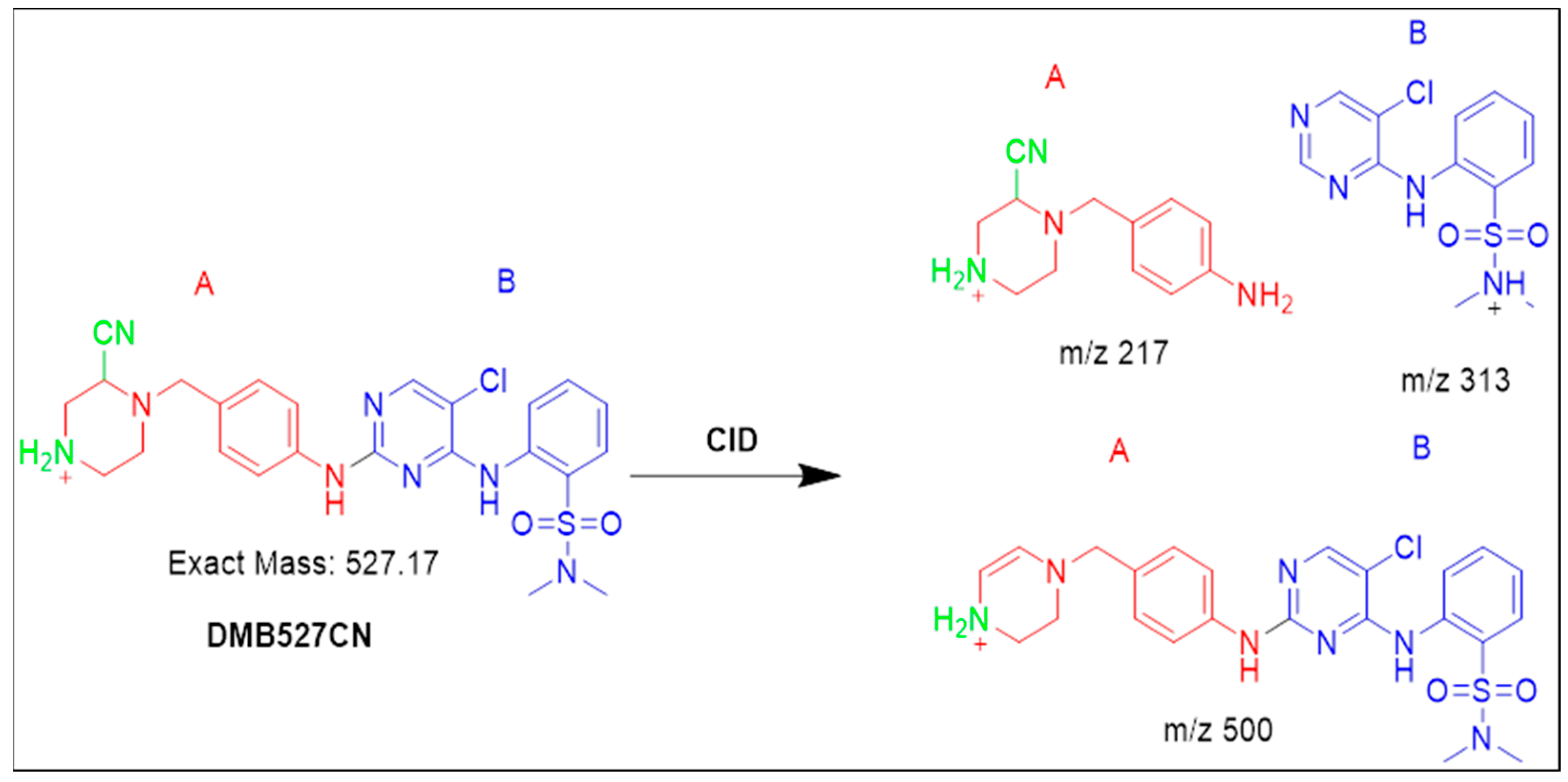
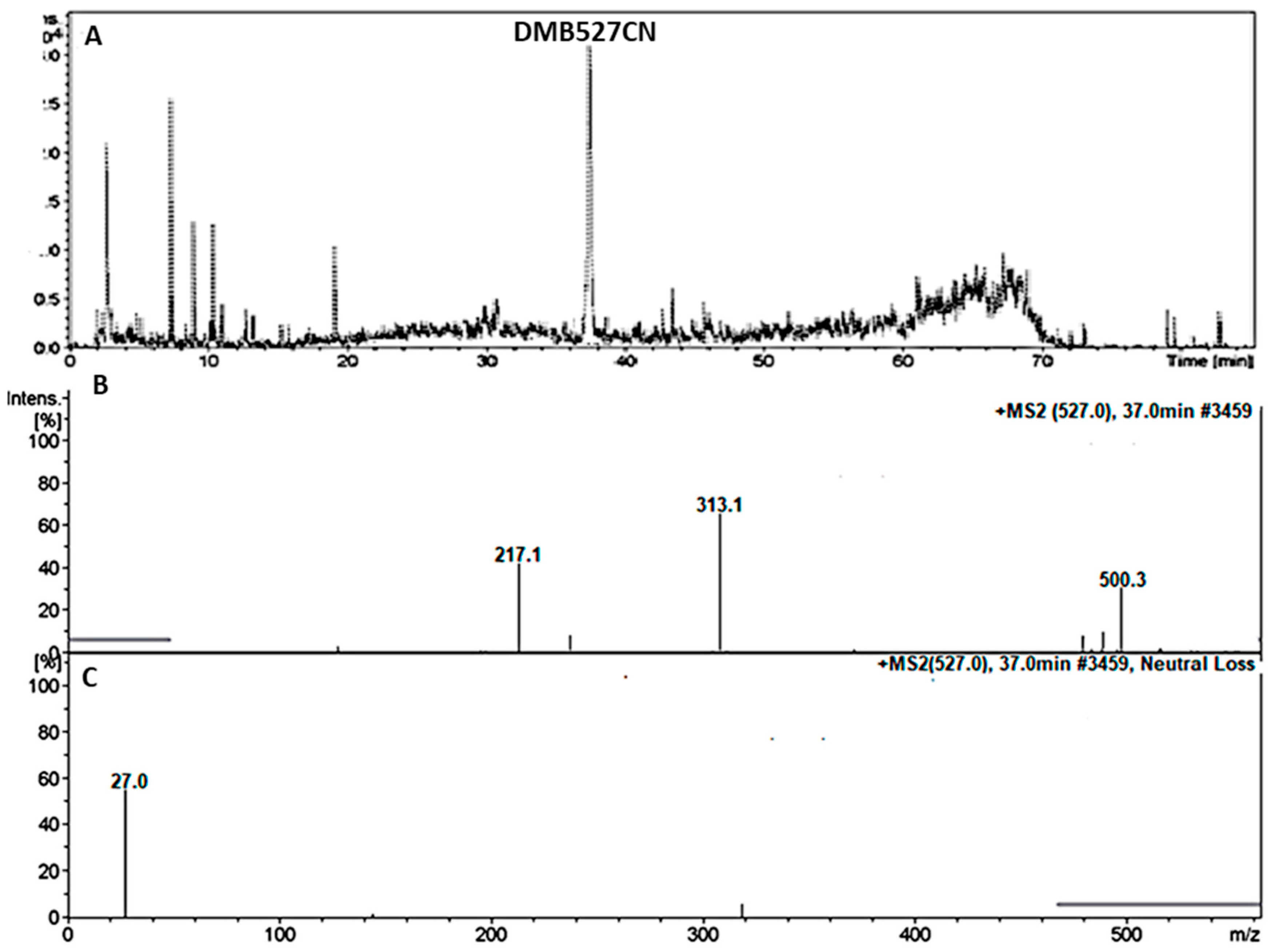
3.4.2. Identification of the DMB527CN Cyano Adduct Reactive Metabolite of DMB
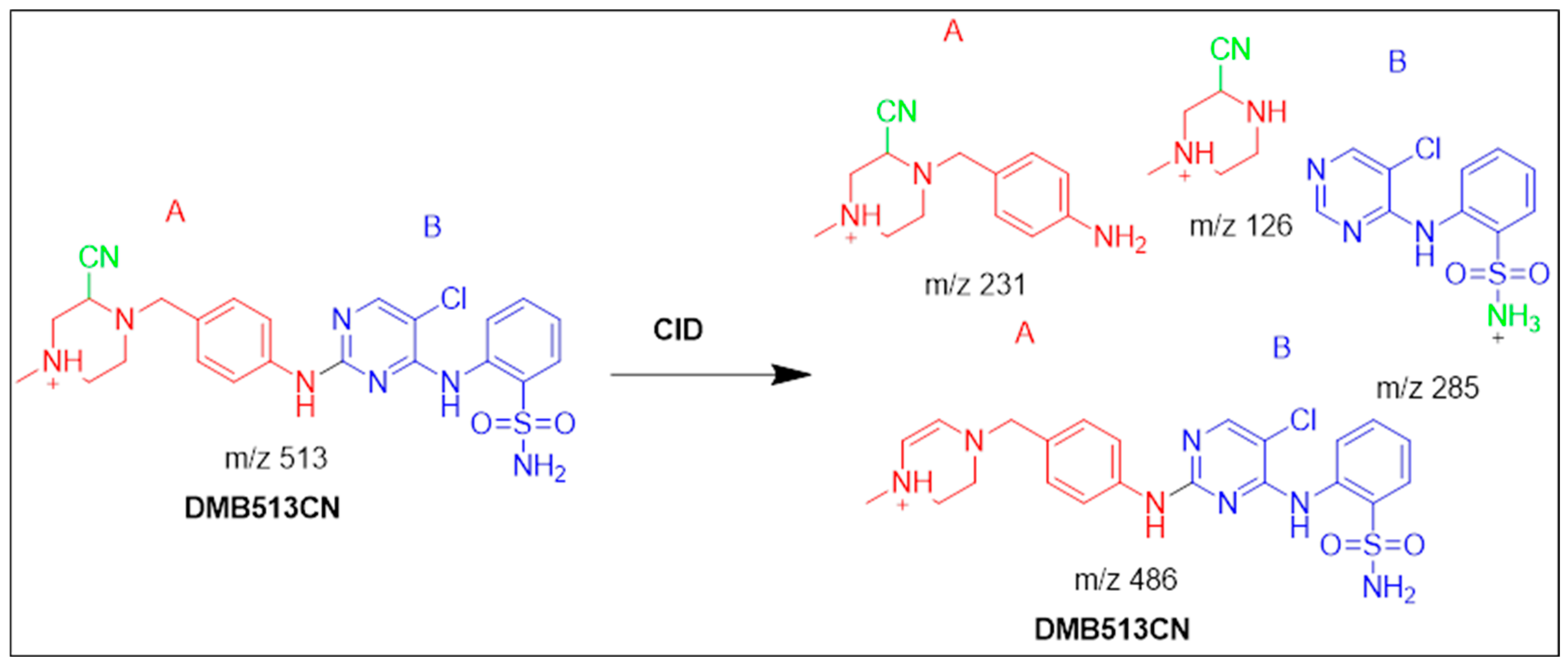
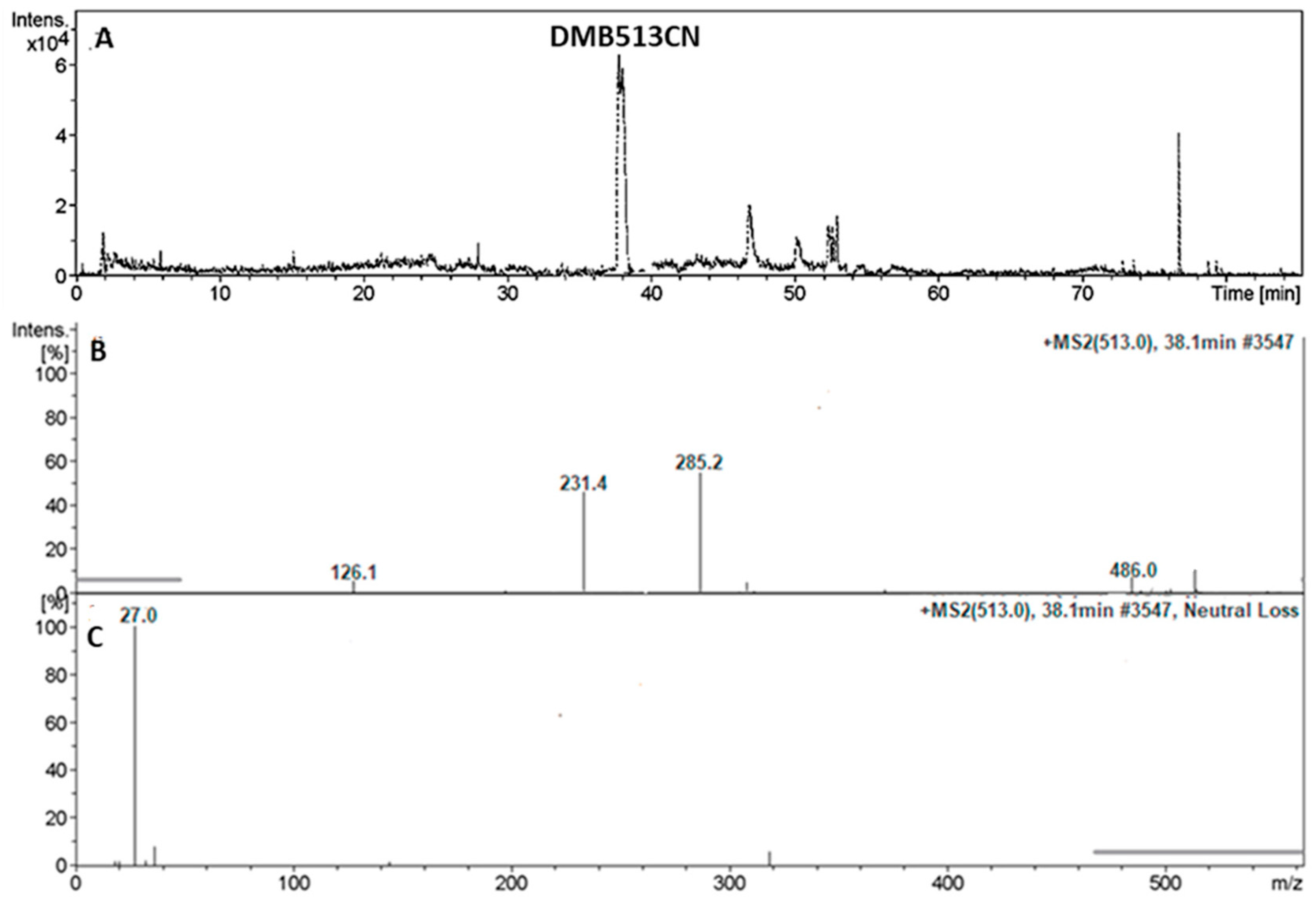
3.4.3. Identification of the DMB513CN Cyano Adduct Reactive Metabolite of DMB
3.5. Identification of DMB GSH Conjugates
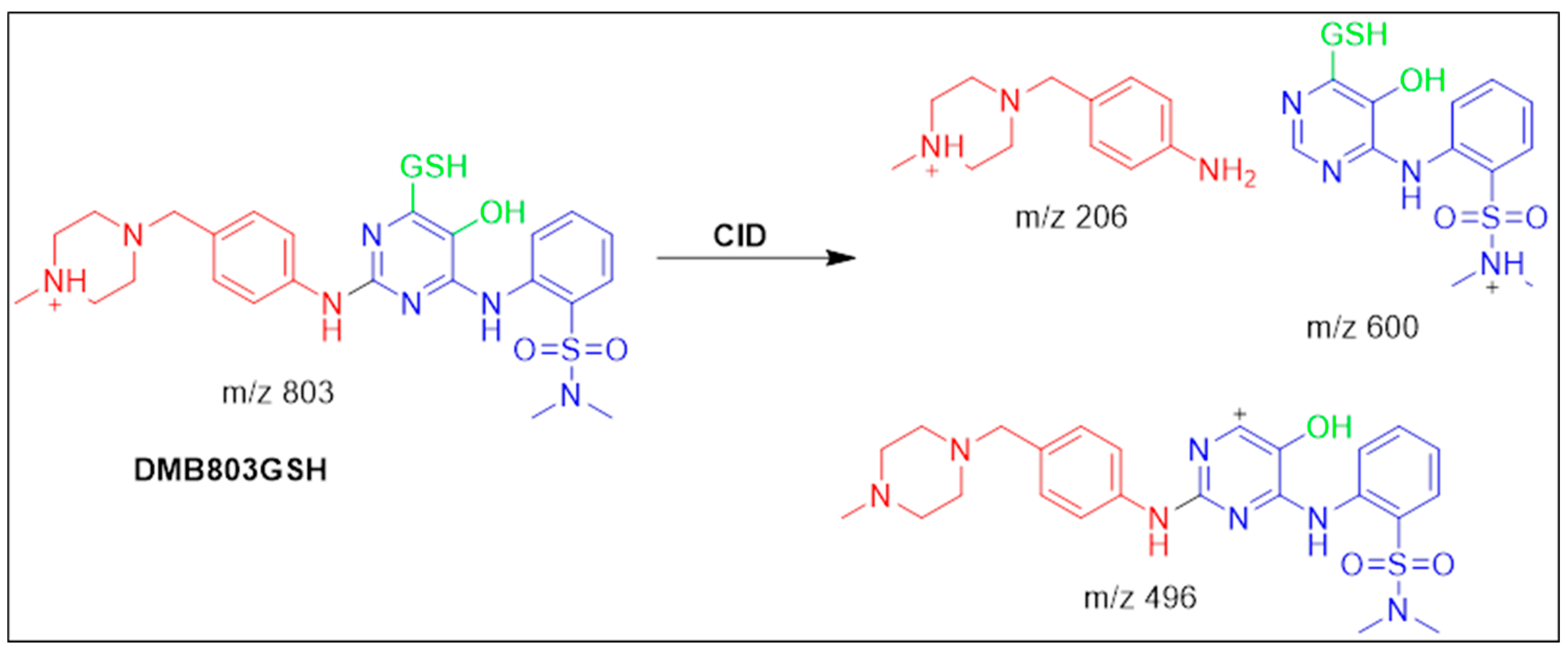
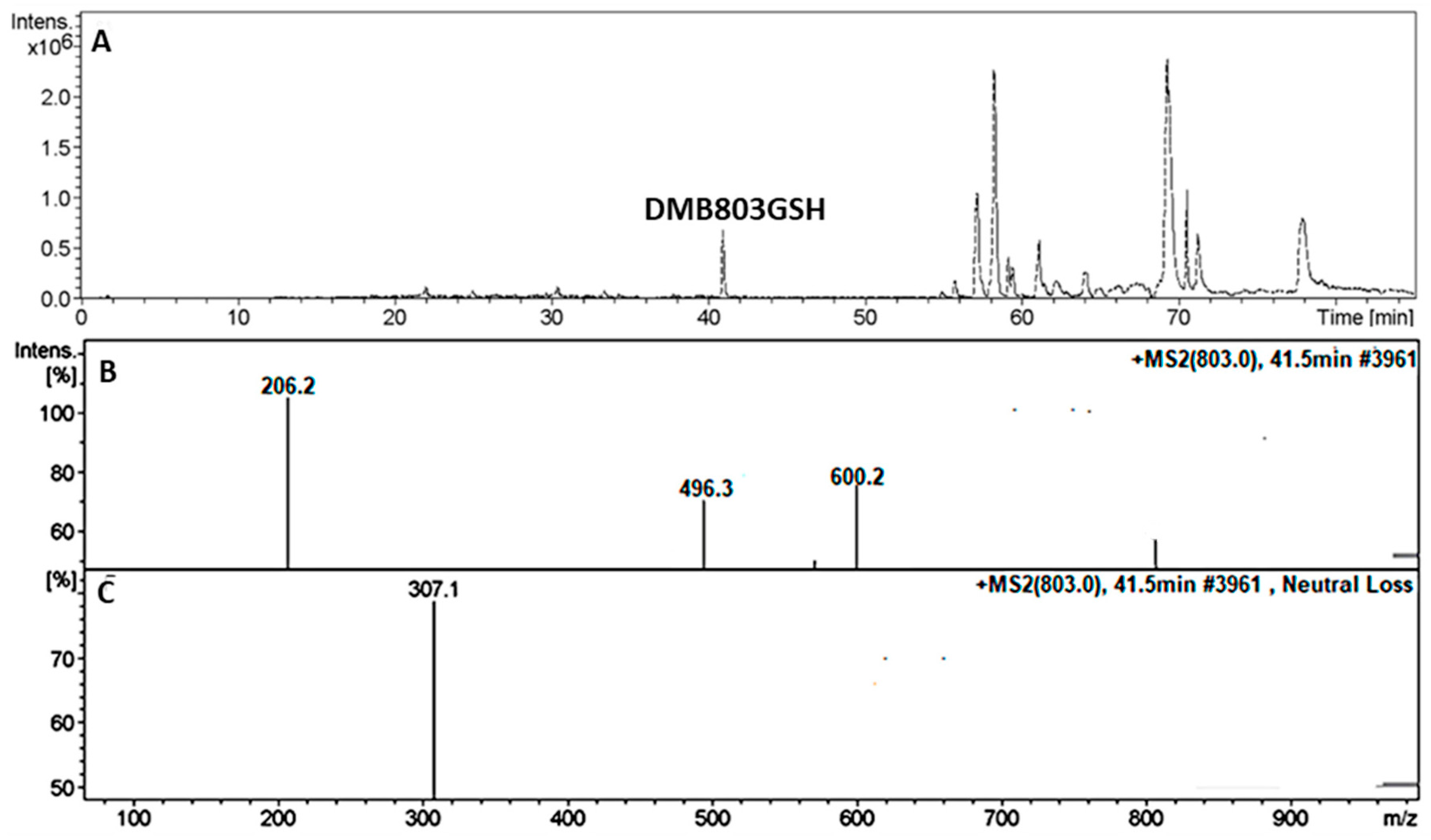
3.5.1. Identification of the DMB803GSH Conjugate DMB
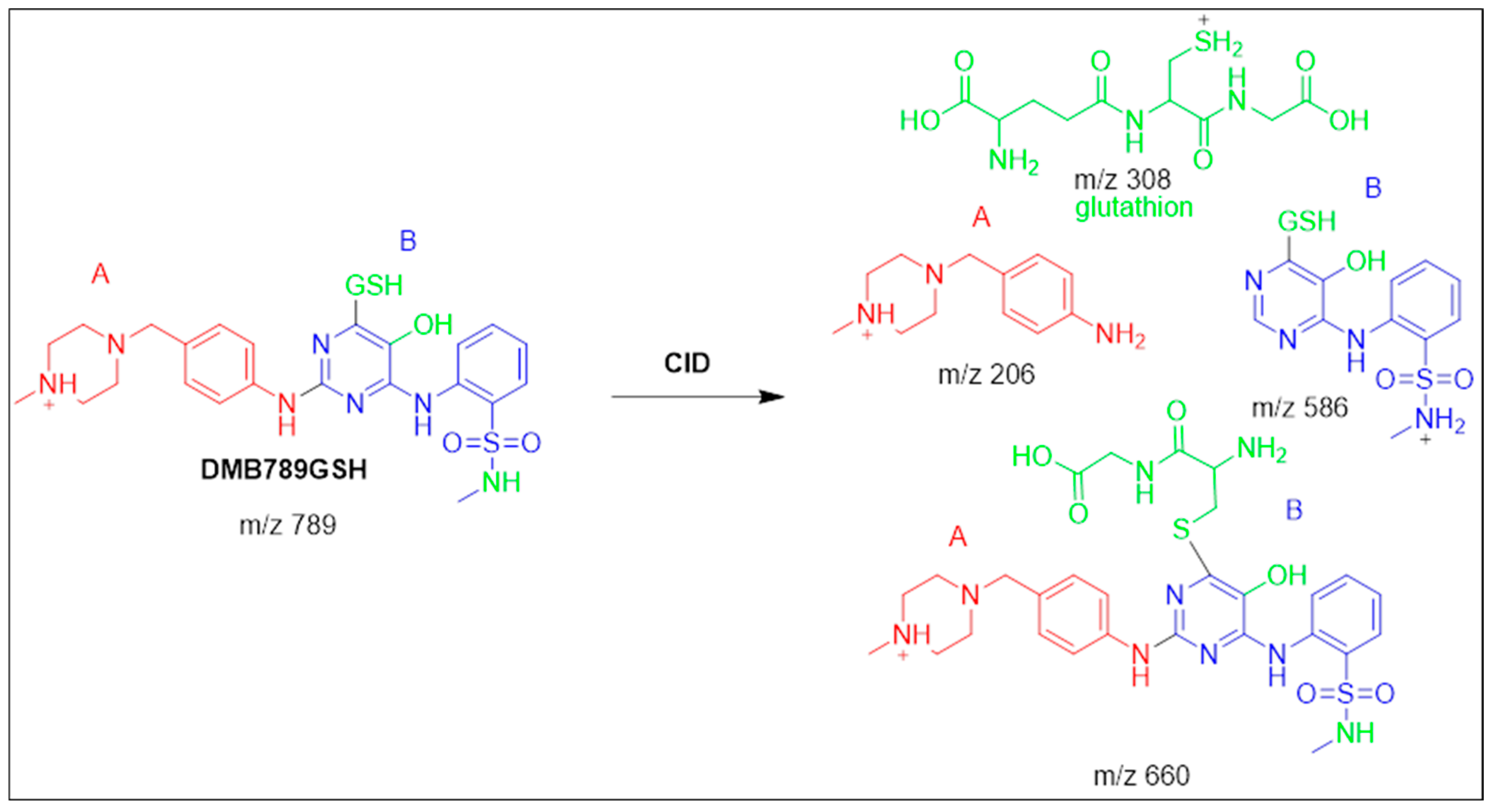
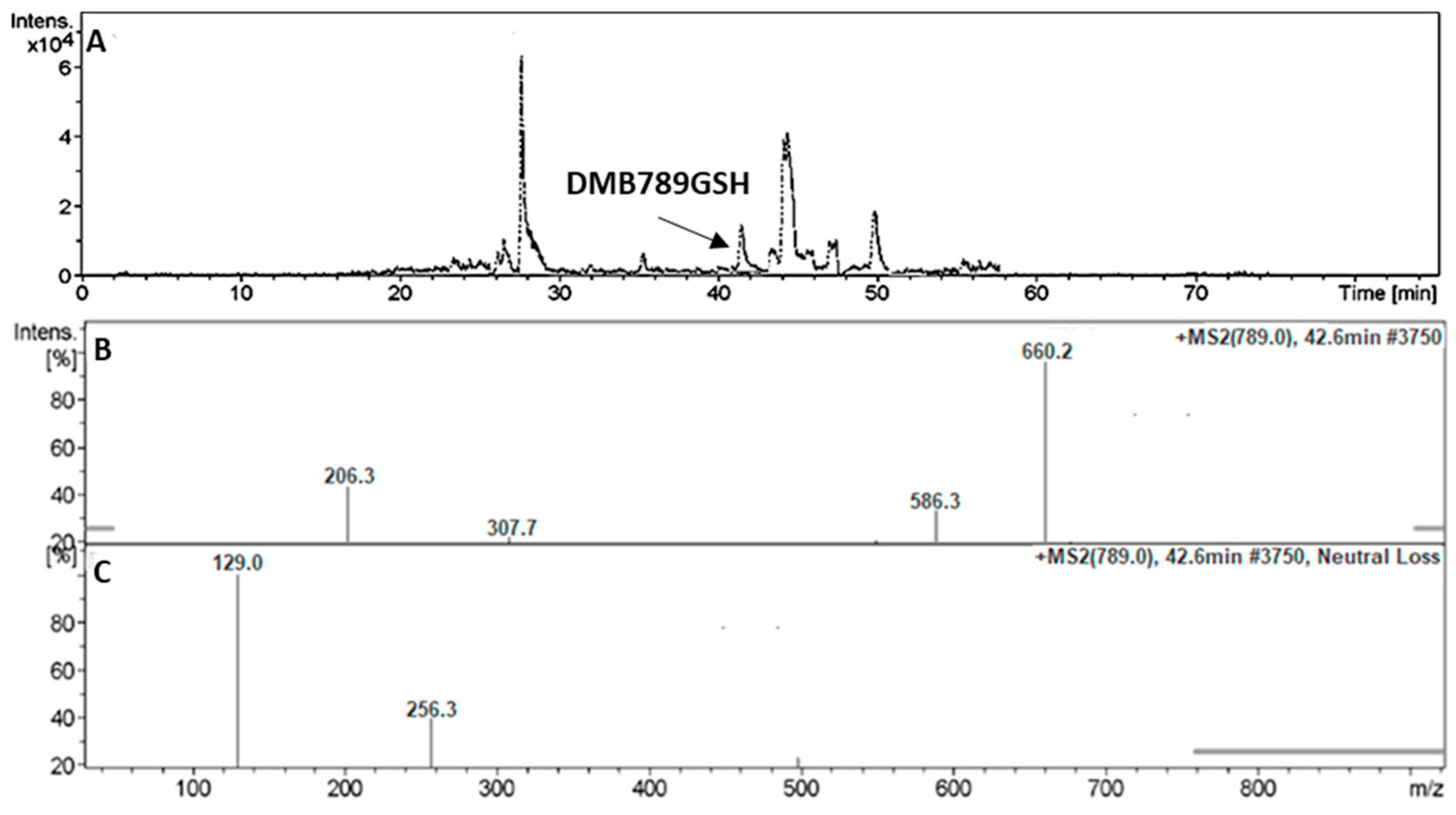
3.5.2. Identification of the DMB789GSH Conjugate DMB
3.6. Proposed Bioactivation Pathways of DMB
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Natoli, C.; Perrucci, B.; Perrotti, F.; Falchi, L.; Iacobelli, S. Tyrosine Kinase Inhibitors. Curr. Cancer Drug Targets 2010, 10, 462–483. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Siemann, D.W. Therapeutic Targeting of the Gas6/Axl Signaling Pathway in Cancer. Int. J. Mol. Sci. 2021, 22, 9953. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Boysen, J.C.; Chaffee, K.G.; Kabat, B.F.; Slager, S.L.; Parikh, S.A.; Secreto, C.R.; Call, T.; Shanafelt, T.D.; Leis, J.F. Chronic Lymphocytic Leukemia Cells from Ibrutinib Treated Patients Are Sensitive to Axl Receptor Tyrosine Kinase Inhibitor Therapy. Oncotarget 2018, 9, 37173. [Google Scholar] [CrossRef]
- Sen, T.; Tong, P.; Diao, L.; Li, L.; Fan, Y.; Hoff, J.; Heymach, J.V.; Wang, J.; Byers, L.A. Targeting AXL and MTOR Pathway Overcomes Primary and Acquired Resistance to WEE1 Inhibition in Small-Cell Lung CancerAXL/MTOR–Mediated WEE1 Inhibitor Resistance in SCLC. Clin. Cancer Res. 2017, 23, 6239–6253. [Google Scholar] [CrossRef]
- Aveic, S.; Corallo, D.; Porcù, E.; Pantile, M.; Boso, D.; Zanon, C.; Viola, G.; Sidarovich, V.; Mariotto, E.; Quattrone, A. TP-0903 Inhibits Neuroblastoma Cell Growth and Enhances the Sensitivity to Conventional Chemotherapy. Eur. J. Pharmacol. 2018, 818, 435–448. [Google Scholar] [CrossRef]
- Li, F.; Lu, J.; Ma, X. Profiling the Reactive Metabolites of Xenobiotics Using Metabolomic Technologies. Chem. Res. Toxicol. 2011, 24, 744–751. [Google Scholar] [CrossRef]
- Al-Shakliah, N.S.; Attwa, M.W.; Kadi, A.A.; AlRabiah, H. Identification and Characterization of In Silico, In Vivo, In Vitro, and Reactive Metabolites of Infigratinib Using LC-ITMS: Bioactivation Pathway Elucidation and in Silico Toxicity Studies of Its Metabolites. RSC Adv. 2020, 10, 16231–16244. [Google Scholar] [CrossRef]
- Bolton, J.L.; Trush, M.A.; Penning, T.M.; Dryhurst, G.; Monks, T.J. Role of Quinones in Toxicology. Chem. Res. Toxicol. 2000, 13, 135–160. [Google Scholar] [CrossRef]
- Ma, S.; Zhu, M. Recent Advances in Applications of Liquid Chromatography–Tandem Mass Spectrometry to the Analysis of Reactive Drug Metabolites. Chem. Biol. Interact. 2009, 179, 25–37. [Google Scholar] [CrossRef]
- Stepan, A.F.; Walker, D.P.; Bauman, J.; Price, D.A.; Baillie, T.A.; Kalgutkar, A.S.; Aleo, M.D. Structural Alert/Reactive Metabolite Concept as Applied in Medicinal Chemistry to Mitigate the Risk of Idiosyncratic Drug Toxicity: A Perspective Based on the Critical Examination of Trends in the Top 200 Drugs Marketed in the United States. Chem. Res. Toxicol. 2011, 24, 1345–1410. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, A.; Turpeinen, M.; Pelkonen, O. Liquid Chromatography–Mass Spectrometry in in Vitro Drug Metabolite Screening. Drug Discov. Today 2009, 14, 120–133. [Google Scholar] [CrossRef]
- Attwa, M.W.; Kadi, A.A.; Darwish, H.W.; Amer, S.M.; Al-Shakliah, N.S. Identification and Characterization of in Vivo, in Vitro and Reactive Metabolites of Vandetanib Using LC–ESI–MS/MS. Chem. Cent. J. 2018, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- DiMasi, J.A.; Hansen, R.W.; Grabowski, H.G. The Price of Innovation: New Estimates of Drug Development Costs. J. Health Econ. 2003, 22, 151–185. [Google Scholar] [CrossRef] [PubMed]
- Raies, A.B.; Bajic, V.B. In Silico Toxicology: Computational Methods for the Prediction of Chemical Toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2016, 6, 147–172. [Google Scholar] [CrossRef]
- Matlock, M.K.; Hughes, T.B.; Swamidass, S.J. XenoSite Server: A Web-Available Site of Metabolism Prediction Tool. Bioinformatics 2015, 31, 1136–1137. [Google Scholar] [CrossRef]
- Zaretzki, J.; Matlock, M.; Swamidass, S.J. XenoSite: Accurately Predicting CYP-Mediated Sites of Metabolism with Neural Networks. J. Chem. Inf. Model. 2013, 53, 3373–3383. [Google Scholar] [CrossRef]
- Dang, N.L.; Hughes, T.B.; Krishnamurthy, V.; Swamidass, S.J. A Simple Model Predicts UGT-Mediated Metabolism. Bioinformatics 2016, 32, 3183–3189. [Google Scholar] [CrossRef]
- Jia, L.; Liu, X. The Conduct of Drug Metabolism Studies Considered Good Practice (II): In Vitro Experiments. Curr. Drug Metab. 2007, 8, 822–829. [Google Scholar] [CrossRef]
- Al-Shakliah, N.S.; Attwa, M.W.; AlRabiah, H.; Kadi, A.A. Identification and Characterization of in Vitro, in Vivo, and Reactive Metabolites of Tandutinib Using Liquid Chromatography Ion Trap Mass Spectrometry. Anal. Methods 2021, 13, 399–410. [Google Scholar] [CrossRef]
- Al-Shakliah, N.; Kadi, A.; Al-Salahi, R.; Rahman, A.F.M. In Vitro Identification of Potential Metabolites of Plinabulin (NPI 2358) in Hepatic Preparations Using Liquid Chromatography–Ion Trap Mass Spectrometry. ACS Omega 2022, 7, 21465–21472. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Liu, H.-F.; Zhao, W.; Jones, E.; Zhu, M. Simultaneous Screening of Glutathione and Cyanide Adducts Using Precursor Ion and Neutral Loss Scans-Dependent Product Ion Spectral Acquisition and Data Mining Tools. J. Am. Soc. Mass Spectrom. 2012, 23, 964–976. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Zhong, D.; Chen, X. A Fragmentation-Based Method for the Differentiation of Glutathione Conjugates by High-Resolution Mass Spectrometry with Electrospray Ionization. Anal. Chim. Acta 2013, 788, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, A.S.; Attwa, M.W.; Kadi, A.A. Characterization of Stable and Reactive Metabolites of the Anticancer Drug, Ensartinib, in Human Liver Microsomes Using LC-MS/MS: An in Silico and Practical Bioactivation Approach. Drug Des. Devel. Ther. 2020, 14, 5259. [Google Scholar] [CrossRef]
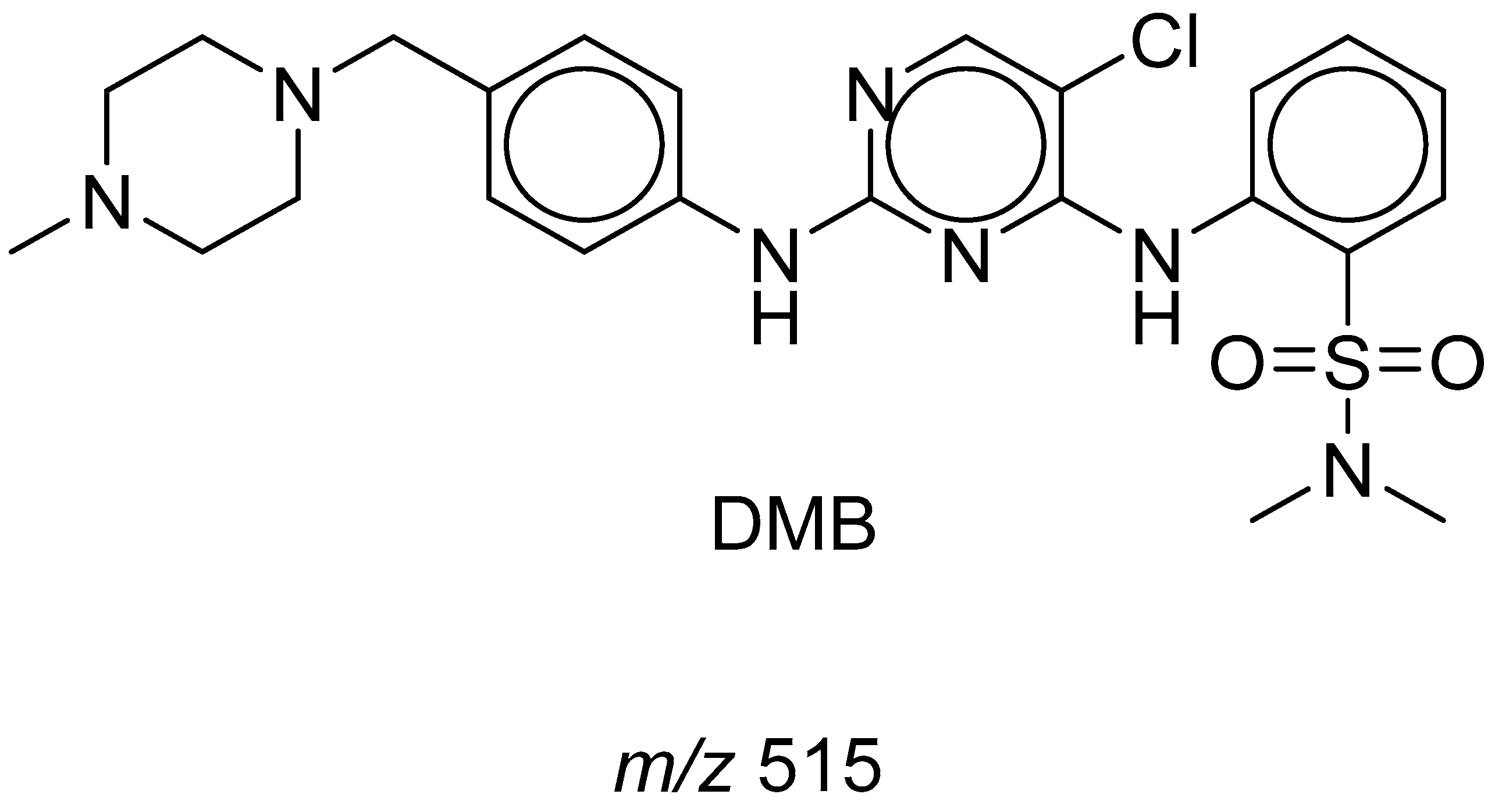
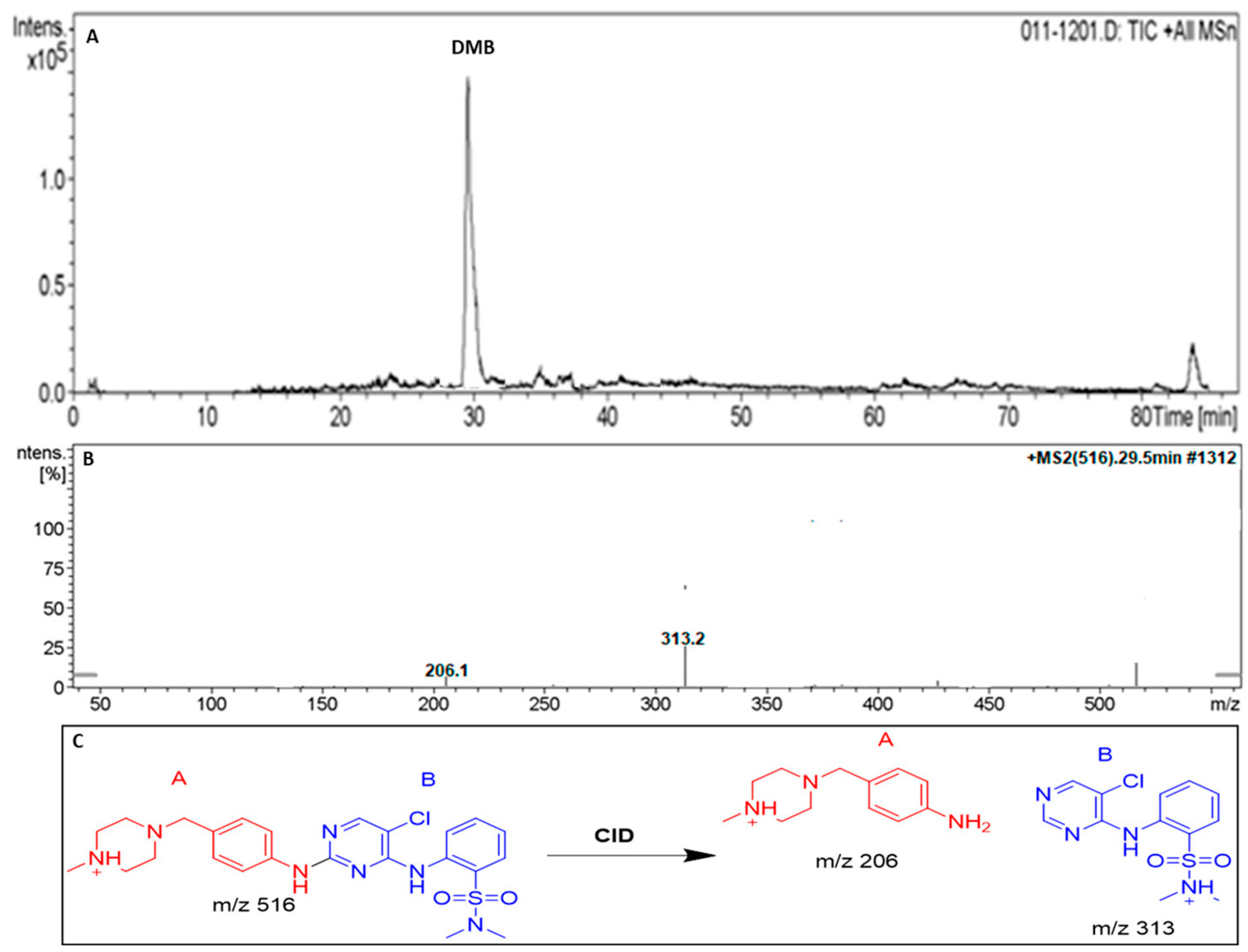
| MS Scan | Major Daughter Ions | tR (min) | Proposed Metabolic Reaction | |
|---|---|---|---|---|
| DMB | 516 | 206,313 | 29.5 | DMB + H |
| M1 | 502 | 313,192 | 22.7 | N-demethylation at piperazine ring |
| M2 | 502 | 299,206 | 23.1 | N-demethylation at sulphonamide group |
| M3 | 474 | 192,285 | 24.4 | N-demethylation at piperazine ring and sulphonamide group |
| M4 | 532 | 313,222,117 | 25.1 | α hydroxylation at piperazine ring |
| M5 | 498 | 295,206,98 | 27.5 | Dechlorination then hydroxylation |
| M6 | 484 | 295.192 | 28.1 | Dechlorination, then hydroxylation and N-demethylation at piperazine ring |
| M7 | 514 | 295,222,117 | 28.6 | Dechlorination followed by hydroxylation at pyrimidine and hydroxylation at piperazine ring |
| MS Scan | Most Abundant Fragment Ions | Rt (min) | Metabolic Reaction | |
|---|---|---|---|---|
| Cyano adducts | ||||
| DMB541CN | 541 | 126, 231, 313, 514 | 35.2 | Attack of KCN at bioactivated N-methyl piperazine ring |
| DMB527CN | 527 | 217, 313, 500 | 37.0 | N-demethylation and attack of KCN at bioactivated N-methyl piperazine ring. |
| DMB513CN | 513 | 126, 231, 285 | 38.1 | Double N-demethylation at sulphonamide group and attack of KCN at bioactivated N-methyl piperazine ring. |
| GSH conjugates | ||||
| DMB803GSH | 803 | 206, 496, 600 | 41.5 | Conjugation of GSH at bioactivated 2,5quinone-imine |
| DMB789GSH | 789 | 206, 308, 586 | 42.6 | N-demethylation at sulphonamide and conjugation of GSH at bioactivated 2,5quinone-imine |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Shakliah, N.S.; A. Kadi, A.; Abuelizz, H.A.; Al-Salahi, R. In Vitro and Reactive Metabolites Investigation of Metabolic Profiling of Tyrosine Kinase Inhibitors Dubermatinib in HLMs by LC–MS/MS. Separations 2023, 10, 353. https://doi.org/10.3390/separations10060353
Al-Shakliah NS, A. Kadi A, Abuelizz HA, Al-Salahi R. In Vitro and Reactive Metabolites Investigation of Metabolic Profiling of Tyrosine Kinase Inhibitors Dubermatinib in HLMs by LC–MS/MS. Separations. 2023; 10(6):353. https://doi.org/10.3390/separations10060353
Chicago/Turabian StyleAl-Shakliah, Nasser S., Adnan A. Kadi, Hatem A. Abuelizz, and Rashad Al-Salahi. 2023. "In Vitro and Reactive Metabolites Investigation of Metabolic Profiling of Tyrosine Kinase Inhibitors Dubermatinib in HLMs by LC–MS/MS" Separations 10, no. 6: 353. https://doi.org/10.3390/separations10060353
APA StyleAl-Shakliah, N. S., A. Kadi, A., Abuelizz, H. A., & Al-Salahi, R. (2023). In Vitro and Reactive Metabolites Investigation of Metabolic Profiling of Tyrosine Kinase Inhibitors Dubermatinib in HLMs by LC–MS/MS. Separations, 10(6), 353. https://doi.org/10.3390/separations10060353








