Leukemic Transdifferentiation: From Pathological Plasticity to Dendritic Cell-Based Immunotherapy
Abstract
1. Introduction
2. Maintenance of the Lineage Identity of Leukemic Cells
3. Transdifferentiation
3.1. The Event
3.2. Evolutionary Conservation
4. Transdifferentiation Is One of the Events Responsible for the Plasticity of Leukemic Precursors
4.1. Partial, Spontaneous Transdifferentiation in Hematopoiesis
4.2. Spontaneous Transdifferentiation in Leukemia
5. Dendritic Cell Subtypes and Their Role in the Anti-Leukemic Response
6. Changing Cell Identity Toward a Dendritic Phenotype
6.1. Experimental Techniques
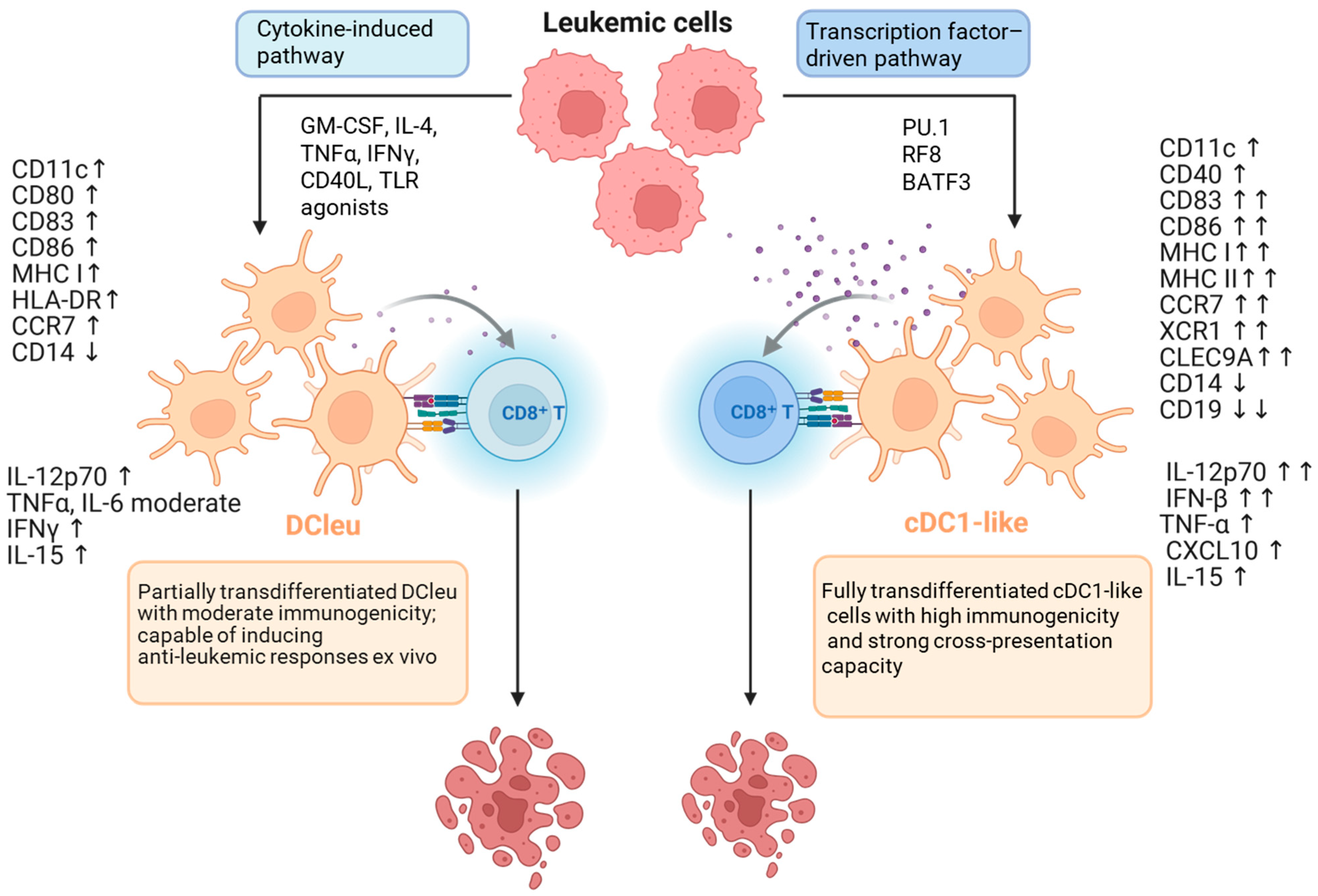
6.2. Transdifferentiation in the Therapies of Leukemias
6.3. Dendritic Cell-Based Immunotherapies
6.4. Clinical Trials in AML
7. Conclusions and Perspectives
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Comprehensive Cancer Network. Available online: https://www.nccn.org (accessed on 10 October 2025).
- Kantarjian, H.M.; Jain, N.; Garcia-Manero, G.; Welch, M.A.; Ravandi, F.; Wierda, W.G.; Jabbour, E.J. The cure of leukemia through the optimist’s prism. Cancer 2022, 128, 240–259. [Google Scholar] [CrossRef]
- Cancer Research UK. Available online: https://find.cancerresearchuk.org/ (accessed on 10 October 2025).
- Kantarjian, H.; Borthakur, G.; Daver, N.; DiNardo, C.D.; Issa, G.; Jabbour, E.; Kadia, T.; Sasaki, K.; Short, N.J.; Yilmaz MRavandi, F. Current status and research directions in acute myeloid leukemia. Blood Cancer J. 2024, 14, 163. [Google Scholar] [CrossRef]
- Shimony, S.; Stahl, M.; Stone, R.M. Acute Myeloid Leukemia: 2025 Update on Diagnosis, Risk-Stratification, and Management. Am. J. Hematol. 2025, 100, 860–891. [Google Scholar] [CrossRef]
- Bidikian, A.; Bewersdorf, J.P.; Kewan, T.; Stahl, M.; Zeidan, A.M. Acute Promyelocytic Leukemia in the Real World: Understanding Outcome Differences and How We Can Improve Them. Cancers 2024, 16, 4092. [Google Scholar] [CrossRef]
- Shimony, S.; Luskin, M.R.; Gangat, N.; LeBoeuf, N.R.; Feraco, A.M.; Lane, A.A. Blastic Plasmacytoid Dendritic Cell Neoplasm (BPDCN): 2025 Update on Diagnosis, Pathophysiology, Risk Assessment, and Management. Am. J. Hematol. 2025, 100, 1408–1422. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2025 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2024, 99, 2191–2212. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M. Chronic Lymphocytic Leukemia: 2025 Update on the Epidemiology, Pathogenesis, Diagnosis, and Therapy. Am. J. Hematol. 2025, 100, 450–480. [Google Scholar] [CrossRef]
- Rai, K.R.; Sawitsky, A.; Cronkite, E.P.; Chanana, A.D.; Levy, R.N.; Pasternack, B.S. Clinical staging of chronic lymphocytic leukemia. Blood 1975, 46, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Binet, J.L.; Auquier, A.; Dighiero, G.; Chastang, C.; Piguet, H.; Goasguen, J.; Vaugier, G.; Potron, G.; Colona, P.; Oberling, F.; et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer 1981, 48, 198–206. [Google Scholar] [CrossRef]
- Orsini, E.M.; Perelas, A.; Southern, B.D.; Grove, L.M.; Olman, M.A.; Scheraga, R.G. Stretching the Function of Innate Immune Cells. Front. Immunol. 2021, 12, 767319. [Google Scholar] [CrossRef]
- Mendez-Ferrer, S. HSCs revive their niche after transplantation. Blood 2020, 136, 2597–2598. [Google Scholar] [CrossRef] [PubMed]
- Riether, C.; Pabst, T.; Hopner, S.; Bacher, U.; Hinterbrandner, M.; Banz, Y.; Muller, R.; Manz, M.G.; Gharib, W.H.; Francisco, D.; et al. Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nat. Med. 2020, 26, 1459–1467. [Google Scholar] [CrossRef]
- Stringaris, K.; Sekine, T.; Khoder, A.; Alsuliman, A.; Razzaghi, B.; Sargeant, R.; Pavlu, J.; Brisley, G.; de Lavallade, H.; Sarvaria, A.; et al. Leukemia-induced phenotypic and functional defects in natural killer cells predict failure to achieve remission in acute myeloid leukemia. Haematologica 2014, 99, 836–847. [Google Scholar] [CrossRef]
- Greaves, M. Evolutionary determinants of cancer. Cancer Discov. 2015, 5, 806–820. [Google Scholar] [CrossRef]
- Huang, F.Y.; Trumpp, A.; Stelmach, P. Resolving leukemic stem cell heterogeneity and plasticity with single-cell multiomics. Semin. Hematol. 2025, 62, 218–225. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Lugthart, S.; Li, Y.; Erpelinck-Verschueren, C.; Deng, X.; Christos, P.J.; Schifano, E.; Booth, J.; van Putten, W.; Skrabanek, L.; et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010, 17, 13–27. [Google Scholar] [CrossRef]
- Li, Y.; Xue, M.; Deng, X.; Dong, L.; Nguyen, L.X.T.; Ren, L.; Han, L.; Li, C.; Xue, J.; Zhao, Z.; et al. TET2-mediated mRNA demethylation regulates leukemia stem cell homing and self-renewal. Cell Stem Cell 2023, 30, 1072–1090.e10. [Google Scholar] [CrossRef]
- Amador, C.; Cook, J.R.; Czader, M.; Duffield, A.; Goodlad, J.; Nejati, R.; Ott, G.; Xiao, W.; Dave, S.; Wasik, M.A.; et al. Transdifferentiation, phenotypic infidelity, progression, and transformation in T/NK-cell neoplasms: Report from the 2021 SH/EAHP Workshop. Am. J. Clin. Pathol. 2023, 159, 626–637. [Google Scholar] [CrossRef]
- Luo, Y.; Luo, J.; Yang, M.; Zhao, X. Mechanisms of T-cell metabolic reprogramming in the microenvironment of acute myeloid leukemia and its therapeutic potential (Review). Oncol. Lett. 2025, 30, 455. [Google Scholar] [CrossRef] [PubMed]
- Higos, R.; Saitoski, K.; Hautefeuille, M.; Marcelin, G.; Clement, K.; Varin-Blank, N.; Breton, C.; Lecoutre, S.; Lambert, M. The Critical Role of Adipocytes in Leukemia. Biology 2025, 14, 624. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Liu, Y.; Shu, X.; Li, Y.; Zhang, X.; Wang, C.; He, S.; Li, J.; Li, T.; Liu, T.; et al. Molecular mechanisms of viral oncogenesis in haematological malignancies: Perspectives from metabolic reprogramming, epigenetic regulation and immune microenvironment remodeling. Exp. Hematol. Oncol. 2025, 14, 69. [Google Scholar] [CrossRef]
- Addanki, S.; Kim, L.; Stevens, A. Understanding and Targeting Metabolic Vulnerabilities in Acute Myeloid Leukemia: An Updated Comprehensive Review. Cancers 2025, 17, 1355. [Google Scholar] [CrossRef]
- Hindes, M.T.; McElligott, A.M.; Best, O.G.; Ward, M.P.; Selemidis, S.; Miles, M.A.; Nturubika, B.D.; Gregory, P.A.; Anderson, P.H.; Logan, J.M.; et al. Metabolic reprogramming, malignant transformation and metastasis: Lessons from chronic lymphocytic leukaemia and prostate cancer. Cancer Lett. 2025, 611, 217441. [Google Scholar] [CrossRef]
- Ko, M.; Rao, A. TET2: Epigenetic safeguard for HSC. Blood 2011, 118, 4501–4503. [Google Scholar] [CrossRef]
- Lu, C.; Thompson, C.B. Metabolic regulation of epigenetics. Cell Metab. 2012, 16, 9–17. [Google Scholar] [CrossRef]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O’Dwyer, K.M.; et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013, 12, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Carracedo, A.; Weiss, D.; Arai, F.; Ala, U.; Avigan, D.E.; Schafer, Z.T.; Evans, R.M.; Suda, T.; Lee, C.H.; et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 2012, 18, 1350–1358. [Google Scholar] [CrossRef]
- Takubo, K.; Nagamatsu, G.; Kobayashi, C.I.; Nakamura-Ishizu, A.; Kobayashi, H.; Ikeda, E.; Goda, N.; Rahimi, Y.; Johnson, R.S.; Soga, T.; et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 2013, 12, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Graf, T.; Enver, T. Forcing cells to change lineages. Nature 2009, 462, 587–594. [Google Scholar] [CrossRef]
- Povinelli, B.J.; Rodriguez-Meira, A.; Mead, A.J. Single cell analysis of normal and leukemic hematopoiesis. Mol. Asp. Med. 2018, 59, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Azagury, M.; Buganim, Y. Unlocking trophectoderm mysteries: In vivo and in vitro perspectives on human and mouse trophoblast fate induction. Dev. Cell 2024, 59, 941–960. [Google Scholar] [CrossRef]
- Azagury, M.; Buganim, Y. Direct Reprogramming of Human Fibroblasts into Fully Functional Trophoblast Stem Cells. In Methods in Molecular Biology; Springer: New York, NY, USA, 2025. [Google Scholar] [CrossRef]
- Hu, T.; Murdaugh, R.; Nakada, D. Transcriptional and Microenvironmental Regulation of Lineage Ambiguity in Leukemia. Front. Oncol. 2017, 7, 268. [Google Scholar] [CrossRef] [PubMed]
- Silbert, S.K.; Rankin, A.W.; Hoang, C.N.; Semchenkova, A.; Myers, R.M.; Zerkalenkova, E.; Wang, H.W.; Kovach, A.E.; Yuan, C.M.; Delgado Colon, D.; et al. Project EVOLVE: An international analysis of postimmunotherapy lineage switch, an emergent form of relapse in leukemia. Blood 2025, 146, 437–455. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, P.; Zhu, H.; Wang, Z.; Zhang, H.; Xu, H.; Li, R.; Sheng, Y.; Peng, H. Single-Cell Transcriptomic Analysis of Myeloid Lineage Evolution from CD19 CAR-T Cell Therapy. Pathobiology 2025, 92, 180–186. [Google Scholar] [CrossRef]
- Schepers, K.; Campbell, T.B.; Passegue, E. Normal and leukemic stem cell niches: Insights and therapeutic opportunities. Cell Stem Cell 2015, 16, 254–267. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Rodriguez, S.; Cao, L.; Parish, J.; Mumaw, C.; Zollman, A.; Kamoka, M.M.; Mu, J.; Chen, D.Z.; et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-kappaB-dependent manner. Cell Stem Cell 2014, 15, 51–65. [Google Scholar] [CrossRef]
- Bhowmik, S.; Bose, A.; Sengupta, A. Innate immune-inflammatory signaling milieu in myeloid leukemia and aging-associated clonal hematopoiesis pathologies. Front. Immunol. 2025, 16, 1660709. [Google Scholar] [CrossRef]
- Cavalcante, C.B.A.; Chaves, A.C.; de Oliveira, V.S.; de Araujo, M.A.S.; Cunha, E.S.T.M.; Goes, J.V.C.; de Oliveira, R.T.G.; Pinheiro, R.F.; Ribeiro-Junior, H.L. Role of Toll-Like Receptors in Myeloid Neoplasms: Focuses on the Molecular Mechanisms and Clinical Impact on Myelodysplastic Syndromes, Acute Myeloid Leukemia, and Chronic Myeloid Leukemia. Apmis 2025, 133, e70065. [Google Scholar] [CrossRef]
- Jopling, C.; Boue, S.; Izpisua Belmonte, J.C. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Cieslar-Pobuda, A.; Knoflach, V.; Ringh, M.V.; Stark, J.; Likus, W.; Siemianowicz, K.; Ghavami, S.; Hudecki, A.; Green, J.L.; Los, M.J. Transdifferentiation and reprogramming: Overview of the processes, their similarities and differences. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, J.; Yue, C.; Song, S.; Tian, L.; Wang, Y. Recent advances in pancreatic alpha-cell transdifferentiation for diabetes therapy. Front. Immunol. 2025, 16, 1551372. [Google Scholar] [CrossRef]
- Ascic, E.; Pereira, C.F. Transcription factor-mediated reprogramming to antigen-presenting cells. Curr. Opin. Genet. Dev. 2025, 90, 102300. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Sudhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef]
- Xie, H.; Ye, M.; Feng, R.; Graf, T. Stepwise reprogramming of B cells into macrophages. Cell 2004, 117, 663–676. [Google Scholar] [CrossRef]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Soufi, A.; Garcia, M.F.; Jaroszewicz, A.; Osman, N.; Pellegrini, M.; Zaret, K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Zhu, J.; Nie, Q.; Xie, B.; Xue, S.; Zhang, A.; Li, Q.; Zhang, Z.; Li, S.; Li, Y.; et al. NLRP3 promotes inflammatory signaling and IL-1beta cleavage in acute lung injury caused by cell wall extract of Lactobacillus casei. Commun. Biol. 2025, 8, 20. [Google Scholar] [CrossRef]
- Stadhouders, R.; Filion, G.J.; Graf, T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature 2019, 569, 345–354. [Google Scholar] [CrossRef]
- Treutlein, B.; Lee, Q.Y.; Camp, J.G.; Mall, M.; Koh, W.; Shariati, S.A.; Sim, S.; Neff, N.F.; Skotheim, J.M.; Wernig, M.; et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 2016, 534, 391–395. [Google Scholar] [CrossRef]
- Mikkelsen, T.S.; Hanna, J.; Zhang, X.; Ku, M.; Wernig, M.; Schorderet, P.; Bernstein, B.E.; Jaenisch, R.; Lander, E.S.; Meissner, A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008, 454, 49–55. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Zepp, J.A.; Alvira, C.M. Nanoparticle Delivery of Angiogenic Gene Therapy. Save the Vessels, Save the Lung! Am. J. Respir. Crit. Care Med. 2020, 202, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.P.; Mostoslavsky, R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol. Cell 2016, 62, 695–711. [Google Scholar] [CrossRef] [PubMed]
- Slack, J.M. Amphibian muscle regeneration--dedifferentiation or satellite cells? Trends Cell Biol. 2006, 16, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, M.; Tsoneva, D.K. Histone variants: Key regulators of inflammation in cell dedifferentiation and transdifferentiation. Front. Immunol. 2025, 16, 1619100. [Google Scholar] [CrossRef]
- Li, W.; George, P.; Azadian, M.M.; Ning, M.; Dhand, A.; Cramer, S.C.; Carmichael, S.T.; Lo, E.H. Changing genes, cells and networks to reprogram the brain after stroke. Nat. Neurosci. 2025, 28, 1130–1145. [Google Scholar] [CrossRef]
- Conrad, C.; Magnen, M.; Tsui, J.; Wismer, H.; Naser, M.; Venkataramani, U.; Samad, B.; Cleary, S.J.; Qiu, L.; Tian, J.J.; et al. Decoding functional hematopoietic progenitor cells in the adult human lung. Blood 2025, 145, 1975–1986. [Google Scholar] [CrossRef]
- Wu, Z.; Shi, Y.; Cui, Y.; Xing, X.; Zhang, L.; Liu, D.; Zhang, Y.; Dong, J.; Jin, L.; Pang, M.; et al. Single-cell analysis reveals an Angpt4-initiated EPDC-EC-CM cellular coordination cascade during heart regeneration. Protein Cell 2023, 14, 350–368. [Google Scholar] [CrossRef]
- Smith, D.; Eichinger, A.; Fennell, E.; Xu-Monette, Z.Y.; Rech, A.; Wang, J.; Esteva, E.; Seyedian, A.; Yang, X.; Zhang, M.; et al. Spatial and single cell mapping of castleman disease reveals key stromal cell types and cytokine pathways. Nat. Commun. 2025, 16, 6009. [Google Scholar] [CrossRef]
- Rodriguez-Meira, A.; Norfo, R.; Wen, S.; Chedeville, A.L.; Rahman, H.; O’Sullivan, J.; Wang, G.; Louka, E.; Kretzschmar, W.W.; Paterson, A.; et al. Single-cell multi-omics identifies chronic inflammation as a driver of TP53-mutant leukemic evolution. Nat. Genet. 2023, 55, 1531–1541. [Google Scholar] [CrossRef]
- Radwan, A.; Eccleston, J.; Sabag, O.; Marcus, H.; Sussman, J.; Ouro, A.; Rahamim, M.; Azagury, M.; Azria, B.; Stanger, B.Z.; et al. Transdifferentiation occurs without resetting development-specific DNA methylation, a key determinant of full-function cell identity. Proc. Natl. Acad. Sci. USA 2024, 121, e2411352121. [Google Scholar] [CrossRef] [PubMed]
- Bosch, T.C. Hydra and the evolution of stem cells. Bioessays 2009, 31, 478–486. [Google Scholar] [CrossRef]
- Reddien, P.W. The Cellular and Molecular Basis for Planarian Regeneration. Cell 2018, 175, 327–345. [Google Scholar] [CrossRef] [PubMed]
- Voss, S.R.; Palumbo, A.; Nagarajan, R.; Gardiner, D.M.; Muneoka, K.; Stromberg, A.J.; Athippozhy, A.T. Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression. Regeneration 2015, 2, 120–136. [Google Scholar] [CrossRef]
- Echeverri, K.; Tanaka, E.M. Electroporation as a tool to study in vivo spinal cord regeneration. Dev. Dyn. 2003, 226, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Eski, S.E.; Mi, J.; Pozo-Morales, M.; Hovhannisyan, G.G.; Perazzolo, C.; Manco, R.; Ez-Zammoury, I.; Barbhaya, D.; Lefort, A.; Libert, F.; et al. Cholangiocytes contribute to hepatocyte regeneration after partial liver injury during growth spurt in zebrafish. Nat. Commun. 2025, 16, 5260. [Google Scholar] [CrossRef]
- Huang, D.; Kapadia, E.H.; Liang, Y.; Shriver, L.P.; Dai, S.; Patti, G.J.; Humbel, B.M.; Laudet, V.; Parichy, D.M. Agouti and BMP signaling drive a naturally occurring fate conversion of melanophores to leucophores in zebrafish. Proc. Natl. Acad. Sci. USA 2025, 122, e2424180122. [Google Scholar] [CrossRef]
- Beaulieu, M.O.; Thomas, E.D.; Raible, D.W. Transdifferentiation is temporally uncoupled from progenitor pool expansion during hair cell regeneration in the zebrafish inner ear. Development 2024, 151, dev202944. [Google Scholar] [CrossRef]
- Tarlow, B.D.; Pelz, C.; Naugler, W.E.; Wakefield, L.; Wilson, E.M.; Finegold, M.J.; Grompe, M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014, 15, 605–618. [Google Scholar] [CrossRef]
- Bernadskaya, Y.; Christiaen, L. Transcriptional Control of Developmental Cell Behaviors. Annu. Rev. Cell Dev. Biol. 2016, 32, 77–101. [Google Scholar] [CrossRef]
- Dobrzycki, T.; Lalwani, M.; Telfer, C.; Monteiro, R.; Patient, R. The roles and controls of GATA factors in blood and cardiac development. IUBMB Life 2020, 72, 39–44. [Google Scholar] [CrossRef]
- Bakri, Y.; Sarrazin, S.; Mayer, U.P.; Tillmanns, S.; Nerlov, C.; Boned, A.; Sieweke, M.H. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood 2005, 105, 2707–2716. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Ahrens, T.D.; Lutz, L.; Lassmann, S.; Werner, M. Turning Skyscrapers into Town Houses: Insights into Barrett’s Esophagus. Pathobiology 2017, 84, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Thorel, F.; Nepote, V.; Avril, I.; Kohno, K.; Desgraz, R.; Chera, S.; Herrera, P.L. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 2010, 464, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Imperato, M.R.; Cauchy, P.; Obier, N.; Bonifer, C. The RUNX1-PU.1 axis in the control of hematopoiesis. Int. J. Hematol. 2015, 101, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.C.; Sher, D.; Eisenstein, M.; Shigesada, K.; Reitzel, A.M.; Marlow, H.; Levanon, D.; Groner, Y.; Finnerty, J.R.; Gat, U. The evolutionary origin of the Runx/CBFbeta transcription factors--studies of the most basal metazoans. BMC Evol. Biol. 2008, 8, 228. [Google Scholar] [CrossRef]
- Gangenahalli, G.U.; Gupta, P.; Saluja, D.; Verma, Y.K.; Kishore, V.; Chandra, R.; Sharma, R.K.; Ravindranath, T. Stem cell fate specification: Role of master regulatory switch transcription factor PU.1 in differential hematopoiesis. Stem Cells Dev. 2005, 14, 140–152. [Google Scholar] [CrossRef]
- Anderson, M.K.; Sun, X.; Miracle, A.L.; Litman, G.W.; Rothenberg, E.V. Evolution of hematopoiesis: Three members of the PU.1 transcription factor family in a cartilaginous fish, Raja eglanteria. Proc. Natl. Acad. Sci. USA 2001, 98, 553–558. [Google Scholar] [CrossRef]
- Avellino, R.; Delwel, R. Expression and regulation of C/EBPalpha in normal myelopoiesis and in malignant transformation. Blood 2017, 129, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- Di Tullio, A.; Graf, T. C/EBPalpha bypasses cell cycle-dependency during immune cell transdifferentiation. Cell Cycle 2012, 11, 2739–2746. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Desbordes, S.C.; Xie, H.; Tillo, E.S.; Pixley, F.; Stanley, E.R.; Graf, T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6057–6062. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Wang, L.; Wang, N.; Li, Y.; Li, H. Transdifferentiation of fibroblasts into adipocyte-like cells by chicken adipogenic transcription factors. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010, 156, 502–508. [Google Scholar] [CrossRef]
- Lyons, S.E.; Shue, B.C.; Oates, A.C.; Zon, L.I.; Liu, P.P. A novel myeloid-restricted zebrafish CCAAT/enhancer-binding protein with a potent transcriptional activation domain. Blood 2001, 97, 2611–2617. [Google Scholar] [CrossRef]
- Kueh, H.Y.; Champhekar, A.; Nutt, S.L.; Elowitz, M.B.; Rothenberg, E.V. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 2013, 341, 670–673. [Google Scholar] [CrossRef]
- Nerlov, C. C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer 2004, 4, 394–400. [Google Scholar] [CrossRef]
- Johnson, C.S.; Williams, M.; Sham, K.; Belluschi, S.; Ma, W.; Wang, X.; Lau, W.W.Y.; Kaufmann, K.B.; Krivdova, G.; Calderbank, E.F.; et al. Adaptation to ex vivo culture reduces human hematopoietic stem cell activity independently of the cell cycle. Blood 2024, 144, 729–741. [Google Scholar] [CrossRef]
- Stadhouders, R. Expanding the toolbox for 3D genomics. Nat. Genet. 2018, 50, 634–635. [Google Scholar] [CrossRef]
- Laurenti, E.; Gottgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 2018, 553, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Sheng, M.; Rong, S.; Zhou, D.; Wang, C.; Wu, W.; Huang, J.; Sun, Y.; Wang, Y.; Chen, P.; et al. STING activation in TET2-mutated hematopoietic stem/progenitor cells contributes to the increased self-renewal and neoplastic transformation. Leukemia 2023, 37, 2457–2467. [Google Scholar] [CrossRef]
- Belluschi, S.; Calderbank, E.F.; Ciaurro, V.; Pijuan-Sala, B.; Santoro, A.; Mende, N.; Diamanti, E.; Sham, K.Y.C.; Wang, X.; Lau, W.W.Y.; et al. Myelo-lymphoid lineage restriction occurs in the human haematopoietic stem cell compartment before lymphoid-primed multipotent progenitors. Nat. Commun. 2018, 9, 4100. [Google Scholar] [CrossRef]
- Zhang, Y.; Blomquist, T.M.; Kusko, R.; Stetson, D.; Zhang, Z.; Yin, L.; Sebra, R.; Gong, B.; Lococo, J.S.; Mittal, V.K.; et al. Deep oncopanel sequencing reveals within block position-dependent quality degradation in FFPE processed samples. Genome Biol. 2022, 23, 141. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.R.; Wang, W.; Gomez, M.; Zhang, T.; Mathew, S.; Furman, R.R.; Knowles, D.M.; Orazi, A.; Tam, W. Transformation of chronic lymphocytic leukemia/small lymphocytic lymphoma to interdigitating dendritic cell sarcoma: Evidence for transdifferentiation of the lymphoma clone. Am. J. Clin. Pathol. 2009, 132, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Xi, L.; Raffeld, M.; Feldman, A.L.; Ketterling, R.P.; Knudson, R.; Rodriguez-Canales, J.; Hanson, J.; Pittaluga, S.; Jaffe, E.S. Clonally related histiocytic/dendritic cell sarcoma and chronic lymphocytic leukemia/small lymphocytic lymphoma: A study of seven cases. Mod. Pathol. 2011, 24, 1421–1432. [Google Scholar] [CrossRef]
- Argyropoulos, K.V.; Aypar, U.; Ewalt, M.D.; Roshal, M.; Dogan, A.; Sen, F. Chronic lymphocytic leukemia transdifferentiated to blastic neoplasm with T/plasmacytoid dendritic cell immunophenotype. Leuk. Lymphoma 2023, 64, 734–737. [Google Scholar] [CrossRef]
- Antherieu, G.; Donzel, M.; Balme, B.; Traverse-Glehen, A.; Heiblig, M. Chronic myelomonocytic leukemia transdifferentiation landscape: From histiocytosis to blastic plasmacytoid dendritic cell neoplasm. Am. J. Hematol. 2023, 98, 1343–1345. [Google Scholar] [CrossRef]
- Jenei, A.; Bedics, G.; Erdelyi, D.J.; Muller, J.; Gyorke, T.; Bodor, C.; Szepesi, A. Potential role of MAP2K1 mutation in the trans-differentiation of interdigitating dendritic cell sarcoma: Case report and literature review. Front. Pediatr. 2022, 10, 959307. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.F.d.O.; Marani, L.O.; Bianco, T.M.; Arantes, A.Q.; Lopes, I.A.; Pereira-Martins, D.A.; Palma, L.C.; Scheucher, P.S.; Schiavinato, J.L.d.S.; Binelli, L.S.; et al. Altered distribution and function of NK-cell subsets lead to impaired tumor surveillance in JAK2V617F myeloproliferative neoplasms. Front. Immunol. 2022, 13, 768592. [Google Scholar] [CrossRef]
- Takeda, R.; Yokoyama, K.; Fukuyama, T.; Kawamata, T.; Ito, M.; Yusa, N.; Kasajima, R.; Shimizu, E.; Ohno, N.; Uchimaru, K.; et al. Repeated Lineage Switches in an Elderly Case of Refractory B-Cell Acute Lymphoblastic Leukemia with MLL Gene Amplification: A Case Report and Literature Review. Front. Oncol. 2022, 12, 799982. [Google Scholar] [CrossRef]
- Yang, D.; Jones, M.G.; Naranjo, S.; Rideout, W.M., 3rd.; Min, K.H.J.; Ho, R.; Wu, W.; Replogle, J.M.; Page, J.L.; Quinn, J.J.; et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell 2022, 185, 1905–1923 e1925. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness. Cancers 2020, 12, 3716. [Google Scholar] [CrossRef] [PubMed]
- Goardon, N.; Marchi, E.; Atzberger, A.; Quek, L.; Schuh, A.; Soneji, S.; Woll, P.; Mead, A.; Alford, K.A.; Rout, R.; et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 2011, 19, 138–152. [Google Scholar] [CrossRef]
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010, 363, 2424–2433. [Google Scholar] [CrossRef]
- Rosenbauer, F.; Tenen, D.G. Transcription factors in myeloid development: Balancing differentiation with transformation. Nat. Rev. Immunol. 2007, 7, 105–117. [Google Scholar] [CrossRef]
- Iacobucci, I.; Mullighan, C.G. Genetic Basis of Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2017, 35, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.N.; Chen, Z.; Braas, D.; Lee, J.W.; Xiao, G.; Geng, H.; Cosgun, K.N.; Hurtz, C.; Shojaee, S.; Cazzaniga, V.; et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 2017, 542, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.N.; Muschen, M. B-cell identity as a metabolic barrier against malignant transformation. Exp. Hematol. 2017, 53, 1–6. [Google Scholar] [CrossRef]
- Wasik, M.A.; Kim, P.M.; Nejati, R. Diverse and reprogrammable mechanisms of malignant cell transformation in lymphocytes: Pathogenetic insights and translational implications. Front. Oncol. 2024, 14, 1383741. [Google Scholar] [CrossRef]
- Newsam, A.D.; Ziccheddu, B.; Gowda Saralamma, V.V.; Coughlin, C.A.; Goretsky, Y.E.; Youssfi, A.A.; Russo, M.V.; Gallego, N.C.; Fattakhov, N.; Coffey, D.G.; et al. RHOA Loss of Function Impairs the IFNgamma Response and Promotes CD19 Antigen Escape to Drive CAR-T Resistance in Diffuse Large B-cell Lymphoma. bioRxiv 2025. bioRxiv:02.27.640687. [Google Scholar] [CrossRef]
- Mills, L.J.; Scott, M.C.; Shah, P.; Cunanan, A.R.; Deshpande, A.; Auch, B.; Curtin, B.; Beckman, K.B.; Spector, L.G.; Sarver, A.L.; et al. Comparative analysis of genome-wide DNA methylation identifies patterns that associate with conserved transcriptional programs in osteosarcoma. Bone 2022, 158, 115716. [Google Scholar] [CrossRef]
- Jacoby, E.; Nguyen, S.M.; Fountaine, T.J.; Welp, K.; Gryder, B.; Qin, H.; Yang, Y.; Chien, C.D.; Seif, A.E.; Lei, H.; et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat. Commun. 2016, 7, 12320. [Google Scholar] [CrossRef]
- Gardner, R.; Wu, D.; Cherian, S.; Fang, M.; Hanafi, L.A.; Finney, O.; Smithers, H.; Jensen, M.C.; Riddell, S.R.; Maloney, D.G.; et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 2016, 127, 2406–2410. [Google Scholar] [CrossRef]
- Feldman, A.L.; Arber, D.A.; Pittaluga, S.; Martinez, A.; Burke, J.S.; Raffeld, M.; Camos, M.; Warnke, R.; Jaffe, E.S. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: Evidence for transdifferentiation of the follicular lymphoma clone. Blood 2008, 111, 5433–5439. [Google Scholar] [CrossRef] [PubMed]
- West, R.R.; Hsu, A.P.; Holland, S.M.; Cuellar-Rodriguez, J.; Hickstein, D.D. Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. Haematologica 2014, 99, 276–281. [Google Scholar] [CrossRef] [PubMed]
- West, D.S.; Dogan, A.; Quint, P.S.; Tricker-Klar, M.L.; Porcher, J.C.; Ketterling, R.P.; Law, M.E.; McPhail, E.D.; Viswanatha, D.S.; Kurtin, P.J.; et al. Clonally related follicular lymphomas and Langerhans cell neoplasms: Expanding the spectrum of transdifferentiation. Am. J. Surg. Pathol. 2013, 37, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Nakamine, H.; Yamakawa, M.; Yoshino, T.; Fukumoto, T.; Enomoto, Y.; Matsumura, I. Langerhans Cell Histiocytosis and Langerhans Cell Sarcoma: Current Understanding and Differential Diagnosis. J. Clin. Exp. Hematop. 2016, 56, 109–118. [Google Scholar] [CrossRef]
- Klauer, L.K.; Rejeski, H.A.; Ugur, S.; Rackl, E.; Abdulmajid, J.; Fischer, Z.; Pepeldjiyska, E.; Frischhut, A.; Schmieder, N.; Volker, A.; et al. Leukemia-Derived Dendritic Cells Induce Anti-Leukemic Effects Ex Vivo in AML Independently of Patients’ Clinical and Biological Features. Int. J. Mol. Sci. 2025, 26, 1700. [Google Scholar] [CrossRef] [PubMed]
- Skala, S.L.; Ye, J.C.; Stumph, J.; Macon, W.R.; Quinones, F.R.; Khachaturov, V.; Ketterling, R.P.; Dewar, R. Combined Tumors in Hematolymphoid Neoplasms: Case Series of Histiocytic and Langerhans Cell Sarcomas Arising From Low-Grade B-Cell Lymphoma. Clin. Pathol. 2019, 12, 2632010X19878410. [Google Scholar] [CrossRef]
- Kazama, S.; Yokoyama, K.; Ueki, T.; Kazumoto, H.; Satomi, H.; Sumi, M.; Ito, I.; Yusa, N.; Kasajima, R.; Shimizu, E.; et al. Case report: Common clonal origin of concurrent langerhans cell histiocytosis and acute myeloid leukemia. Front. Oncol. 2022, 12, 974307. [Google Scholar] [CrossRef]
- Auer, F.; Ingenhag, D.; Bhatia, S.; Enczmann, J.; Cobaleda, C.; Sanchez-Garcia, I.; Borkhardt, A.; Hauer, J. GEMMs addressing Pax5 loss-of-function in childhood pB-ALL. Eur. J. Med. Genet. 2016, 59, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Rapino, F.; Robles, E.F.; Richter-Larrea, J.A.; Kallin, E.M.; Martinez-Climent, J.A.; Graf, T. C/EBPalpha induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 2013, 3, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, J.P.; e Sousa, C.R. The Role of Type 1 Conventional Dendritic Cells in Cancer Immunity. Trends Cancer 2018, 4, 784–792. [Google Scholar] [CrossRef]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef]
- Del Prete, A.; Salvi, V.; Soriani, A.; Laffranchi, M.; Sozio, F.; Bosisio, D.; Sozzani, S. Dendritic cell subsets in cancer immunity and tumor antigen sensing. Cell Mol. Immunol. 2023, 20, 432–447. [Google Scholar] [CrossRef]
- Murphy, K.M.; Reiner, S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002, 2, 933–944. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Collazo, O.; Azagra, A.; Fernandez-Duran, I.; Serra-Musach, J.; Islam, A.B.; Vega-Garcia, N.; Malatesta, R.; Camos, M.; Gomez, A.; et al. The transcriptional repressor HDAC7 promotes apoptosis and c-Myc downregulation in particular types of leukemia and lymphoma. Cell Death Dis. 2015, 6, e1635. [Google Scholar] [CrossRef]
- Cirovic, B.; Schonheit, J.; Kowenz-Leutz, E.; Ivanovska, J.; Klement, C.; Pronina, N.; Begay, V.; Leutz, A. C/EBP-Induced Transdifferentiation Reveals Granulocyte-Macrophage Precursor-like Plasticity of B Cells. Stem Cell Rep. 2017, 8, 346–359. [Google Scholar] [CrossRef]
- van Oevelen, C.; Collombet, S.; Vicent, G.; Hoogenkamp, M.; Lepoivre, C.; Badeaux, A.; Bussmann, L.; Sardina, J.L.; Thieffry, D.; Beato, M.; et al. C/EBPalpha Activates Pre-existing and De Novo Macrophage Enhancers during Induced Pre-B Cell Transdifferentiation and Myelopoiesis. Stem Cell Rep. 2015, 5, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Klonizakis, A.; Alcoverro-Bertran, M.; Masso, P.; Thomas, J.; de Andres-Aguayo, L.; Wei, X.; Varamogianni-Mamatsi, V.; Nikolaou, C.; Graf, T. Synergistic and antagonistic activities of IRF8 and FOS enhancer pairs during an immune-cell fate switch. EMBO J. 2025, 44, 2025–2055. [Google Scholar] [CrossRef]
- McClellan, J.S.; Dove, C.; Gentles, A.J.; Ryan, C.E.; Majeti, R. Reprogramming of primary human Philadelphia chromosome-positive B cell acute lymphoblastic leukemia cells into nonleukemic macrophages. Proc. Natl. Acad. Sci. USA 2015, 112, 4074–4079. [Google Scholar] [CrossRef]
- Pittet, M.J.; Di Pilato, M.; Garris, C.; Mempel, T.R. Dendritic cells as shepherds of T cell immunity in cancer. Immunity 2023, 56, 2218–2230. [Google Scholar] [CrossRef]
- Breton, G.; Lee, J.; Liu, K.; Nussenzweig, M.C. Defining human dendritic cell progenitors by multiparametric flow cytometry. Nat. Protoc. 2015, 10, 1407–1422. [Google Scholar] [CrossRef] [PubMed]
- Fogg, D.K.; Sibon, C.; Miled, C.; Jung, S.; Aucouturier, P.; Littman, D.R.; Cumano, A.; Geissmann, F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 2006, 311, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Welch, B.M.; Parikh, S.A.; Kay, N.E.; Medina, K.L. Profound phenotypic deficiencies in mature blood and bone marrow progenitor dendritic cells in chronic lymphocytic leukemia patients. Leukemia 2025, 39, 1915–1927. [Google Scholar] [CrossRef]
- Schreibelt, G.; Duiveman-de Boer, T.; Pots, J.M.; van Oorschot, T.G.M.; de Boer, A.J.; Scharenborg, N.M.; van de Rakt, M.; Bos, K.; de Goede, A.L.; Petry, K.; et al. Fully closed and automated enrichment of primary blood dendritic cells for cancer immunotherapy. Methods Cell Biol. 2024, 183, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Barcelos, E.C.S.; Ricciuti, D.; Mondanelli, G.; Gargaro, M. Rewriting the dendritic cell code in cancer-from subset identity to immunotherapeutic design. FEBS Lett. 2025, 599, 2060–2083. [Google Scholar] [CrossRef]
- Monti, M.; Ferrari, G.; Gazzurelli, L.; Bugatti, M.; Facchetti, F.; Vermi, W. Plasmacytoid dendritic cells at the forefront of anti-cancer immunity: Rewiring strategies for tumor microenvironment remodeling. J. Exp. Clin. Cancer Res. 2024, 43, 196. [Google Scholar] [CrossRef]
- Sesti-Costa, R.; Cervantes-Barragan, L.; Swiecki, M.K.; Fachi, J.L.; Cella, M.; Gilfillan, S.; Silva, J.S.; Colonna, M. Leukemia Inhibitory Factor Inhibits Plasmacytoid Dendritic Cell Function and Development. J. Immunol. 2020, 204, 2257–2268. [Google Scholar] [CrossRef]
- Giampazolias, E. Boosting Dendritic Cell Function in Cancer. Cancer Med. 2025, 14, e71062. [Google Scholar] [CrossRef]
- Liu, P.; Zhao, L.; Kroemer, G.; Kepp, O. Conventional type 1 dendritic cells (cDC1) in cancer immunity. Biol. Direct 2023, 18, 71. [Google Scholar] [CrossRef]
- Roberts, E.W.; Broz, M.L.; Binnewies, M.; Headley, M.B.; Nelson, A.E.; Wolf, D.M.; Kaisho, T.; Bogunovic, D.; Bhardwaj, N.; Krummel, M.F. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 2016, 30, 324–336. [Google Scholar] [CrossRef]
- Barry, K.C.; Hsu, J.; Broz, M.L.; Cueto, F.J.; Binnewies, M.; Combes, A.J.; Nelson, A.E.; Loo, K.; Kumar, R.; Rosenblum, M.D.; et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat. Med. 2018, 24, 1178–1191. [Google Scholar] [CrossRef]
- Garg, A.D.; Vara Perez, M.; Schaaf, M.; Agostinis, P.; Zitvogel, L.; Kroemer, G.; Galluzzi, L. Trial watch: Dendritic cell-based anticancer immunotherapy. Oncoimmunology 2017, 6, e1328341. [Google Scholar] [CrossRef] [PubMed]
- Krysko, D.V.; Garg, A.D.; Kaczmarek, A.; Krysko, O.; Agostinis, P.; Vandenabeele, P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer 2012, 12, 860–875. [Google Scholar] [CrossRef]
- Garris, C.S.; Arlauckas, S.P.; Kohler, R.H.; Trefny, M.P.; Garren, S.; Piot, C.; Engblom, C.; Pfirschke, C.; Siwicki, M.; Gungabeesoon, J.; et al. Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-gamma and IL-12. Immunity 2018, 49, 1148–1161.e1147. [Google Scholar] [CrossRef] [PubMed]
- Heras-Murillo, I.; Mananes, D.; Munne, P.; Nunez, V.; Herrera, J.; Catala-Montoro, M.; Alvarez, M.; Del Pozo, M.A.; Melero, I.; Wculek, S.K.; et al. Immunotherapy with conventional type-1 dendritic cells induces immune memory and limits tumor relapse. Nat. Commun. 2025, 16, 3369. [Google Scholar] [CrossRef]
- Hildner, K.; Edelson, B.T.; Purtha, W.E.; Diamond, M.; Matsushita, H.; Kohyama, M.; Calderon, B.; Schraml, B.U.; Unanue, E.R.; Diamond, M.S.; et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008, 322, 1097–1100. [Google Scholar] [CrossRef]
- Murphy, T.L.; Grajales-Reyes, G.E.; Wu, X.; Tussiwand, R.; Briseno, C.G.; Iwata, A.; Kretzer, N.M.; Durai, V.; Murphy, K.M. Transcriptional Control of Dendritic Cell Development. Annu. Rev. Immunol. 2016, 34, 93–119. [Google Scholar] [CrossRef]
- Zimmermannova, O.; Ferreira, A.G.; Ascic, E.; Santiago, M.V.; Kurochkin, I.; Hansen, M.; Met, O.; Caiado, I.; Shapiro, I.E.; Michaux, J.; et al. Restoring tumor immunogenicity with dendritic cell reprogramming. Sci. Immunol. 2023, 8, eadd4817. [Google Scholar] [CrossRef] [PubMed]
- Ascic, E.; Akerstrom, F.; Nair, M.S.; Rosa, A.; Kurochkin, I.; Zimmermannova, O.; Catena, X.; Rotankova, N.; Veser, C.; Rudnik, M.; et al. In vivo dendritic cell reprogramming for cancer immunotherapy. Science 2024, 386, eadn9083. [Google Scholar] [CrossRef]
- Mazzoccoli, L.; Liu, B. Dendritic Cells in Shaping Anti-Tumor T Cell Response. Cancers 2024, 16, 2211. [Google Scholar] [CrossRef]
- Tadepalli, S.; Clements, D.R.; Raquer-McKay, H.M.; Ludtke, A.; Saravanan, S.; Seong, D.; Vitek, L.; Richards, C.M.; Carette, J.E.; Mack, M.; et al. CD301b+ monocyte-derived dendritic cells mediate resistance to radiotherapy. J. Exp. Med. 2025, 222, e20231717. [Google Scholar] [CrossRef]
- Roddie, P.H.; Horton, Y.; Turner, M.L. Primary acute myeloid leukaemia blasts resistant to cytokine-induced differentiation to dendritic-like leukaemia cells can be forced to differentiate by the addition of bryostatin-1. Leukemia 2002, 16, 84–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohty, M.; Olive, D.; Gaugler, B. Leukemic dendritic cells: Potential for therapy and insights towards immune escape by leukemic blasts. Leukemia 2002, 16, 2197–2204. [Google Scholar] [CrossRef] [PubMed]
- Bueno-Costa, A.; Pineyro, D.; Soler, M.; Javierre, B.M.; Raurell-Vila, H.; Subirana-Granes, M.; Pasquali, L.; Martinez-Climent, J.A.; Esteller, M. B-cell leukemia transdifferentiation to macrophage involves reconfiguration of DNA methylation for long-range regulation. Leukemia 2020, 34, 1158–1162. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, Y.; Fan, Y.; Jiang, N.; Meng, Y.; Li, Y.; Xue, M.; Xu, C.; Guo, W.; Liu, W. Development and application of a sensitive droplet digital PCR-based method to detect tilapia parvovirus. J. Fish. Dis. 2023, 46, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Fennell, K.A.; Bell, C.C.; Dawson, M.A. Epigenetic therapies in acute myeloid leukemia: Where to from here? Blood 2019, 134, 1891–1901. [Google Scholar] [CrossRef]
- Murakami, K.; Sasaki, H.; Nishiyama, A.; Kurotaki, D.; Kawase, W.; Ban, T.; Nakabayashi, J.; Kanzaki, S.; Sekita, Y.; Nakajima, H.; et al. A RUNX-CBFbeta-driven enhancer directs the Irf8 dose-dependent lineage choice between DCs and monocytes. Nat. Immunol. 2021, 22, 301–311. [Google Scholar] [CrossRef]
- Woller, S.A.; Ravula, S.B.; Tucci, F.C.; Beaton, G.; Corr, M.; Isseroff, R.R.; Soulika, A.M.; Chigbrow, M.; Eddinger, K.A.; Yaksh, T.L. Systemic TAK-242 prevents intrathecal LPS evoked hyperalgesia in male, but not female mice and prevents delayed allodynia following intraplantar formalin in both male and female mice: The role of TLR4 in the evolution of a persistent pain state. Brain Behav. Immun. 2016, 56, 271–280. [Google Scholar] [CrossRef]
- Marciscano, A.E.; Anandasabapathy, N. The role of dendritic cells in cancer and anti-tumor immunity. Semin. Immunol. 2021, 52, 101481. [Google Scholar] [CrossRef]
- Zevini, A.; Palermo, E.; Di Carlo, D.; Alexandridi, M.; Rinaldo, S.; Paone, A.; Cutruzzola, F.; Etna, M.P.; Coccia, E.M.; Olagnier, D.; et al. Inhibition of Glycolysis Impairs Retinoic Acid-Inducible Gene I-Mediated Antiviral Responses in Primary Human Dendritic Cells. Front. Cell Infect. Microbiol. 2022, 12, 910864. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, C.M.; Holowka, T.; Sun, J.; Blagih, J.; Amiel, E.; DeBerardinis, R.J.; Cross, J.R.; Jung, E.; Thompson, C.B.; Jones, R.G.; et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010, 115, 4742–4749. [Google Scholar] [CrossRef] [PubMed]
- Basit, F.; van Oorschot, T.; van Buggenum, J.; Derks, R.J.E.; Kostidis, S.; Giera, M.; de Vries, I.J.M. Metabolomic and lipidomic signatures associated with activation of human cDC1 (BDCA3(+) /CD141(+)) dendritic cells. Immunology 2022, 165, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Linde, M.H.; Fan, A.C.; Kohnke, T.; Trotman-Grant, A.C.; Gurev, S.F.; Phan, P.; Zhao, F.; Haddock, N.L.; Nuno, K.A.; Gars, E.J.; et al. Reprogramming Cancer into Antigen-Presenting Cells as a Novel Immunotherapy. Cancer Discov. 2023, 13, 1164–1185. [Google Scholar] [CrossRef]
- Zimmermannova, O.; Ferreira, A.G.; Pereira, C.F. Orchestrating an immune response to cancer with cellular reprogramming. Genes. Immun. 2024, 25, 95–97. [Google Scholar] [CrossRef]
- Rosa, F.F.; Pires, C.F.; Kurochkin, I.; Ferreira, A.G.; Gomes, A.M.; Palma, L.G.; Shaiv, K.; Solanas, L.; Azenha, C.; Papatsenko, D.; et al. Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci. Immunol. 2018, 3, eaau4292. [Google Scholar] [CrossRef]
- Palomares, F.; Pina, A.; Dakhaoui, H.; Leiva-Castro, C.; Munera-Rodriguez, A.M.; Cejudo-Guillen, M.; Granados, B.; Alba, G.; Santa-Maria, C.; Sobrino, F. Dendritic Cells as a Therapeutic Strategy in Acute Myeloid Leukemia: Vaccines. Vaccines 2024, 12, 165. [Google Scholar] [CrossRef]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 2017, 31, 711–723.e714. [Google Scholar] [CrossRef]
- Zhang, X.; Goedegebuure, S.P.; Chen, M.Y.; Mishra, R.; Zhang, F.; Yu, Y.Y.; Singhal, K.; Li, L.; Gao, F.; Myers, N.B.; et al. Neoantigen DNA vaccines are safe, feasible, and induce neoantigen-specific immune responses in triple-negative breast cancer patients. Genome Med. 2024, 16, 131. [Google Scholar] [CrossRef]
- Van Tendeloo, V.F.; Van de Velde, A.; Van Driessche, A.; Cools, N.; Anguille, S.; Ladell, K.; Gostick, E.; Vermeulen, K.; Pieters, K.; Nijs, G.; et al. Induction of complete and molecular remissions in acute myeloid leukemia by Wilms’ tumor 1 antigen-targeted dendritic cell vaccination. Proc. Natl. Acad. Sci. USA 2010, 107, 13824–13829. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Hansson, L.; Choudhury, A.; Nasman-Glaser, B.; Eriksson, I.; Adamson, L.; Rossmann, E.; Widen, K.; Horvath, R.; Kokhaei, P.; et al. Vaccination with dendritic cells loaded with tumor apoptotic bodies (Apo-DC) in patients with chronic lymphocytic leukemia: Effects of various adjuvants and definition of immune response criteria. Cancer Immunol. Immunother. 2012, 61, 865–879. [Google Scholar] [CrossRef]
- Stroopinsky, D.; Liegel, J.; Bhasin, M.; Cheloni, G.; Thomas, B.; Bhasin, S.; Panchal, R.; Ghiasuddin, H.; Rahimian, M.; Nahas, M.; et al. Leukemia vaccine overcomes limitations of checkpoint blockade by evoking clonal T cell responses in a murine acute myeloid leukemia model. Haematologica 2021, 106, 1330–1342. [Google Scholar] [CrossRef]
- van de Ven, R.; Reurs, A.W.; Wijnands, P.; van Wetering, S.; Kruisbeek, A.M.; Hooijberg, E.; Scheffer, G.L.; Scheper, R.J.; de Gruijl, T.D. Exposure of CD34+ precursors to cytostatic anthraquinone-derivatives induces rapid dendritic cell differentiation: Implications for cancer immunotherapy. Cancer Immunol. Immunother. 2012, 61, 181–191. [Google Scholar] [CrossRef]
- Geng, G.; Xu, Y.; Hu, Z.; Wang, H.; Chen, X.; Yuan, W.; Shu, Y. Viral and non-viral vectors in gene therapy: Current state and clinical perspectives. EBioMedicine 2025, 118, 105834. [Google Scholar] [CrossRef] [PubMed]
- Egorova, T.; Starikova, A.; Polikarpova, A. Emerging Technologies Tackling Adeno-Associated Viruses (AAV) Immunogenicity in Gene Therapy Applications. Pharmaceutics 2025, 17, 1492. [Google Scholar] [CrossRef]
- Pardhi, E.; Yadav, R.; Chaurasiya, A.; Madan, J.; Guru, S.K.; Singh, S.B.; Mehra, N.K. Multifunctional targetable liposomal drug delivery system in the management of leukemia: Potential, opportunities, and emerging strategies. Life Sci. 2023, 325, 121771. [Google Scholar] [CrossRef]
- Matsudaira, N.; Honda, Y.; Kinoh, H.; Liu, X.; Nagao, S.; Matsutomo-Nitta, S.; Haochen, G.; Hayashita-Kinoh, H.; Aizawa, M.; Muguruma, K.; et al. Tumor-targeted adeno associated virus-loaded complexes comprising tannic acid and phenylboronic acid-polymers for orthotopic glioblastoma therapy. J. Control Release 2025, 389, 114477. [Google Scholar] [CrossRef]
- Inano, S.; Morita, H.; Nakagawa, D.; Takaori-Kondo, A.; Nakajima, T. Antibody-guided AAV vectors for antigen-specific delivery of suicide genes. Gene Ther. 2025, Online Ahead of Print. [Google Scholar] [CrossRef]
- Meissner, J.M.; Toporkiewicz, M.; Czogalla, A.; Matusewicz, L.; Kuliczkowski, K.; Sikorski, A.F. Novel antisense therapeutics delivery systems: In vitro and in vivo studies of liposomes targeted with anti-CD20 antibody. J. Control. Release 2015, 220, 515–528. [Google Scholar] [CrossRef]
- Matusewicz, L.; Filip-Psurska, B.; Psurski, M.; Tabaczar, S.; Podkalicka, J.; Wietrzyk, J.; Ziolkowski, P.; Czogalla, A.; Sikorski, A.F. EGFR-targeted immunoliposomes as a selective delivery system of simvastatin, with potential use in treatment of triple-negative breast cancers. Int. J. Pharm. 2019, 569, 118605. [Google Scholar] [CrossRef] [PubMed]
- Toporkiewicz, M.; Meissner, J.; Matusewicz, L.; Czogalla, A.; Sikorski, A.F. Toward a magic or imaginary bullet? Ligands for drug targeting to cancer cells: Principles, hopes, and challenges. Int. J. Nanomed. 2015, 10, 1399–1414. [Google Scholar] [CrossRef]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef]
- Song, D.; Zhao, Y.; Wang, Z.; Xu, Q. Tuning Lipid Nanoparticles for RNA Delivery to Extrahepatic Organs. Adv. Mater. 2024, 36, e2401445. [Google Scholar] [CrossRef]
- Palucka, K.; Banchereau, J. Cancer immunotherapy via dendritic cells. Nat. Rev. Cancer 2012, 12, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Wang, J.; He, S.; Wang, L.; Yang, J.; Li, W.; Ma, K.; Zhou, Y.; Liu, X.; Wang, S.; et al. Engineered T cells stimulate dendritic cell recruitment and antigen spreading for potent anti-tumor immunity. Cell Rep. Med. 2025, 6, 102307. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.P.; Patente, T.A.; Flatow, E.A.; Sallusto, F.; Barbuto, J.A.M. Frequency determination of breast tumor-reactive CD4 and CD8 T cells in humans: Unveiling the antitumor immune response. Oncoimmunology 2019, 8, 1607674. [Google Scholar] [CrossRef] [PubMed]
- Higano, C.S.; Small, E.J.; Schellhammer, P.; Yasothan, U.; Gubernick, S.; Kirkpatrick, P.; Kantoff, P.W. Sipuleucel-T. Nat. Rev. Drug Discov. 2010, 9, 513–514. [Google Scholar] [CrossRef]
- Kantoff, P.W.; Higano, C.S.; Shore, N.D.; Berger, E.R.; Small, E.J.; Penson, D.F.; Redfern, C.H.; Ferrari, A.C.; Dreicer, R.; Sims, R.B.; et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010, 363, 411–422. [Google Scholar] [CrossRef]
- Small, E.J.; Carducci, M.A.; Burke, J.M.; Rodriguez, R.; Fong, L.; van Ummersen, L.; Yu, D.C.; Aimi, J.; Ando, D.; Working, P.; et al. A phase I trial of intravenous CG7870, a replication-selective, prostate-specific antigen-targeted oncolytic adenovirus, for the treatment of hormone-refractory, metastatic prostate cancer. Mol. Ther. 2006, 14, 107–117. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Chavez, J.C.; Sehgal, A.R.; Epperla, N.; Ulrickson, M.; Bachy, E.; Munshi, P.N.; Casulo, C.; Maloney, D.G.; de Vos, S.; et al. Three-year follow-up analysis of axicabtagene ciloleucel in relapsed/refractory indolent non-Hodgkin lymphoma (ZUMA-5). Blood 2024, 143, 496–506. [Google Scholar] [CrossRef]
- Janssen, L.L.G.; Westers, T.M.; Rovers, J.; Valk, P.J.M.; Cloos, J.; de Gruijl, T.D.; van de Loosdrecht, A.A. Durable Responses and Survival in High-risk Myelodysplastic Syndrome and Acute Myeloid Leukemia Patients Receiving the Allogeneic Leukemia-derived Dendritic Cell Vaccine DCP-001. Hemasphere 2023, 7, e968. [Google Scholar] [CrossRef]
- Zuo, H.; van Lierop, M.C.; Kaspers, J.; Bos, R.; Reurs, A.; Sarkar, S.; Konry, T.; Kamermans, A.; Kooij, G.; de Vries, H.E.; et al. Transfer of Cellular Content from the Allogeneic Cell-Based Cancer Vaccine DCP-001 to Host Dendritic Cells Hinges on Phosphatidylserine and Is Enhanced by CD47 Blockade. Cells 2021, 10, 3233. [Google Scholar] [CrossRef]
- van de Loosdrecht, A.A.; van Wetering, S.; Santegoets, S.; Singh, S.K.; Eeltink, C.M.; den Hartog, Y.; Koppes, M.; Kaspers, J.; Ossenkoppele, G.J.; Kruisbeek, A.M.; et al. A novel allogeneic off-the-shelf dendritic cell vaccine for post-remission treatment of elderly patients with acute myeloid leukemia. Cancer Immunol. Immunother. 2018, 67, 1505–1518. [Google Scholar] [CrossRef]
- Alzaaqi, S.; Naka, N.; Hamada, K.; Hosen, N.; Kanegae, M.; Outani, H.; Adachi, M.; Imanishi, R.; Morii, E.; Iwai, M.; et al. WT1 epitope-specific IgG and IgM antibodies for immune-monitoring in patients with advanced sarcoma treated with a WT1 peptide cancer vaccine. Oncol. Lett. 2022, 23, 65. [Google Scholar] [CrossRef]
- Van Leeuwen-Kerkhoff, N.; Westers, T.M.; Poddighe, P.J.; Povoleri, G.A.M.; Timms, J.A.; Kordasti, S.; De Gruijl, T.D.; Van de Loosdrecht, A.A. Reduced frequencies and functional impairment of dendritic cell subsets and non-classical monocytes in myelodysplastic syndromes. Haematologica 2022, 107, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Yokota, C.; Kagawa, N.; Takano, K.; Chiba, Y.; Kinoshita, M.; Kijima, N.; Oji, Y.; Oka, Y.; Sugiyama, H.; Tsuboi, A.; et al. Maintenance of WT1 expression in tumor cells is associated with a good prognosis in malignant glioma patients treated with WT1 peptide vaccine immunotherapy. Cancer Immunol. Immunother. 2022, 71, 189–201. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen-Kerkhoff, N.; Lundberg, K.; Westers, T.M.; Kordasti, S.; Bontkes, H.J.; Lindstedt, M.; de Gruijl, T.D.; van de Loosdrecht, A.A. Human Bone Marrow-Derived Myeloid Dendritic Cells Show an Immature Transcriptional and Functional Profile Compared to Their Peripheral Blood Counterparts and Separate from Slan+ Non-Classical Monocytes. Front. Immunol. 2018, 9, 1619. [Google Scholar] [CrossRef]
- Dhodapkar, K.M.; Cohen, A.D.; Kaushal, A.; Garfall, A.L.; Manalo, R.J.; Carr, A.R.; McCachren, S.S.; Stadtmauer, E.A.; Lacey, S.F.; Melenhorst, J.J.; et al. Changes in Bone Marrow Tumor and Immune Cells Correlate with Durability of Remissions Following BCMA CAR T Therapy in Myeloma. Blood Cancer Discov. 2022, 3, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Anassi, E.; Ndefo, U.A. Sipuleucel-T (provenge) injection: The first immunotherapy agent (vaccine) for hormone-refractory prostate cancer. Pharm. Ther. 2011, 36, 197–202. [Google Scholar]
- Anguille, S.; Van de Velde, A.L.; Smits, E.L.; Van Tendeloo, V.F.; Juliusson, G.; Cools, N.; Nijs, G.; Stein, B.; Lion, E.; Van Driessche, A.; et al. Dendritic cell vaccination as postremission treatment to prevent or delay relapse in acute myeloid leukemia. Blood 2017, 130, 1713–1721. [Google Scholar] [CrossRef]
- Liegel, J.; Cheloni, G.; Bisharat, L.; Karagkouni, D.; Gerhard, G.; Chedid, G.; Saldarriaga, I.; Logan, E.; Narcis, M.; Ankita, S.; et al. Final Results of Phase 1 Trial of Dendritic Cell/AML Fusion Cell Vaccine after Allogeneic Stem Cell Transplant in Acute Myeloid Leukemia. Transplant. Cell. Ther. 2025, 31, S109. [Google Scholar] [CrossRef]
- van de Loosdrecht, A.A.; Drouet, E.-M.W.; Platzbecker, U.; Holderried, T.A.; Van Elssen, C.; Giagounidis, A.; Lehmann, S.; Cloos, J.; Van Zeeburg, H.; Rovers, J.; et al. Active Immunotherapy with Vididencel As Maintenance Treatment in MRD+ AML Patients in CR1 Results in Strong Anti-Tumor Immune Responses and Durable Long-Term Survival in Patients with an Immune Competent Immune Profile. Blood 2024, 144, 2875. [Google Scholar] [CrossRef]
| Therapeutic Strategy (Clinical Trial ID) | Clinical Setting | Phase/Recruitment Status | Efficacy (Key Endpoints) | Safety | References |
|---|---|---|---|---|---|
| WT1 mRNA–electroporated dendritic cell vaccine (NCT00965224) | AML in complete remission after chemotherapy | Phase II; recruitment completed | Induction of WT1-specific CD4+ and CD8+ T cell responses; molecular and immunological remission in a subset of patients; signals of prolonged relapse-free survival in immunological responders | Predominantly grade 1–2 local and flu-like adverse events; no consistent vaccine-related ≥ grade 3 toxicities reported | [174,204] |
| DC/AML fusion vaccine post allo-HCT (NCT03679650) | AML after allogeneic hematopoietic stem cell transplantation | Phase I; recruitment completed (follow-up ongoing) | Induction of leukemia-specific T cell responses; the majority of vaccinated patients remained in remission during the observation period | Acceptable safety profile; mainly grade 1–2 adverse events (injection-site reactions); cases of acute and chronic GVHD reported, including events assessed as possibly related to vaccination | [205] |
| Allogeneic DC vaccine DCP-001 (NCT01373515) | AML in complete remission or at high risk of relapse | Phase I; recruitment completed | Broad immunogenicity; improvement in residual disease-related parameters in a subset of patients; clinical signals of prolonged relapse-free survival in patients with low disease burden | Well tolerated; predominantly grade 1–2 adverse events; no vaccine-related ≥ grade 3 toxicities reported | [197] |
| Allogeneic DC vaccine DCP-001—observational follow-up (NCT01373515) | High-risk AML/MDS | Observational long-term follow-up; completed | Durable immune responses; clinical signals of improved overall and relapse-free survival | No new safety signals identified during long-term follow-up | [195] |
| Allogeneic DC vaccine DCP-001—ADVANCE II) | AML in complete remission with measurable residual disease | Phase II/IIb; recruitment ongoing | Primary endpoints include relapse-free survival and MRD dynamics (analysis ongoing) | Safety evaluation ongoing; no unexpected safety signals reported to date | [206] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubis, J.; Czogalla, A.; Kuliczkowski, K.; Sikorski, A.F. Leukemic Transdifferentiation: From Pathological Plasticity to Dendritic Cell-Based Immunotherapy. Biomedicines 2025, 13, 3099. https://doi.org/10.3390/biomedicines13123099
Dubis J, Czogalla A, Kuliczkowski K, Sikorski AF. Leukemic Transdifferentiation: From Pathological Plasticity to Dendritic Cell-Based Immunotherapy. Biomedicines. 2025; 13(12):3099. https://doi.org/10.3390/biomedicines13123099
Chicago/Turabian StyleDubis, Joanna, Aleksander Czogalla, Kazimierz Kuliczkowski, and Aleksander F. Sikorski. 2025. "Leukemic Transdifferentiation: From Pathological Plasticity to Dendritic Cell-Based Immunotherapy" Biomedicines 13, no. 12: 3099. https://doi.org/10.3390/biomedicines13123099
APA StyleDubis, J., Czogalla, A., Kuliczkowski, K., & Sikorski, A. F. (2025). Leukemic Transdifferentiation: From Pathological Plasticity to Dendritic Cell-Based Immunotherapy. Biomedicines, 13(12), 3099. https://doi.org/10.3390/biomedicines13123099






